From Isolation to Application: Designing a Multi-Target Phage Cocktail for Bivalve Safety
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Phage Isolation, Host Range and Purification
2.3. Bacterial Killing Curves
2.4. Statistical Analysis
3. Results
3.1. Phage Isolation and Purification
3.2. Phage Screening Assays
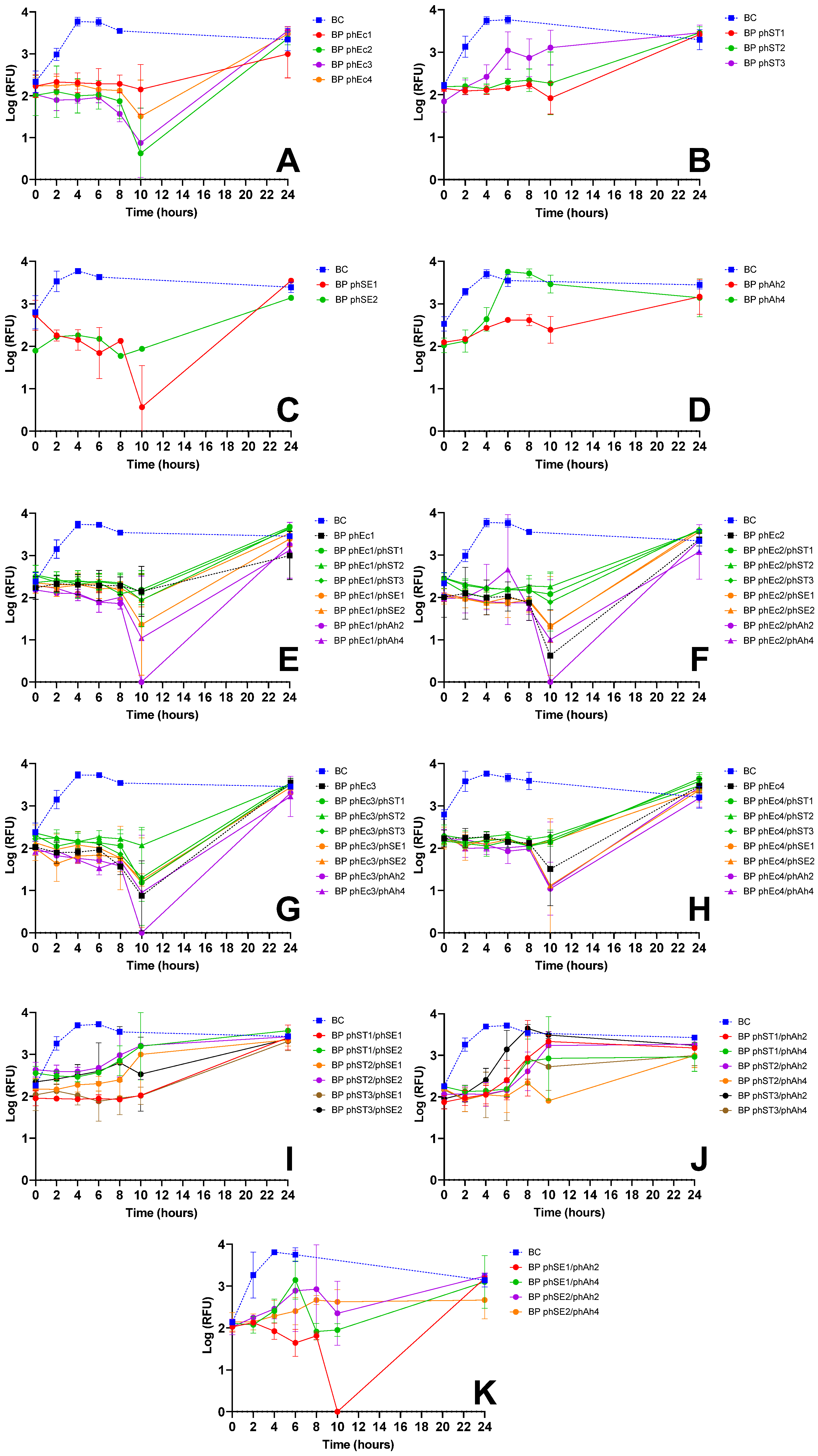
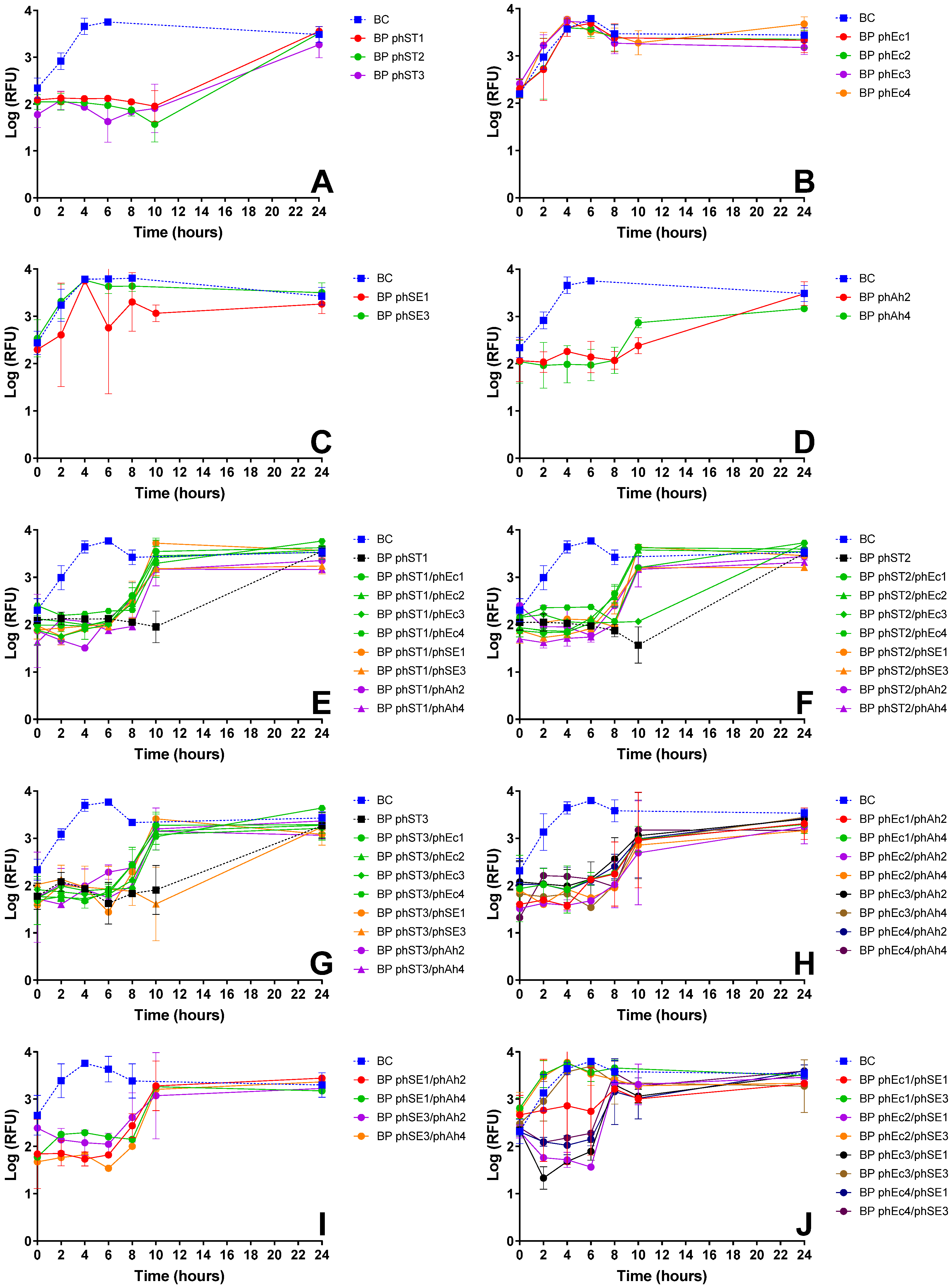
3.2.1. E. coli Inactivation Screening
3.2.2. S. Typhimurium Inactivation Screening
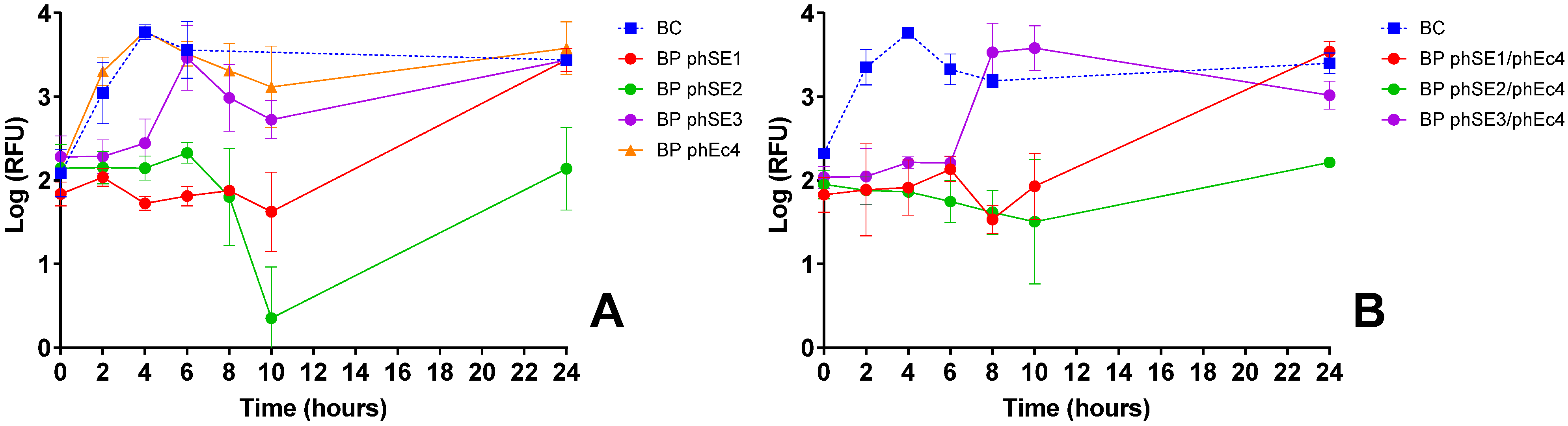
3.2.3. S. Enteritidis Inactivation Screening
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CFU | Colony-Forming Unit |
| EOP | Efficiency of Plating |
| MOI | Multiplicity Of Infection |
| OD | Optical Density |
| PFU | Plaque-Forming Unit |
| RFU | Relative Fluorescent Unit |
| TSA | Tryptic Soy Agar |
| TSB | Tryptic Soy Broth |
Appendix A
| Phages | phEc1 | phEc2 | phEc3 | phEc4 | phST1 | phST2 | phST3 | phSE1 | phSE2 | phAh2 | phAh4 |
| phEc1 | 10 | - | - | - | - | - | - | - | - | - | - |
| phEc2 | ND | 10 | - | - | - | - | - | - | - | - | - |
| phEc3 | ND | ND | 10 | - | - | - | - | - | - | - | - |
| phEc4 | ND | ND | ND | 10 | - | - | - | - | - | - | - |
| phST1 | 10 | 10 | 10 | 10 | 10 | - | - | - | - | - | - |
| phST2 | 10 | 10 | 10 | 10 | ND | 10 | - | - | - | - | - |
| phST3 | 10 | 10 | 10 | 10 | ND | ND | 8 | - | - | - | - |
| phSE1 | 10 | 10 | 10 | 10 | 10 | 8 | 10 | 10 | - | - | - |
| phSE2 | 10 | 10 | 10 | 10 | 8 | 8 | 10 | ND | 10 | - | - |
| phAh2 | 10 | 10 | 10 | 10 | 8 | 8 | 6 | 10 | 10 | 10 | - |
| phAh4 | 10 | 10 | 10 | 10 | 8 | 10 | 8 | 10 | 10 | ND | 4 |
 Hold Time (Hours).
Hold Time (Hours).| Phages | phST1 | phST2 | phST3 | phEc1 | phEc2 | phEc3 | phEc4 | phSE1 | phSE3 | phAh2 | phAh4 |
| phST1 | 10 | - | - | - | - | - | - | - | - | - | - |
| phST2 | ND | 10 | - | - | - | - | - | - | - | - | - |
| phST3 | ND | ND | 10 | - | - | - | - | - | - | - | - |
| phEc1 | 8 | 8 | 8 | 0 | - | - | - | - | - | - | - |
| phEc2 | 8 | 8 | 8 | ND | 0 | - | - | - | - | - | - |
| phEc3 | 8 | 10 | 8 | ND | ND | 0 | - | - | - | - | - |
| phEc4 | 8 | 8 | 8 | ND | ND | ND | 0 | - | - | - | - |
| phSE1 | 8 | 8 | 8 | 6 | 6 | 6 | 6 | 2 | - | - | - |
| phSE3 | 8 | 8 | 10 | 0 | 6 | 0 | 6 | ND | 0 | - | - |
| phAh2 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 10 | - |
| phAh4 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | ND | 10 |
 Hold Time (Hours).
Hold Time (Hours).| Phages | phSE1 | phSE2 | phSE3 | phEc4 |
| phSE1 | 10 | - | - | - |
| phSE2 | ND | 10 | - | - |
| phSE3 | ND | ND | 4 | - |
| phEc4 | 10 | 10 | 6 | ND |
 Hold Time (Hours).
Hold Time (Hours).References
- Chan, B.K.; Abedon, S.T.; Loc-Carrillo, C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013, 8, 769–783. [Google Scholar] [CrossRef] [PubMed]
- Curtright, A.J.; Abedon, S.T. Phage therapy: Emergent property pharmacology. J. Bioanal. Biomed. 2012, s6, 2. [Google Scholar] [CrossRef]
- Abedon, S.; Thomas-Abedon, C. Phage therapy pharmacology. Curr. Pharm. Biotechnol. 2010, 11, 28–47. [Google Scholar] [CrossRef] [PubMed]
- Loc-Carrillo, C.; Abedon, S. Pros and cons of phage therapy. Bacteriophage 2011, 1, 111–114. [Google Scholar] [CrossRef]
- Olszowska-Zaremba, N.; Borysowski, J.; Dąbrowska, K.; Górski, A. Phage translocation, safety and immunomodulation. In Bacteriophages in Health and Disease; Hyman, P., Abedon, S.T., Eds.; CABI: Wallingford, UK, 2012; pp. 168–184. [Google Scholar]
- Sulakvelidze, A.; Alavidze, Z.; Morris, J.G. Bacteriophage Therapy. Antimicrob. Agents Chemother. 2001, 45, 649–659. [Google Scholar] [CrossRef]
- Abedon, S.; García, P.; Mullany, P.; Aminov, R. Editorial: Phage therapy: Past, present and future. Front. Microbiol. 2017, 8, 981. [Google Scholar] [CrossRef]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 162. [Google Scholar] [CrossRef] [PubMed]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage applications for food production and processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef]
- Pereira, C.; Costa, P.; Duarte, J.; Balcão, V.M.; Almeida, A. Phage therapy as a potential approach in the biocontrol of pathogenic bacteria associated with shellfish consumption. Int. J. Food Microbiol. 2021, 338, 108995. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yuan, S.; Liu, Q.; Mai, G.; Yang, J.; Deng, D.; Zhang, B.; Liu, C.; Ma, Y. In vitro design and evaluation of phage cocktails against Aeromonas salmonicida. Front. Microbiol. 2018, 9, 1476. [Google Scholar] [CrossRef]
- Hyman, P.; Abedon, S. Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 2010, 70, 217–248. [Google Scholar] [CrossRef]
- Regeimbal, J.M.; Jacobs, A.C.; Corey, B.W.; Henry, M.S.; Thompson, M.G.; Pavlicek, R.L.; Quinones, J.; Hannah, R.M.; Ghebremedhin, M.; Crane, N.J.; et al. Personalized therapeutic cocktail of wild environmental phages rescues mice from Acinetobacter baumannii wound infections. Antimicrob. Agents Chemother. 2016, 60, 5806–5816. [Google Scholar] [CrossRef]
- Valappil, S.K.; Shetty, P.; Deim, Z.; Terhes, G.; Urbán, E.; Váczi, S.; Patai, R.; Polgár, T.; Pertics, B.Z.; Schneider, G.; et al. Survival comes at a cost: A coevolution of phage and its host leads to phage resistance and antibiotic sensitivity of Pseudomonas aeruginosa multidrug resistant strains. Front. Microbiol. 2021, 12, 783722. [Google Scholar] [CrossRef]
- Pirnay, J.-P.; De Vos, D.; Verbeken, G.; Merabishvili, M.; Chanishvili, N.; Vaneechoutte, M.; Zizi, M.; Laire, G.; Lavigne, R.; Huys, I.; et al. The phage therapy paradigm: Prêt-à-Porter or Sur-mesure? Pharm. Res. 2011, 28, 934–937. [Google Scholar] [CrossRef]
- Castillo, D.; Christiansen, R.H.; Dalsgaard, I.; Madsen, L.; Middelboe, M. Bacteriophage resistance mechanisms in the fish pathogen Flavobacterium psychrophilum: Linking genomic mutations to changes in bacterial virulence factors. Appl. Environ. Microbiol. 2015, 81, 1157–1167. [Google Scholar] [CrossRef] [PubMed]
- Melo, L.D.R.; Veiga, P.; Cerca, N.; Kropinski, A.M.; Almeida, C.; Azeredo, J.; Sillankorva, S. Development of a phage cocktail to control Proteus mirabilis catheter-associated urinary tract infections. Front. Microbiol. 2016, 7, 1024. [Google Scholar] [CrossRef] [PubMed]
- Yen, M.; Cairns, L.S.; Camilli, A. A cocktail of three virulent bacteriophages prevents Vibrio cholerae infection in animal models. Nat. Commun. 2017, 8, 14187. [Google Scholar] [CrossRef]
- Abedon, S.T.; Danis-Wlodarczyk, K.M.; Wozniak, D.J. Phage cocktail development for bacteriophage therapy: Toward improving spectrum of activity breadth and depth. Pharmaceuticals 2021, 14, 1019. [Google Scholar] [CrossRef] [PubMed]
- Butt, A.A.; Aldridge, K.E.; Sanders, C. V Infections related to the ingestion of seafood Part I: Viral and bacterial infections. Lancet Infect. Dis. 2004, 4, 201–212. [Google Scholar] [CrossRef]
- Feldhusen, F. The role of seafood in bacterialfoodborne diseases. Microbes Infect. 2000, 2, 1651–1660. [Google Scholar] [CrossRef]
- Costa, P.; Pereira, C.; Romalde, J.L.; Almeida, A. A game of resistance: War between bacteria and phages and how phage cocktails can be the solution. Virology 2024, 599, 110209. [Google Scholar] [CrossRef]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef]
- Yoo, S.; Lee, K.-M.; Kim, N.; Vu, T.N.; Abadie, R.; Yong, D. Designing phage cocktails to combat the emergence of bacteriophage-resistant mutants in multidrug-resistant Klebsiella pneumoniae. Microbiol. Spectr. 2024, 12, e0125823. [Google Scholar] [CrossRef]
- Mateus, L.; Costa, L.; Silva, Y.J.; Pereira, C.; Cunha, A.; Almeida, A. Efficiency of phage cocktails in the inactivation of Vibrio in aquaculture. Aquaculture 2014, 424–425, 167–173. [Google Scholar] [CrossRef]
- Costa, P.; Pereira, C.; Gomes, A.T.P.C.; Almeida, A. Efficiency of single phage suspensions and phage cocktail in the inactivation of Escherichia coli and Salmonella Typhimurium: An in vitro preliminary study. Microorganisms 2019, 7, 94. [Google Scholar] [CrossRef] [PubMed]
- Duarte, J.; Pereira, C.; Moreirinha, C.; Salvio, R.; Lopes, A.; Wang, D.; Almeida, A. New insights on phage efficacy to control Aeromonas salmonicida in aquaculture systems: An in vitro preliminary study. Aquaculture 2018, 495, 970–982. [Google Scholar] [CrossRef]
- Chen, H.; Liu, H.; Gong, Y.; Dunstan, R.A.; Ma, Z.; Zhou, C.; Zhao, D.; Tang, M.; Lithgow, T.; Zhou, T. A Klebsiella-phage cocktail to broaden the host range and delay bacteriophage resistance both in vitro and in vivo. NPJ Biofilms Microbiomes 2024, 10, 127. [Google Scholar] [CrossRef] [PubMed]
- Burrowes, B.H.; Molineux, I.J.; Fralick, J.A. Directed in vitro evolution of therapeutic bacteriophages: The appelmans protocol. Viruses 2019, 11, 241. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Batra, H.; Dong, J.; Chen, C.; Rao, V.B.; Tao, P. Genetic engineering of bacteriophages against infectious diseases. Front. Microbiol. 2019, 10, 954. [Google Scholar] [CrossRef]
- Merabishvili, M.; Pirnay, J.-P.; De Vos, D. Guidelines to Compose an Ideal Bacteriophage Cocktail. Methods Mol. Biol. 2018, 1693, 99–110. [Google Scholar]
- Haines, M.E.K.; Hodges, F.E.; Nale, J.Y.; Mahony, J.; van Sinderen, D.; Kaczorowska, J.; Alrashid, B.; Akter, M.; Brown, N.; Sauvageau, D.; et al. Analysis of selection methods to develop novel phage therapy cocktails against antimicrobial resistant clinical isolates of bacteria. Front. Microbiol. 2021, 12, 613529. [Google Scholar] [CrossRef]
- Merril, C.R.; Scholl, D.; Adhya, S.L. The prospect for bacteriophage therapy in Western medicine. Nat. Rev. Drug Discov. 2003, 2, 489–497. [Google Scholar] [CrossRef]
- Koskella, B.; Meaden, S. Understanding bacteriophage specificity in natural microbial communities. Viruses 2013, 5, 806–823. [Google Scholar] [CrossRef] [PubMed]
- de Jonge, P.A.; Nobrega, F.L.; Brouns, S.J.J.; Dutilh, B.E. Molecular and evolutionary determinants of bacteriophage host range. Trends Microbiol. 2019, 27, 51–63. [Google Scholar] [CrossRef]
- Ross, A.; Ward, S.; Hyman, P. More is better: Selecting for broad host range bacteriophages. Front. Microbiol. 2016, 7, 1352. [Google Scholar] [CrossRef]
- Hyman, P. Phages for phage therapy: Isolation, characterization, and host range breadth. Pharmaceuticals 2019, 12, 35. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, W.; Zhong, Q.; Chen, Q.; He, X.; Baker, J.L.; Xiong, K.; Jin, X.; Wang, J.; Hu, F.; et al. Development of a bacteriophage cocktail to constrain the emergence of phage-resistant Pseudomonas aeruginosa. Front. Microbiol. 2020, 11, 327. [Google Scholar] [CrossRef]
- Costa, P.; Gomes, A.T.P.C.; Braz, M.; Pereira, C.; Almeida, A. Application of the resazurin cell viability assay to monitor Escherichia coli and Salmonella Typhimurium inactivation mediated by phages. Antibiotics 2021, 10, 974. [Google Scholar] [CrossRef] [PubMed]
- Adams, M. Bacteriophages; John Wiley and Sons Inc.: New York, NY, USA, 1959. [Google Scholar]
- Abedon, S.T. Kinetics of phage-mediated biocontrol of bacteria. Foodborne Pathog. Dis. 2009, 6, 807–815. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 2010, 23, 35–73. [Google Scholar] [CrossRef] [PubMed]
- Tuttle, A.R.; Trahan, N.D.; Son, M.S. Growth and maintenance of Escherichia coli laboratory strains. Curr. Protoc. 2021, 1, e20. [Google Scholar] [CrossRef]
- Futoma-Kołoch, B.; Małaszczuk, M.; Korzekwa, K.; Steczkiewicz, M.; Gamian, A.; Bugla-Płoskońska, G. The prolonged treatment of Salmonella enterica strains with human serum effects in phenotype related to virulence. Int. J. Mol. Sci. 2023, 24, 883. [Google Scholar] [CrossRef] [PubMed]
- Meysman, P.; Sánchez-Rodríguez, A.; Fu, Q.; Marchal, K.; Engelen, K. Expression divergence between Escherichia coli and Salmonella enterica serovar Typhimurium reflects their lifestyles. Mol. Biol. Evol. 2013, 30, 1302–1314. [Google Scholar] [CrossRef]
- Jensen, E.C.; Schrader, H.S.; Rieland, B.; Thompson, T.L.; Lee, K.W.; Nickerson, K.W.; Kokjohn, T.A. Prevalence of broad-host-range lytic bacteriophages of Sphaerotilus natans, Escherichia coli, and Pseudomonas aeruginosa. Appl. Environ. Microbiol. 1998, 64, 575–580. [Google Scholar] [CrossRef]
- Yu, P.; Mathieu, J.; Li, M.; Dai, Z.; Alvarez, P.J.J. Isolation of polyvalent bacteriophages by sequential multiple-host approaches. Appl. Environ. Microbiol. 2016, 82, 808–815. [Google Scholar] [CrossRef]
- Burrowes, B.; Harper, D.R.; Anderson, J.; McConville, M.; Enright, M.C. Bacteriophage therapy: Potential uses in the control of antibiotic-resistant pathogens. Expert Rev. Anti. Infect. Ther. 2011, 9, 775–785. [Google Scholar] [CrossRef]
- Wandro, S.; Oliver, A.; Gallagher, T.; Weihe, C.; England, W.; Martiny, J.B.H.; Whiteson, K. Predictable molecular adaptation of coevolving Enterococcus faecium and lytic phage EfV12-phi1. Front. Microbiol. 2019, 9, 3192. [Google Scholar] [CrossRef] [PubMed]
- Costa, P.; Pereira, C.; Oliveira, V.; Gomes, N.C.M.; Romalde, J.L.; Almeida, A. Characterising phages for the control of pathogenic bacteria associated with bivalve consumption. Int. J. Food Microbiol. 2025, 432, 111096. [Google Scholar] [CrossRef]
- Shkoporov, A.N.; Khokhlova, E.V.; Fitzgerald, C.B.; Stockdale, S.R.; Draper, L.A.; Ross, R.P.; Hill, C. ΦCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nat. Commun. 2018, 9, 4781. [Google Scholar] [CrossRef]
- Mulla, Y.; Müller, J.; Trimcev, D.; Bollenbach, T. Extreme diversity of phage amplification rates and phage–antibiotic interactions revealed by PHORCE. PLoS Biol. 2025, 23, e3003065. [Google Scholar] [CrossRef] [PubMed]
- Vieira-da-Silva, B.; Castanho, M.A.R.B. Resazurin reduction-based assays revisited: Guidelines for accurate reporting of relative differences on metabolic status. Molecules 2023, 28, 2283. [Google Scholar] [CrossRef]
- Schmerer, M.; Molineux, I.J.; Bull, J.J. Synergy as a rationale for phage therapy using phage cocktails. PeerJ 2014, 2, e590. [Google Scholar] [CrossRef]
- Oechslin, F. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses 2018, 10, 351. [Google Scholar] [CrossRef]
- Molina, F.; Simancas, A.; Ramírez, M.; Tabla, R.; Roa, I.; Rebollo, J.E. A new pipeline for designing phage cocktails based on phage-bacteria infection networks. Front. Microbiol. 2021, 12, 564532. [Google Scholar] [CrossRef]
- Costa, P.; Pereira, C.; Barja, J.L.; Romalde, J.L.; Almeida, A. Enhancing bivalve depuration using a phage cocktail: An in vitro and in vivo study. Food Control 2025, 177, 111442. [Google Scholar] [CrossRef]
- Attrill, E.L.; Łapińska, U.; Westra, E.R.; Harding, S.V.; Pagliara, S. Slow growing bacteria survive bacteriophage in isolation. ISME Commun. 2023, 3, 95. [Google Scholar] [CrossRef] [PubMed]
- Fister, S.; Robben, C.; Witte, A.K.; Schoder, D.; Wagner, M.; Rossmanith, P. Influence of environmental factors on phage–bacteria interaction and on the efficacy and infectivity of phage P100. Front. Microbiol. 2016, 7, 1152. [Google Scholar] [CrossRef] [PubMed]
- Šivec, K.; Podgornik, A. Determination of bacteriophage growth parameters under cultivating conditions. Appl. Microbiol. Biotechnol. 2020, 104, 8949–8960. [Google Scholar] [CrossRef] [PubMed]
| Phages | E. coli | S. Typhimurium | S. Enteritidis | A. hydrophila |
|---|---|---|---|---|
| phEc1 | H | ● | X | X |
| phEc2 | H | ● | X | X |
| phEc3 | H | ● | X | X |
| phEc4 | H | ● | ● | X |
| phST1 | ● | H | X | X |
| phST2 | ● | H | X | X |
| phST3 | ● | H | X | X |
| phSE1 | ● | ● | H | X |
| phSE2 | ● | X | H | X |
| phSE3 | X | ● | H | X |
| phAh2 | ● | ● | X | H |
| phAh4 | ● | ● | X | H |
| Target Bacterium | Selected Phage | Efficacy Highlights | Host Range Justification | Reason for Selection over Alternatives |
|---|---|---|---|---|
| E. coli | phEc3 | Strong inactivation alone (Figure 1A) and in cocktails (e.g., phEc3/phAh2); no antagonism observed | Good specificity without cross-host antagonism | More effective than other E. coli phages; synergetic in cocktails |
| S. Typhimurium | phST1 | Robust suppression; maintained efficacy in multiple cocktails (e.g., phST1/phSE1); did not induce early regrowth | Functional in both single and combination treatments | Preferred over phST2, which showed antagonism with phEc3 (Figure 1G) |
| S. Enteritidis | phSE1 | Effective in single and combo assays (Figure 1C,E,K); e.g., phEc1/phSE1 and phSE1/phAh2 showed enhanced activity | Broader host range than phSE2 or phSE3, aligning with multi-pathogen coverage goal | Better synergy and coverage than more potent but narrower phSE2 |
| A. hydrophila | phAh2 | Strong enhancement in cocktails with phEc3 and phSE1 (Figure 1E–G); consistent inactivation across targets | Broader efficacy across host bacteria compared to phAh4 | Chosen over phAh4 for broader effectiveness with fewer phage–phage antagonisms |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, P.; Pereira, C.; Romalde, J.L.; Almeida, A. From Isolation to Application: Designing a Multi-Target Phage Cocktail for Bivalve Safety. Microorganisms 2025, 13, 2708. https://doi.org/10.3390/microorganisms13122708
Costa P, Pereira C, Romalde JL, Almeida A. From Isolation to Application: Designing a Multi-Target Phage Cocktail for Bivalve Safety. Microorganisms. 2025; 13(12):2708. https://doi.org/10.3390/microorganisms13122708
Chicago/Turabian StyleCosta, Pedro, Carla Pereira, Jesús L. Romalde, and Adelaide Almeida. 2025. "From Isolation to Application: Designing a Multi-Target Phage Cocktail for Bivalve Safety" Microorganisms 13, no. 12: 2708. https://doi.org/10.3390/microorganisms13122708
APA StyleCosta, P., Pereira, C., Romalde, J. L., & Almeida, A. (2025). From Isolation to Application: Designing a Multi-Target Phage Cocktail for Bivalve Safety. Microorganisms, 13(12), 2708. https://doi.org/10.3390/microorganisms13122708









