Morphology and Molecular Phylogeny of Four Anaerobic Ciliates (Protista, Ciliophora, Armophorea), with Report of a New Species and a Unique Arrangement Pattern of Dikinetids in Family Metopidae
Abstract
1. Introduction
2. Materials and Methods
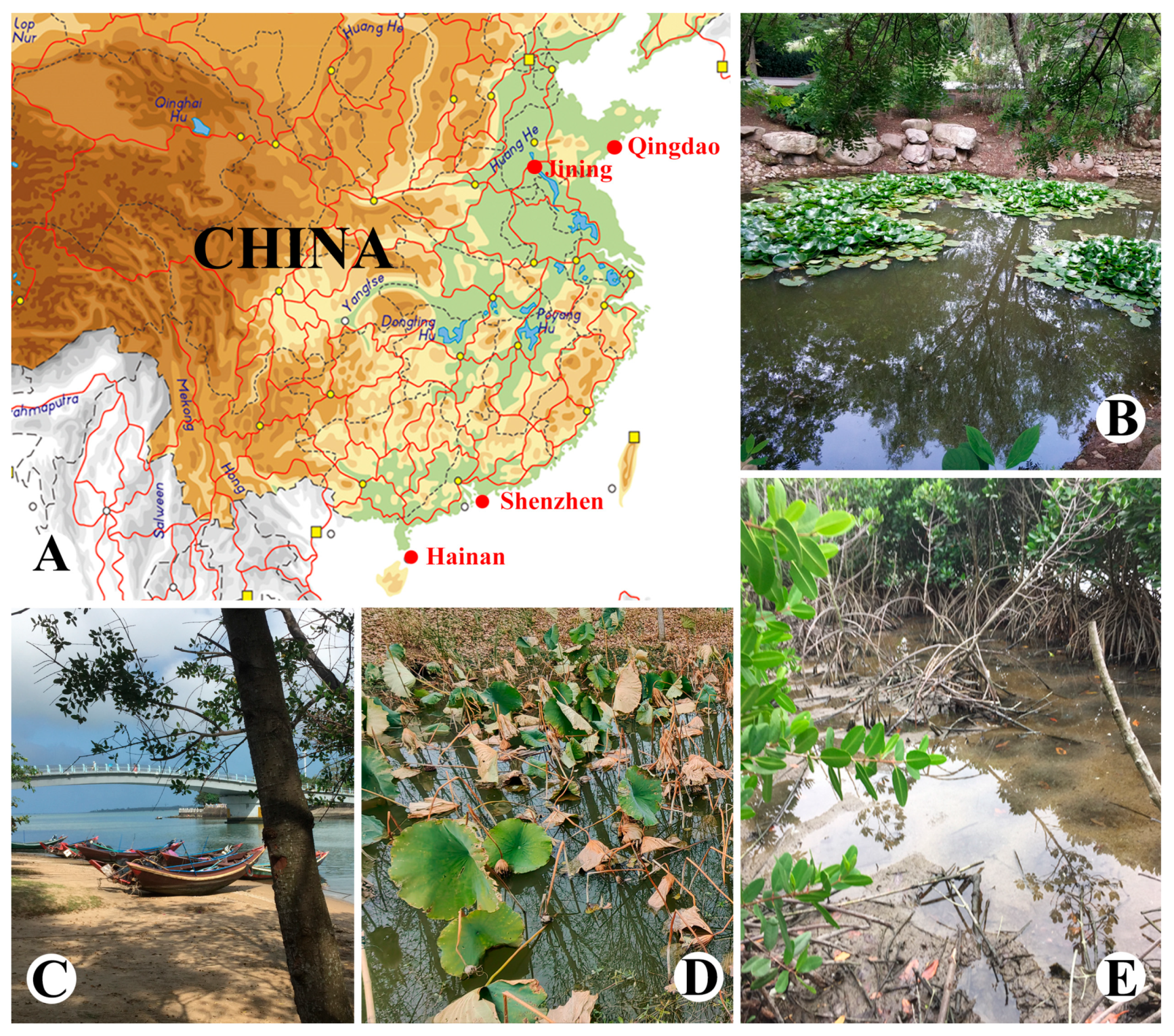
2.1. Sample Collection
2.2. Morphological Observations
2.3. DNA Extraction and Gene Sequencing
2.4. Phylogenetic Analyses
3. Results
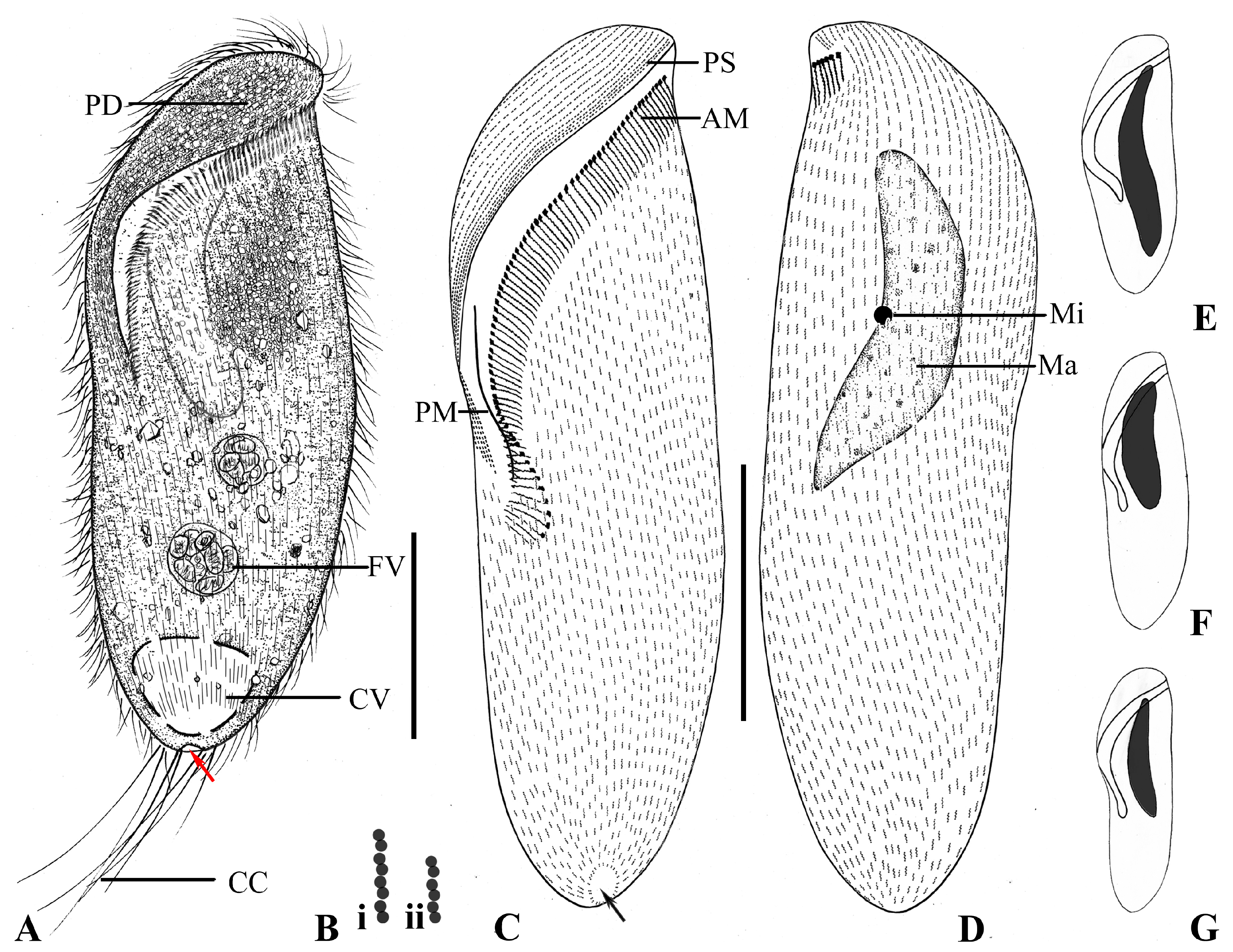
3.1. Brachonella abnormalis sp. nov.
3.1.1. Diagnosis
3.1.2. Type Material
3.1.3. Type Locality
3.1.4. Etymology
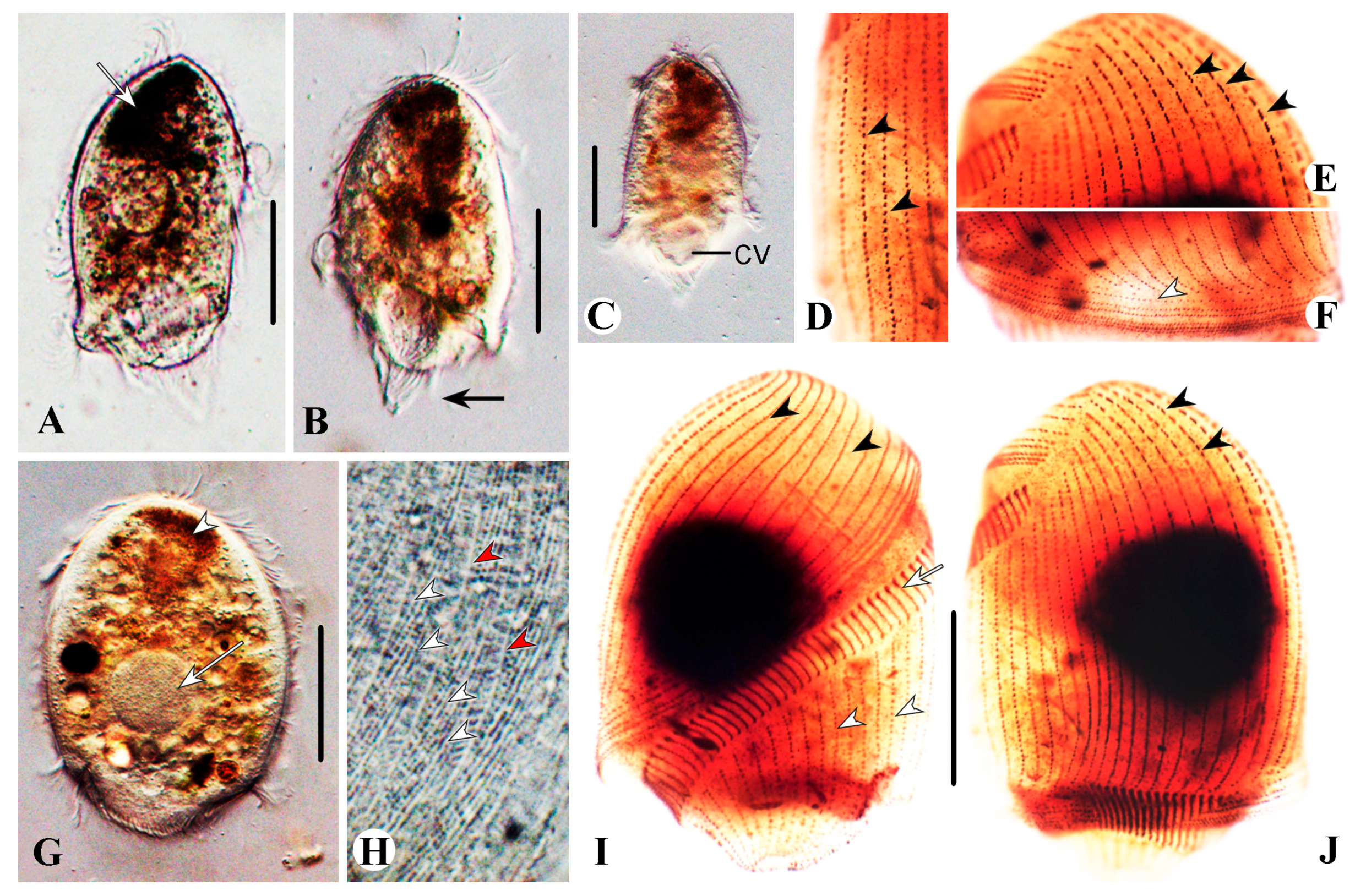
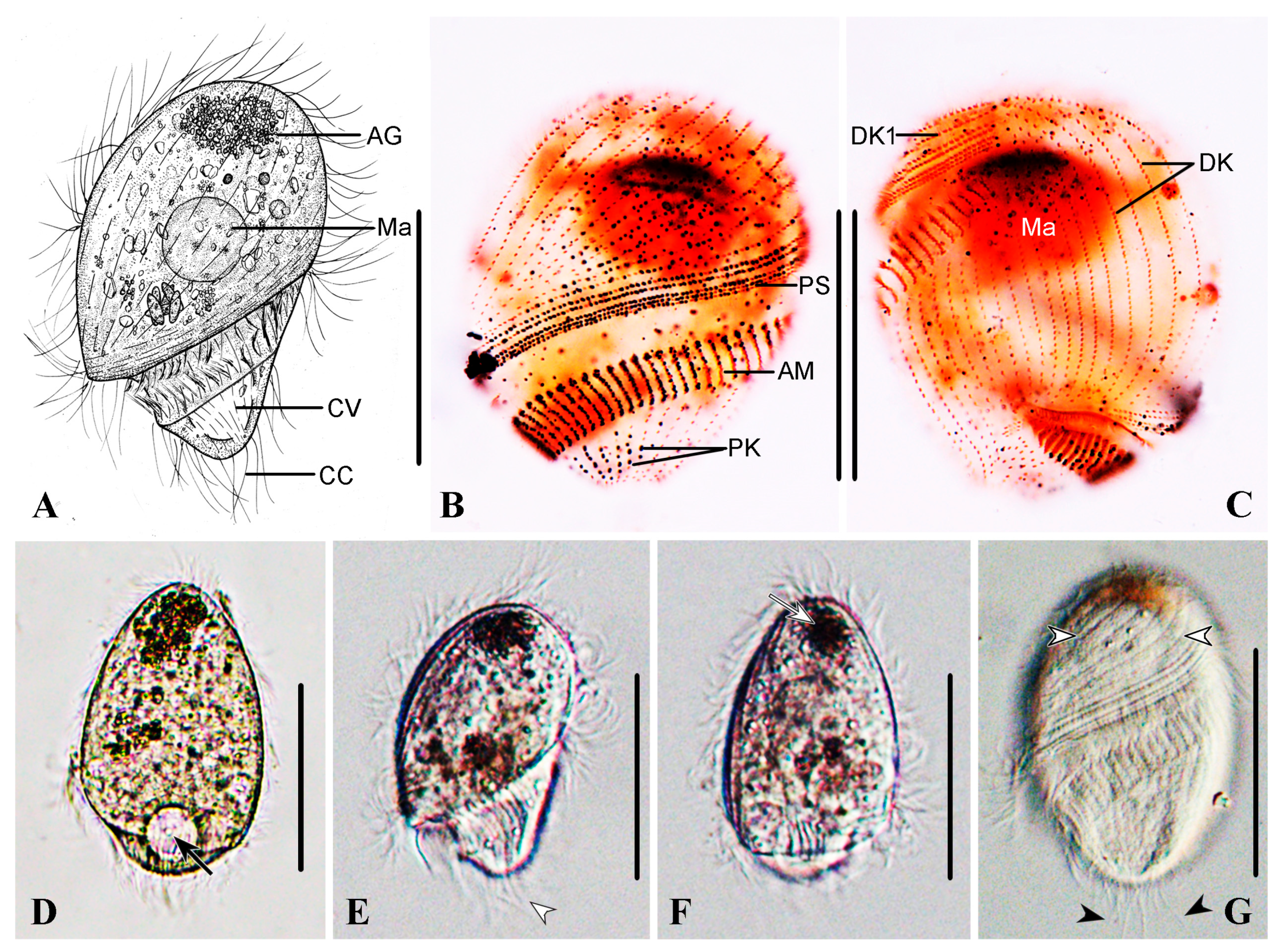
3.1.5. Description
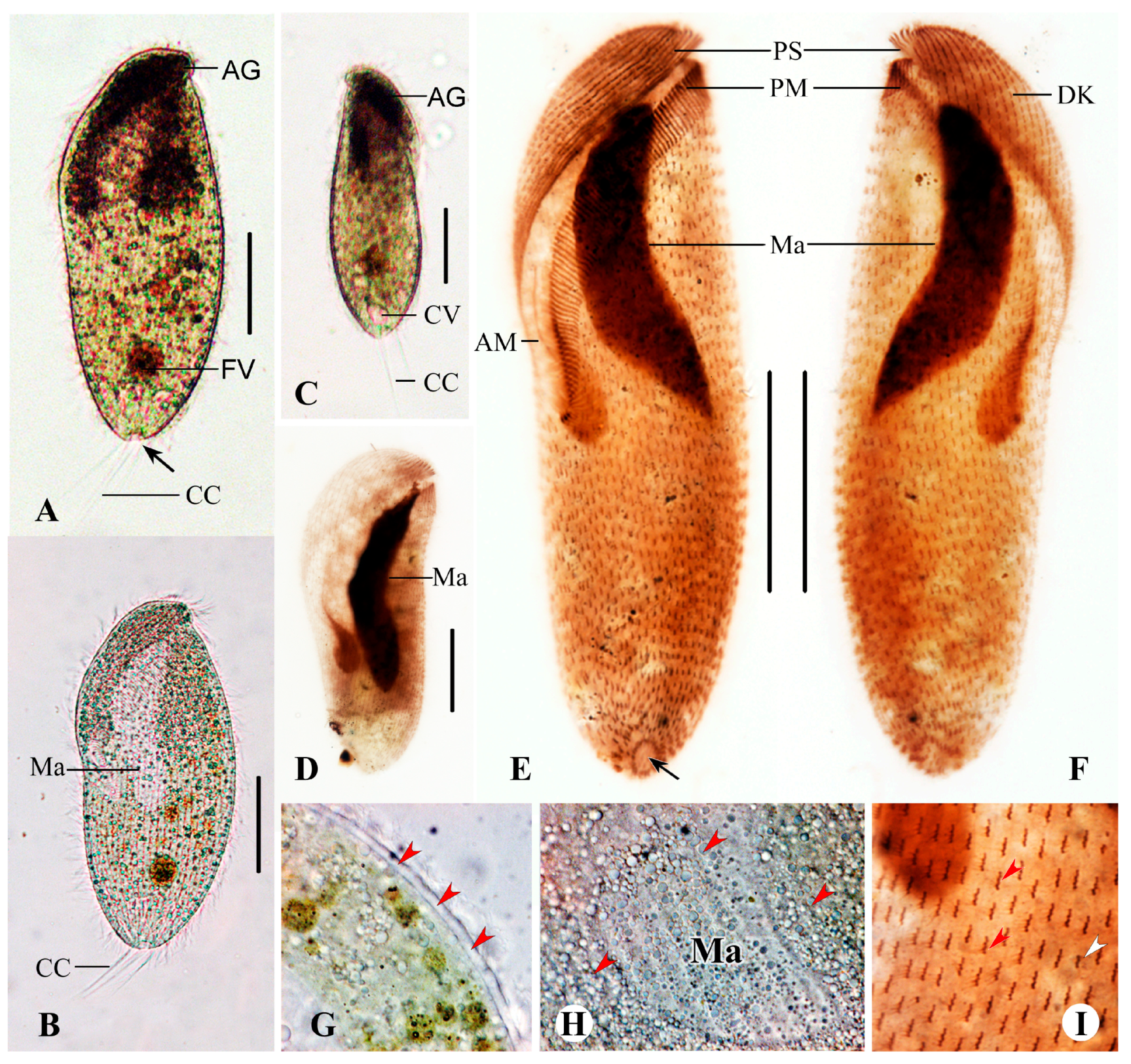
3.2. Brachonella contorta (Levander, 1894) Jankowski, 1964 [28]
Description Based on Qingdao Population
3.3. Metopus contortus (Quennnerstedt, 1867) Kahl, 1932 [31]
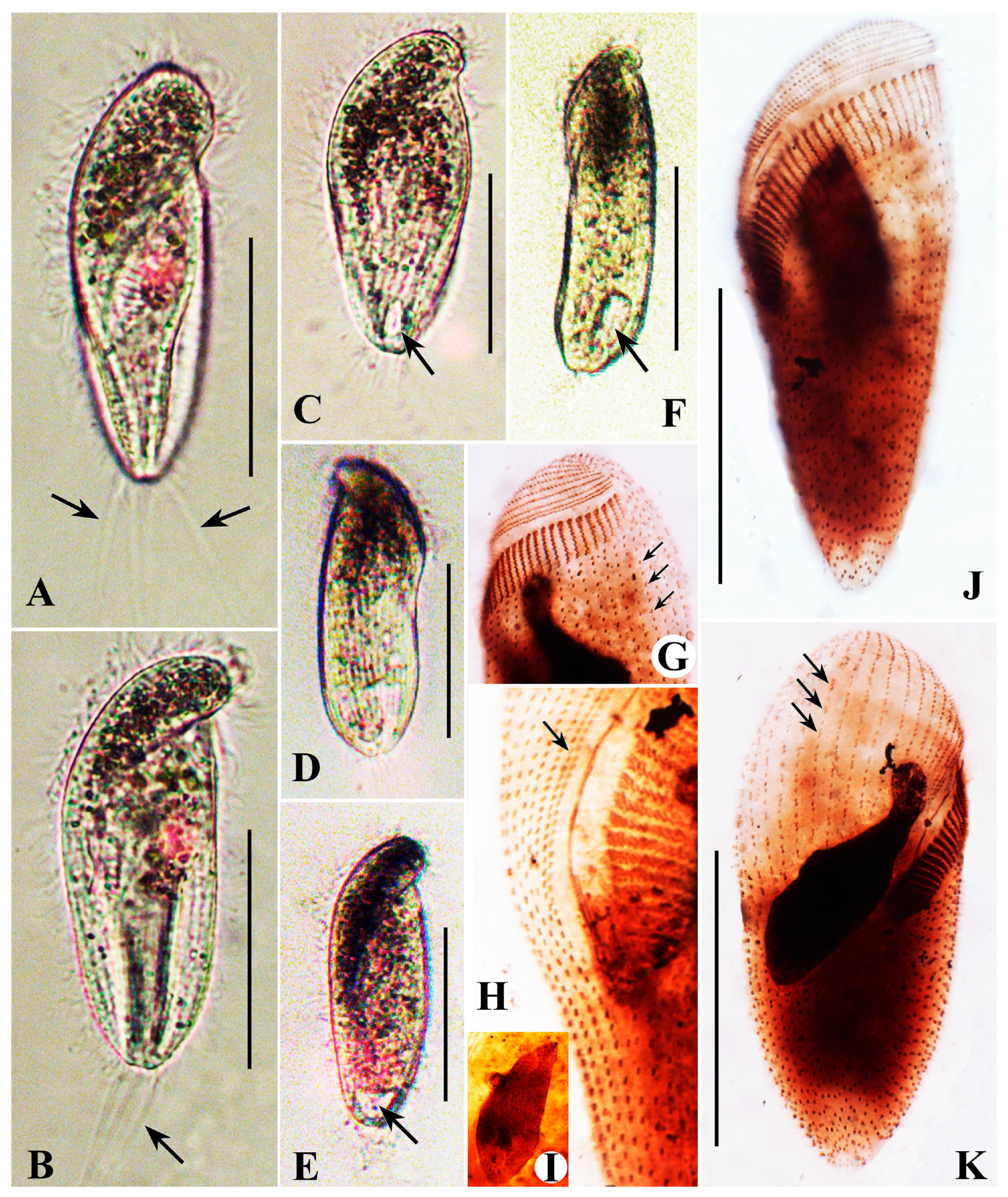
Description Base on Qingdao and Jining Populations
3.4. Metopus major Kahl, 1932 [31]
Description Base on Haikou Population
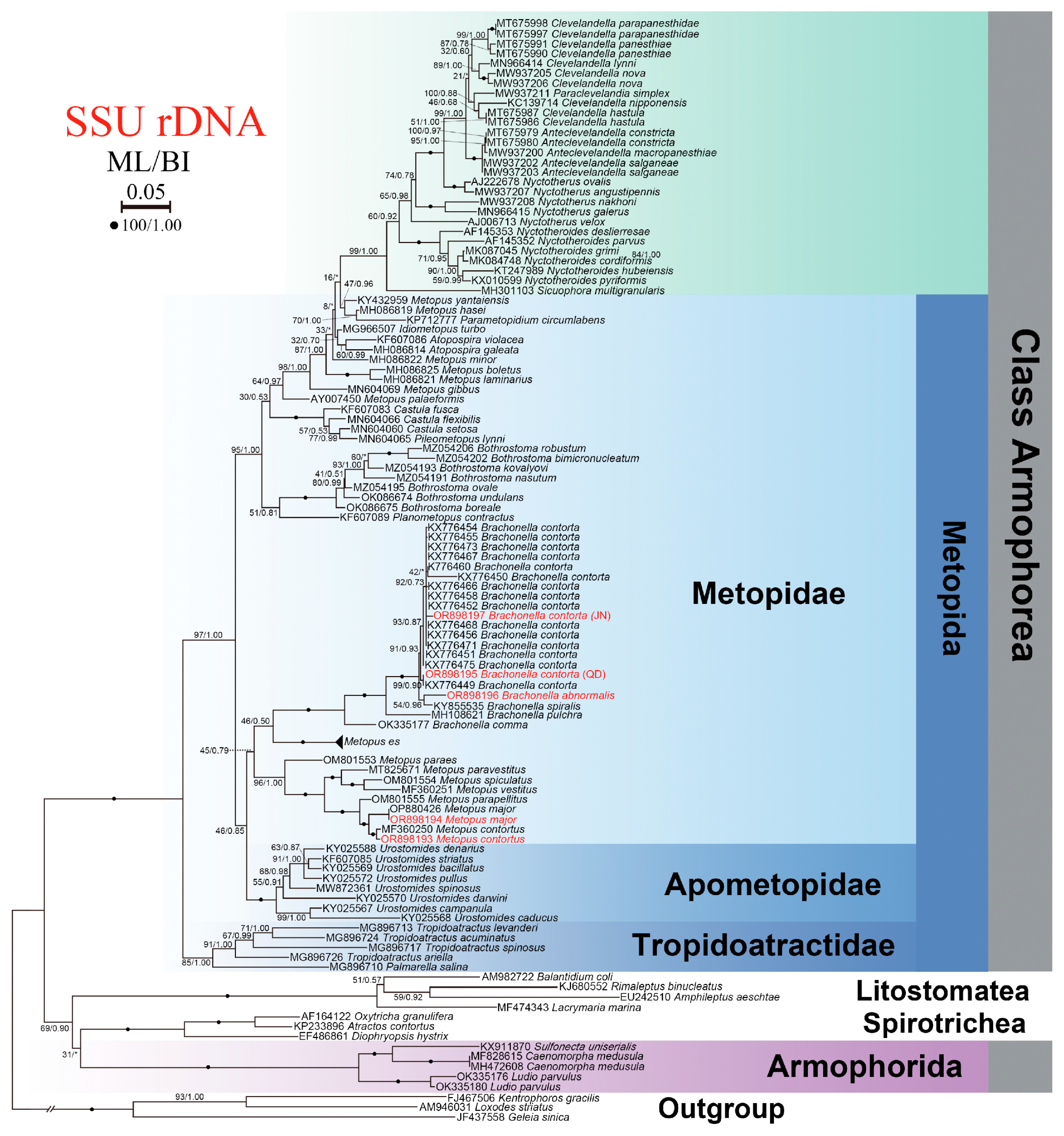
3.5. SSU rDNA Sequence and Phylogenetic Analyses
4. Discussion
4.1. Comparison of Brachonella abnormalis sp. nov. with Congeners
4.2. Comparison of Metopus major with Related Forms
4.3. Comparison of Metopus contortus with Related Forms
4.4. Uneven Distribution of Dikinetids in Dome Kineties in Brachonella abnormalis sp. nov., Metopus major and Metopus contortus
4.5. Phylogeny of Metopus and Brachonella Species
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rassoulzadegan, F.; Sheldon, R.W. Predator-prey interactions of nanozooplankton and bacteria in an oligotrophic marine environment. Limnol. Oceanogr. 1986, 31, 1010–1021. [Google Scholar] [CrossRef]
- Sherr, E.B.; Sherr, B.F.; Fallon, R.D.; Newell, S.Y. Small, aloricate ciliates as a major component of the marine heterotrophic nanoplankton. Limnol. Oceanogr. 1986, 31, 177–183. [Google Scholar] [CrossRef]
- Sherr, E.B.; Sherr, B.F. High rates of consumption of bacteria by pelagic ciliates. Nature 1987, 325, 710–711. [Google Scholar] [CrossRef]
- Lynn, D.H. The Ciliated Protozoa: Characterization, Classification and Guide to the Literature, 3rd ed.; Springer: Dordrecht, The Netherlands, 2008. [Google Scholar]
- Bourland, W.A.; Rotterová, J.; Čepička, I. Redescription and molecular phylogeny of the type species for two main metopid genera, Metopus es (Müller, 1776) Lauterborn, 1916 and Brachonella contorta (Levander, 1894) Jankowski, 1964 (Metopida, Ciliophora), based on broad geographic sampling. Eur. J. Protistol. 2017, 59, 133–154. [Google Scholar] [CrossRef] [PubMed]
- Gijzen, H.J.; Broers, C.A.; Barughare, M.; Stumm, C.K. Methanogenic bacteria as endosymbionts of the ciliate Nyctotherus ovalis in the cockroach hindgut. Appl. Environ. Microbiol. 1991, 57, 1630–1634. [Google Scholar] [CrossRef]
- Roger, A.J.; Muñoz-Gómez, S.A.; Kamikawa, R. The origin and diversification of mitochondria. Curr. Biol. 2017, 27, R1177–R1192. [Google Scholar] [CrossRef] [PubMed]
- Van Hoek, A.H.; Van Alen, T.A.; Sprakel, V.S.; Leunissen, J.A.; Brigge, T.; Vogels, G.D.; Hackstein, J.H. Multiple acquisition of methanogenic archaeal symbionts by anaerobic ciliates. Mol. Biol. Evol. 2000, 17, 251–258. [Google Scholar] [CrossRef]
- Lewis, W.H.; Lind, A.E.; Sendra, K.M.; Onsbring, H.; Williams, T.A.; Esteban, G.F.; Hirt, R.P.; Ettema, T.J.G.; Embley, T.M. Convergent evolution of hydrogenosomes from mitochondria by gene transfer and loss. Mol. Biol. Evol. 2020, 37, 524–539. [Google Scholar] [CrossRef] [PubMed]
- Rotterová, J.; Salomaki, E.; Pánek, T.; Bourland, W.; Žihala, D.; Táborský, P.; Edgcomb, V.P.; Beinart, R.A.; Kolísko, M.; Čepička, I. Genomics of New Ciliate Lineages Provides Insight into the Evolution of Obligate Anaerobiosis. Curr. Biol. 2020, 30, 2037–2050.e6. [Google Scholar] [CrossRef]
- Bernhard, J.M.; Buck, K.R.; Farmer, M.A.; Bowser, S.S. The Santa Barbara Basin is a symbiosis oasis. Nature 2000, 403, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Esteban, G.F.; Fenchel, T.; Finlay, B. Diversity of free-living morphospecies in the ciliate genus Metopus. Arch. Protistenkd. 1995, 146, 137–164. [Google Scholar] [CrossRef]
- Foissner, W. Protists as bioindicators in activated sludge: Identification, ecology and future needs. Eur. J. Protistol. 2016, 55, 75–94. [Google Scholar] [CrossRef] [PubMed]
- Rotterová, J.; Bourland, W.; Čepička, I. Tropidoatractidae fam. nov., a deep branching lineage of Metopida (Armophorea, Ciliophora) found in diverse habitats and possessing prokaryotic symbionts. Protist 2018, 169, 362–405. [Google Scholar] [CrossRef]
- Bourland, W.A.; Rotterová, J.; Luo, X.; Čepička, I. The little-known freshwater metopid ciliate, Idiometopus turbo (Dragesco and Dragesco-Kernéis, 1986) nov. gen., nov. comb., originally discovered in Africa, found on the Micronesian island of Guam. Protist 2018, 169, 494–506. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhuang, W.; Pérez-Uz, B.; Zhang, Q.; Hu, X. Two anaerobic ciliates (Ciliophora, Armophorea) from China: Morphology and SSU rDNA sequence, with report of a new species, Metopus paravestitus sp. nov. J. Eukaryot. Microbiol. 2021, 68, e12822. [Google Scholar] [CrossRef] [PubMed]
- Vďačný, P.; Foissner, W. A huge diversity of metopids (Ciliophora, Armophorea) in soil from the Murray River floodplain, Australia. I. Description of five new species and redescription of Metopus setosus Kahl, 1927. Eur. J. Protistol. 2017, 57, 35–76. [Google Scholar] [CrossRef]
- Vďačný, P.; Foissner, W. A huge diversity of metopids (Ciliophora, Armophorea) in soil from the Murray River floodplain, Australia. II. Morphology and morphogenesis of Lepidometopus platycephalus nov. gen., sp. nov. Acta Protozool. 2017, 56, 39–57. [Google Scholar] [CrossRef]
- Bourland, W.A.; Wendell, L.; Hampikian, G. Morphologic and molecular description of Metopus fuscus Kahl from North America and new rDNA sequences from seven metopids (Armophorea, Metopidae). Eur. J. Protistol. 2014, 50, 213–230. [Google Scholar] [CrossRef] [PubMed]
- Wilbert, N. Eine verbesserte Technik der Protargolimprägnation für Ciliaten. Mikrokosmos 1975, 64, 171–179. [Google Scholar]
- Pan, X.M.; Bourland, W.A.; Song, W.B. Protargol synthesis: An in-house protocol. J. Eukaryot. Microbiol. 2013, 60, 609–614. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wang, C.; Zhuang, W.; Li, S.; Hu, X. Taxonomy, phylogeny, and geographical distribution of the little-known Helicoprorodon multinucleatum Dragesco, 1960 (Ciliophora, Haptorida) and key to species within the genus. Eur. J. Protistol. 2021, 78, 125769. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post–Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Nylander, J. MrModeltest v2. Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Jankowski, A.W. Morphology and evolution of Ciliophora. III. Diagnoses and phylogenesis of 53 sapropelebionts, mainly of the oder Heterotrichida. Protist 1964, 107, 185–294. [Google Scholar]
- Dai, R.; Xu, K.; Song, W. Redescription and neotypification of Metopus contortus (Quennerstedt, 1867) Kahl, 1932 (Protozoa, Ciliophora) from the intertidal sediment of the Jiaozhou Bay. Acta Hydrobiol. Sin. 2008, 32, 90–96, (In Chinese with English abstract). [Google Scholar]
- Omar, A.; Zhang, Q.; Zou, S.; Gong, J. Morphology and phylogeny of the soil ciliate Metopus yantaiensis n. sp. (Ciliophora, Metopida), with identification of the intracellular bacteria. J. Eukaryot. Microbiol. 2017, 64, 792–805. [Google Scholar] [CrossRef] [PubMed]
- Kahl, A. Urtiere oder Protozoa I: Wimpertiere oder Ciliata (Infusoria) 3. Spirotricha; Die Tierwelt Deutschlands; Gustav Fischer Verlag: Stuttgart, Germany, 1932; pp. 399–650. [Google Scholar]
- Bourland, W.; Pomahač, O.; Čepička, I. Morphology and phylogeny of two anaerobic freshwater ciliates: Brachonella comma sp. nov. and the widely distributed but little–known caenomorphid, Ludio parvulus Penard, 1922. J. Eukaryot. Microbiol. 2022, 69, e12892. [Google Scholar] [CrossRef] [PubMed]
- Bourland, W.A.; Rotterová, J.; Čepička, I. Morphologic and molecular characterization of Brachonella pulchra (Kahl, 1927) comb. nov. (Armophorea, Ciliophora) with comments on cyst structure and formation. Int. J. Syst. Evol. Microbiol. 2018, 68, 3052–3065. [Google Scholar] [CrossRef]
- Dragesco, J.; Dragesco-Kernéis, A. Ciliés Libres de L’Afrique Intertropicale. Introduction á la Connaissance et á l’Étude des Ciliés; Faune Tropicale; Éditions de l’ORSTOM: Paris, France, 1986; 559p. [Google Scholar]
- Alekperov, I.K.h. New species of ciliates (Heterotrichida) from artificial reservoirs of Azerbaijan. Zool. J. 1984, 63, 1731–1733. [Google Scholar]
- Foissner, W. Taxonomische Studien über die Ciliaten des Grossglocknergebietes IX. Ordnungen Heterotrichida und Hypotrichida. Ber. Nat.-Med. Ver Salzbg. 1980, 5, 71–117. [Google Scholar]
- Foissner, W.; Agatha, S.; Berger, H. Soil Ciliates (Protozoa, Ciliophora) from Namibia (Southwest Africa), with Emphasis on Two Contrasting Environments, the Etosha Region and the Namib Desert; Biologiezentrum der Oberösterreichischen Landesmuseums: Linz, Austria, 2002; Volume 5, 1459p. [Google Scholar]
- Borror, A.C. Morphology and ecology of the benthic ciliated protozoa of Alligator Harbor, Florida. Arch. Protistenk. 1963, 106, 465–534. [Google Scholar]
- Dragesco, J. Infraciliature et morphométrie de cinq espèces de ciliés mésopsammiques méditerranéens. Cah. Biol. Mar. 1996, 37, 261–293. [Google Scholar]
- Czapik, A.; Fyda, J. Contribution à la connaissance des ciliés psammophiles de la Baltique. Acta Protozool. 1992, 31, 109–114. [Google Scholar]
- Al-Rasheid, K. Records of Marine Interstitial Heterotrichida (Ciliata) from the Saudi Arabian Jubail Marine Wildlife Sanctuary in the Arabian Gulf. Arab. Gulf. J. Sci. Res. 1999, 17, 127–141. [Google Scholar]
- Schulze, E. Morphologische, cytologische und ökologisch-physiologische Untersuchungen an Faulschlammciliaten (Meto-pus sigmoides Clap et Lachm. und Metopus contortus Lev.). Arch. Protistenkd. 1959, 103, 371–426. [Google Scholar]
- da Silva Paiva, T.; Küppers, G.C.; Lahr, D.J.G.; Schweikert, M.; Silva–Neto, I.D. Discomorphella pedroeneasi sp. nov. (Ciliophora, Odontostomatida): An anaerobic ciliate hosting multiple cytoplasmic and macronuclear endocytobionts. Eur. J. Protistol. 2017, 58, 103–134. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, W.; Li, R.; Feng, X.; Al-Farraj, A.; Hu, X. New contribution to the diversity of the anaerobic genus Metopus (Ciliophora, Armophorea), with descriptions of three new marine species. Front. Mar. Sci. 2022, 9, 884834. [Google Scholar] [CrossRef]


| Characteristics | Mean ± SD | M | CV | Min–Max | n |
|---|---|---|---|---|---|
| Body, length | 136.0 ± 12.3 | 136.1 | 9.1 | 121–176 | 20 |
| 71.7 ± 10.3 | 74 | 14.4 | 50–88 | 20 | |
| 52.5 ± 4.4 | 52 | 8.4 | 45–62 | 24 | |
| Body, width | 92.9 ± 7.3 | 92.5 | 7.9 | 86–112 | 20 |
| 49.8 ± 6.7 | 51.5 | 13.4 | 33–60 | 20 | |
| 43.6 ± 4.5 | 45 | 10.3 | 35–54 | 24 | |
| Body length-to-width ratio | 1.5 ± 0.1 | 1.5 | 8.5 | 1.3–1.7 | 20 |
| 1.4 ± 0.1 | 1.4 | 5.6 | 1.2–1.6 | 20 | |
| 1.2 ± 0.1 | 1 | 5.2 | 1.1–1.3 | 24 | |
| Anterior body end to proximal end of | 116.5 ± 13.2 | 118.6 | 11.3 | 97–144 | 20 |
| adoral zone, distance | 64.6 ± 8.2 | 65.1 | 12.7 | 51–76 | 20 |
| 44.2 ± 4.2 | 44 | 9.6 | 37–54 | 24 | |
| Distance anterior body end to proximal | 84.3 ± 3.1 | 84.8 | 3.6 | 80–90 | 20 |
| end adoral zone–body length, ratio in % | 84.1 ± 3.0 | 84 | 3.5 | 80–89 | 20 |
| 84.2 ± 4.1 | 83.7 | 4.8 | 77–94 | 24 | |
| Macronucleus, length | 49.7 ± 10.3 | 50 | 20.7 | 34–69 | 9 |
| 29.1 ± 4.4 | 29 | 15.1 | 22–36 | 20 | |
| 26.8 ± 4.7 | 228 | 17.4 | 17–34 | 17 | |
| Macronucleus, width | 41.0 ± 8.5 | 40 | 20.8 | 32–54 | 9 |
| 24.3 ± 5.3 | 24.5 | 21.9 | 15–34 | 20 | |
| 20.2 ± 3.8 | 20 | 18.8 | 13–25 | 17 | |
| Adoral polykinetids, number | 94.0 ± 6.8 | 92 | 7.2 | 87–107 | 9 |
| 46.0 ± 3.7 | 46 | 8.1 | 40–54 | 19 | |
| 41.3 ± 3.8 | 42 | 9.2 | 33–49 | 26 | |
| Somatic kineties, number | 47.3 ± 5.6 | 48 | 11.8 | 39–55 | 10 |
| 26.6 ± 3.4 | 26 | 12.9 | 21–32 | 17 | |
| 37.5 ± 2.7 | 38 | 7.1 | 33–43 | 26 | |
| Preoral dome kineties, number | 34.3 ± 2.2 | 35 | 6.3 | 29–37 | 10 |
| 20.3 ± 1.7 | 20.5 | 8.1 | 17–23 | 17 | |
| 15.6 ± 1.3 | 16 | 8.4 | 12–18 | 26 | |
| Perizonal ciliary stripe rows, number | 5.0 ± 0.0 | 5 | 0 | 5 | 10 |
| 5.0 ± 0.0 | 5 | 0 | 5 | 20 | |
| 5.0 ± 0.0 | 5 | 0 | 5 | 26 | |
| Macronucleus, number | 1.0 ± 0.0 | 1 | 0 | 1 | 10 |
| 1.0 ± 0.0 | 1 | 0 | 1 | 20 | |
| 1.0 ± 0.0 | 1 | 0 | 1 | 26 | |
| Micronucleus, number | 1.0 ± 0.0 | 1 | 0 | 1 | 6 |
| 1.0 ± 0.0 | 1 | 0 | 1 | 16 | |
| 1.0 ± 0.0 | 1 | 0 | 1 | 15 |
| Characteristics | Mean ± SD | M | CV | Min–Max | n |
|---|---|---|---|---|---|
| Body, length | 91.0 ± 8.4 | 90 | 9.2 | 76–106 | 22 |
| 174.6 ± 24.8 | 174 | 14.2 | 134–228 | 20 | |
| Body, width | 37.6 ± 5.5 | 38 | 14.6 | 28–51 | 22 |
| 57.3 ± 8.0 | 58 | 13.9 | 44–73 | 20 | |
| Body length-to-width ratio | 2.5 ± 0.4 | 2.4 | 17 | 1.8–3.3 | 22 |
| 3.1 ± 0.3 | 3.2 | 9.8 | 2.5 –3.5 | 20 | |
| Anterior body end to proximal end of | 50.4 ± 6.1 | 50 | 12.1 | 37–59 | 22 |
| adoral zone, distance | 107.1 ± 11.4 | 105.5 | 10.7 | 89–132 | 20 |
| Distance anterior body end to proximal | 55.5 ± 5.4 | 55 | 9.7 | 46–65 | 22 |
| end adoral zone–body length, ratio in % | 61.9 ± 6.1 | 61.9 | 9.9 | 51–72 | 20 |
| Macronucleus, length | 37.5 ± 6.5 | 39.5 | 17.4 | 22–47 | 22 |
| 87.3 ± 12.17 | 86.5 | 13.9 | 72–111 | 20 | |
| Macronucleus, width | 13.1 ± 2.6 | 12 | 19.6 | 10–18 | 22 |
| 20.3 ± 5.8 | 19 | 28.8 | 15–37 | 20 | |
| Adoral polykinetids, number | 37.5 ± 3.9 | 36.5 | 10.5 | 33–45 | 16 |
| 98.1 ± 6.6 | 97 | 6.8 | 89–112 | 19 | |
| Somatic kineties, number | 41.8 ± 2.6 | 42 | 6.3 | 37–47 | 22 |
| 58.9 ± 4.4 | 58 | 7.5 | 54–74 | 19 | |
| Preoral dome kineties, number | 17.3 ± 2.4 | 17 | 13.8 | 12–23 | 22 |
| 24.7 ± 2.0 | 25 | 8.1 | 22–29 | 19 | |
| Perizonal ciliary stripe rows, number | 4.6 ± 0.5 | 5 | 10.9 | 5 | 11 |
| 5.0 ± 0.0 | 5 | 0 | 5 | 20 | |
| Macronucleus, number | 1.0 ± 0.0 | 1 | 0 | 1 | 22 |
| 1.0 ± 0.0 | 1 | 0 | 1 | 20 | |
| Micronucleus, number | 1.0 ± 0.0 | 1 | 0 | 1 | 18 |
| 1.0 ± 0.0 | 1 | 0 | 1 | 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Zhuang, W.; Feng, X.; Warren, A.; Gong, J. Morphology and Molecular Phylogeny of Four Anaerobic Ciliates (Protista, Ciliophora, Armophorea), with Report of a New Species and a Unique Arrangement Pattern of Dikinetids in Family Metopidae. Microorganisms 2025, 13, 240. https://doi.org/10.3390/microorganisms13020240
Li S, Zhuang W, Feng X, Warren A, Gong J. Morphology and Molecular Phylogeny of Four Anaerobic Ciliates (Protista, Ciliophora, Armophorea), with Report of a New Species and a Unique Arrangement Pattern of Dikinetids in Family Metopidae. Microorganisms. 2025; 13(2):240. https://doi.org/10.3390/microorganisms13020240
Chicago/Turabian StyleLi, Song, Wenbao Zhuang, Xiaochen Feng, Alan Warren, and Jun Gong. 2025. "Morphology and Molecular Phylogeny of Four Anaerobic Ciliates (Protista, Ciliophora, Armophorea), with Report of a New Species and a Unique Arrangement Pattern of Dikinetids in Family Metopidae" Microorganisms 13, no. 2: 240. https://doi.org/10.3390/microorganisms13020240
APA StyleLi, S., Zhuang, W., Feng, X., Warren, A., & Gong, J. (2025). Morphology and Molecular Phylogeny of Four Anaerobic Ciliates (Protista, Ciliophora, Armophorea), with Report of a New Species and a Unique Arrangement Pattern of Dikinetids in Family Metopidae. Microorganisms, 13(2), 240. https://doi.org/10.3390/microorganisms13020240







