Integrated Metagenomic and Lipidomic Profiling Reveals Dysregulation of Facial Skin Microbiome in Moderate Acne Vulgaris
Abstract
1. Introduction
2. Materials and Methods
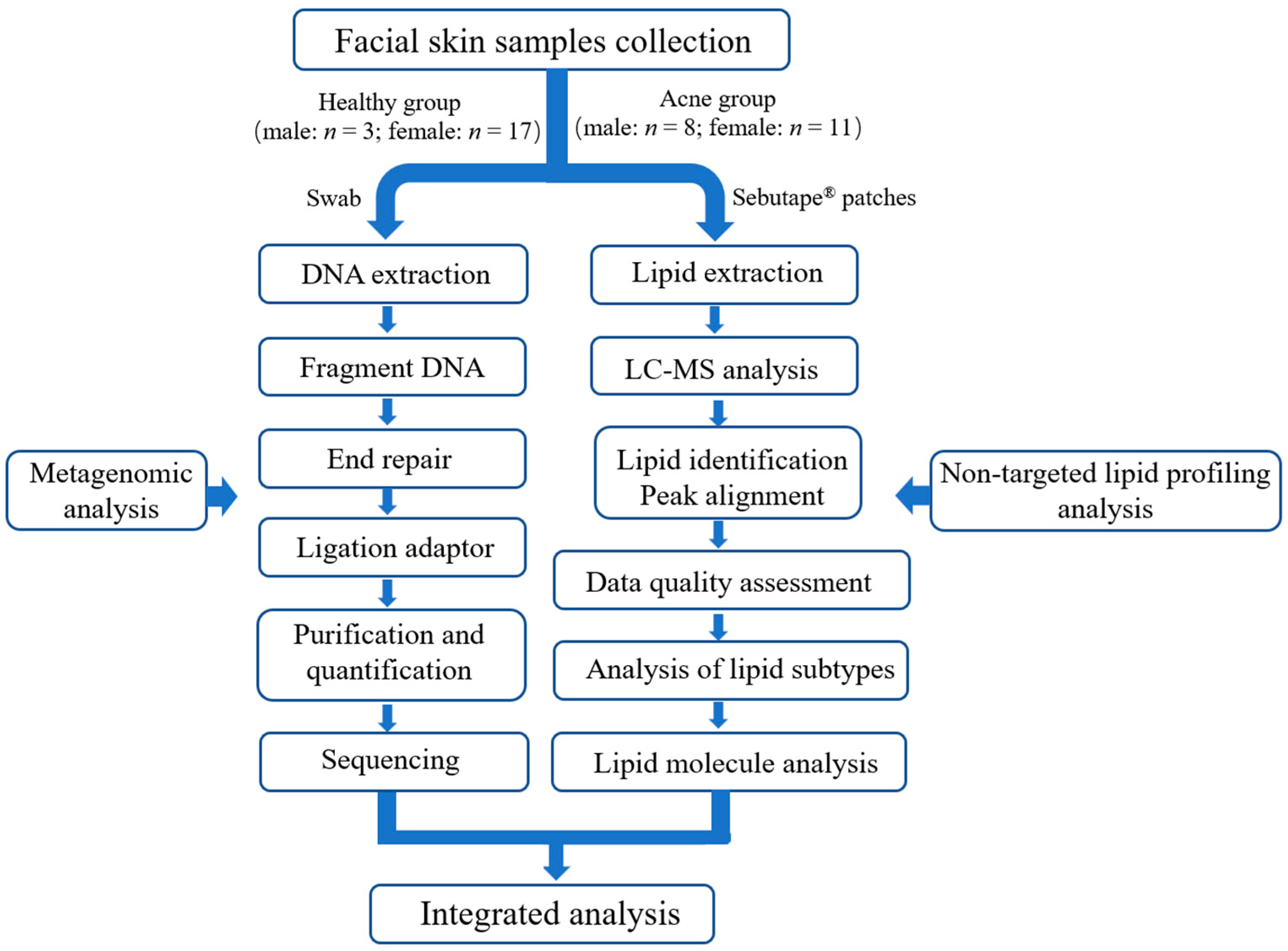
2.1. Study Design
2.2. Recruitment of Participants
2.3. Skin Sample Collection
2.4. Metagenomic Sequencing
2.5. Untargeted Lipidomic Analysis
2.6. Statistical Data Analysis
3. Results
3.1. Facial Skin Physicochemical Indexes
3.2. Abundance of Facial Microbiota Sequencing
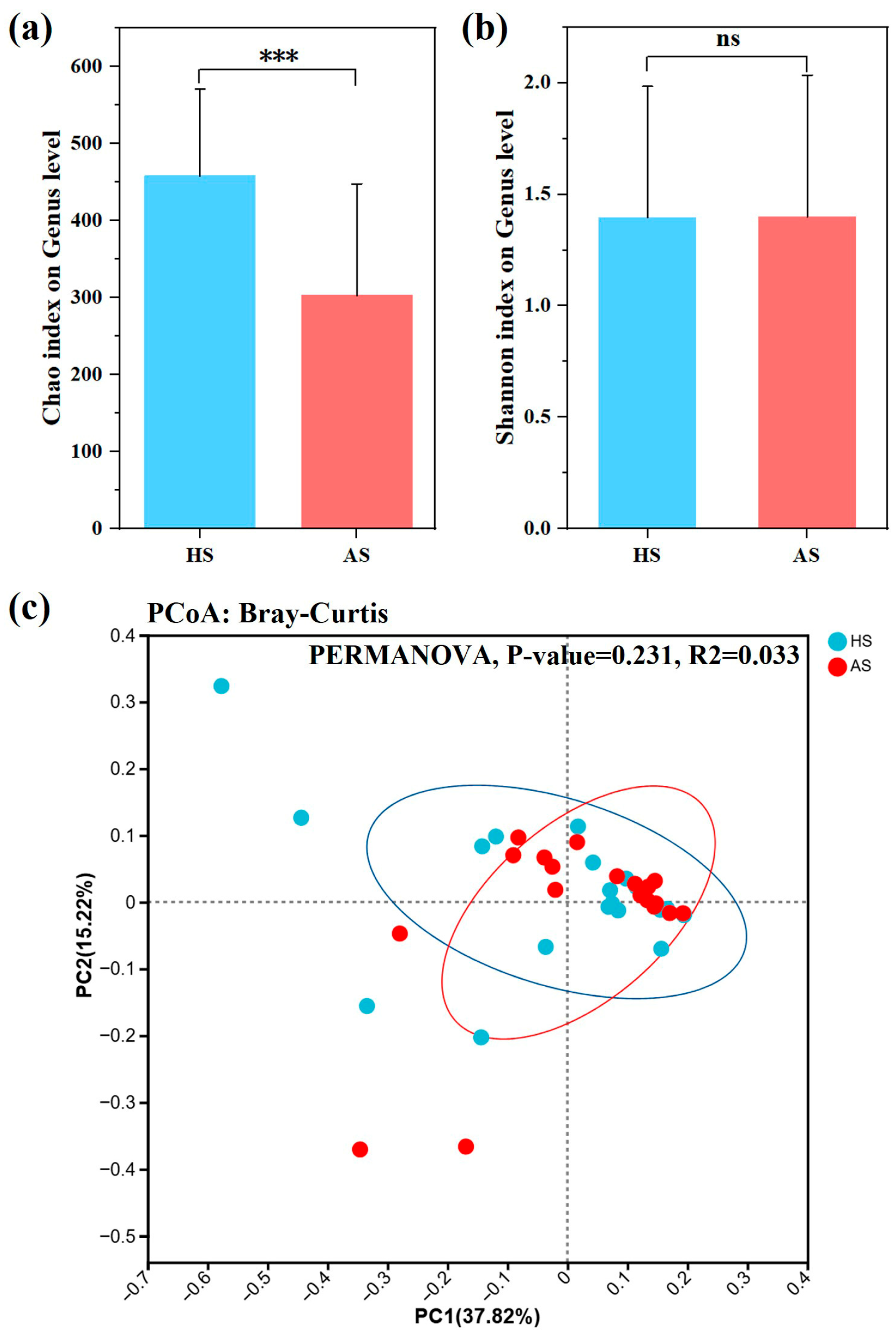
3.3. Analysis of Facial Microbiota Diversity
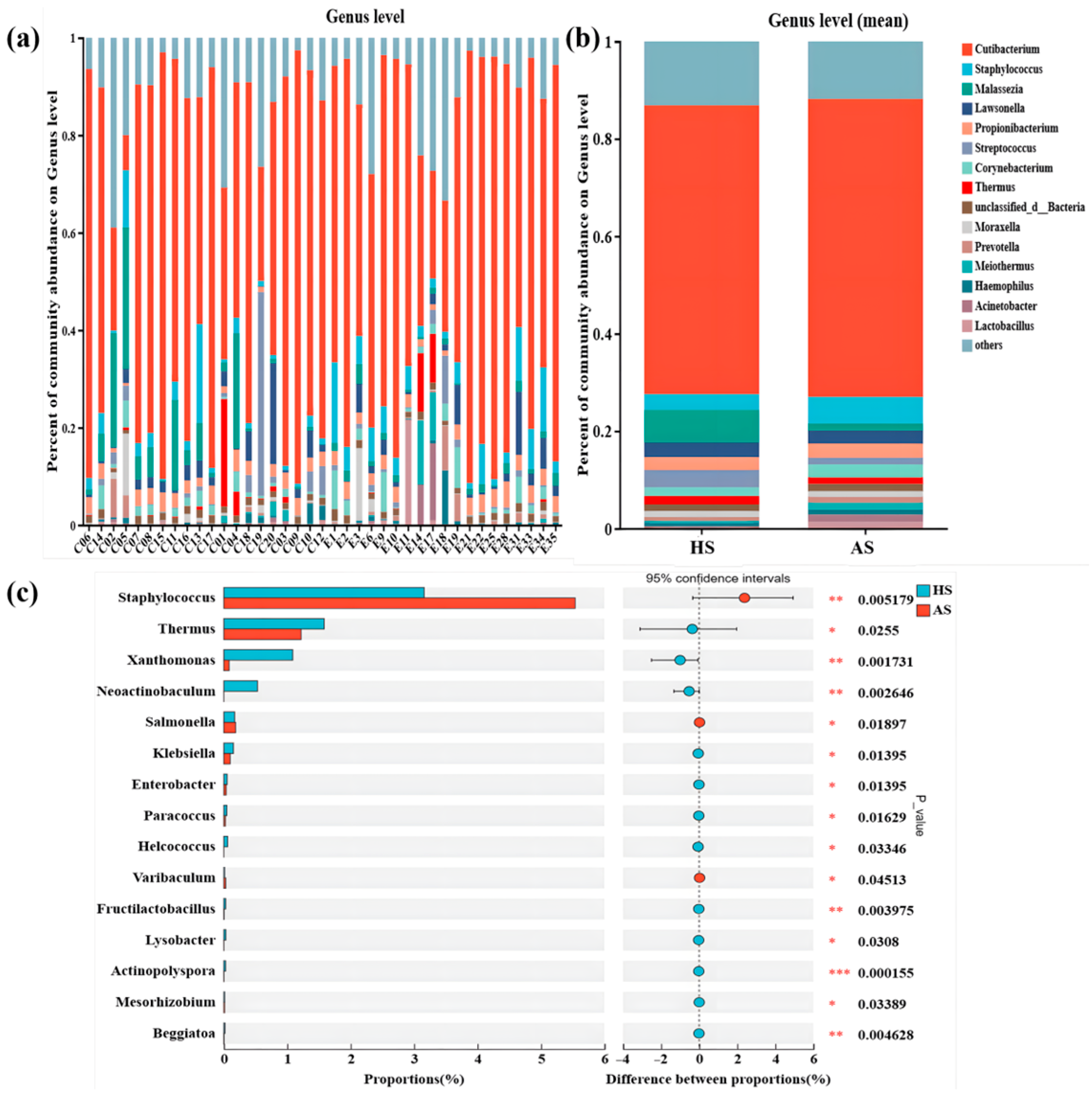
3.4. Taxonomic Composition of the Facial Microbiota in the Acne and Healthy Groups
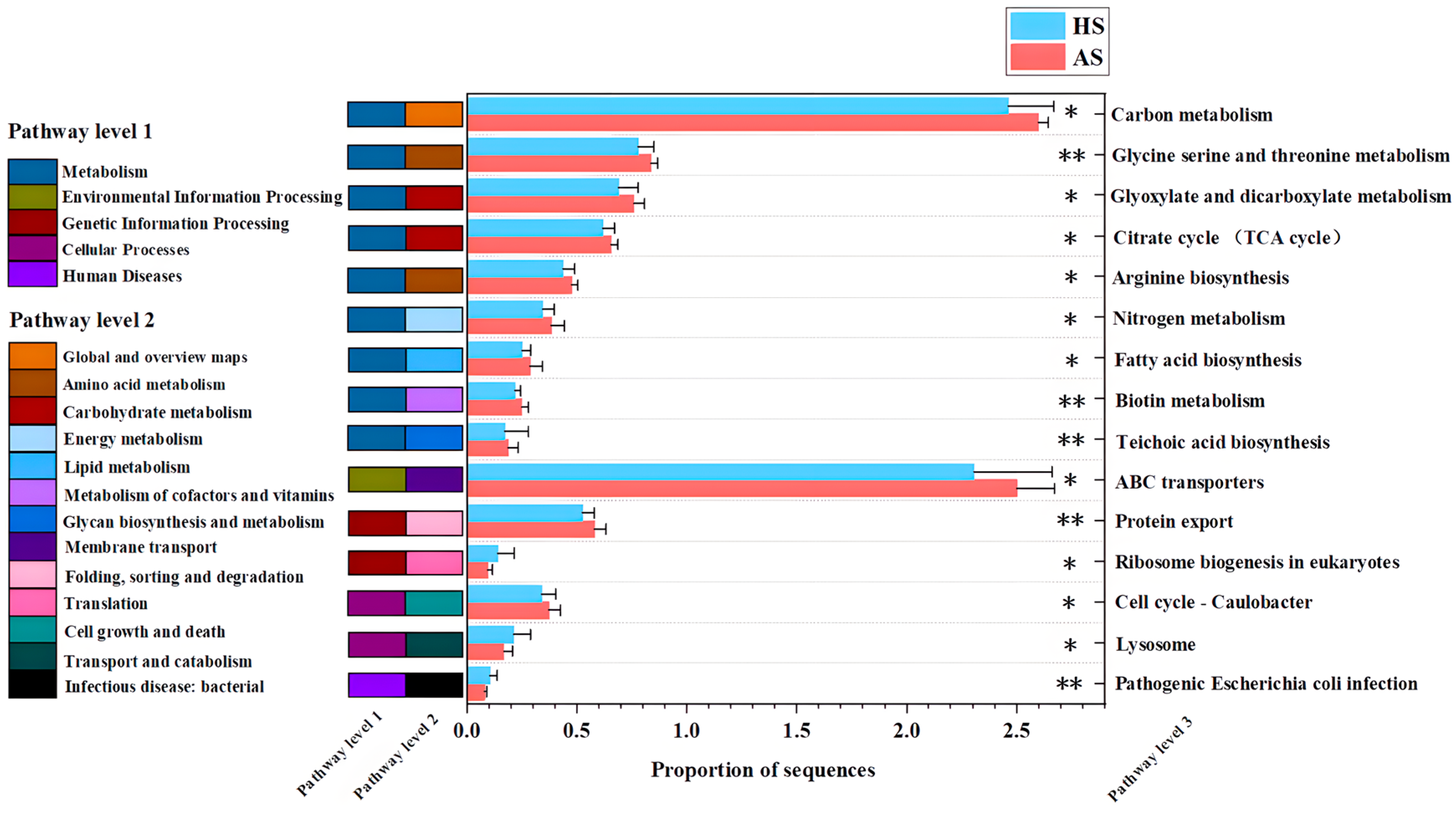
3.5. Functional Analysis of Facial Microbiota Genes in the Acne and Healthy Groups
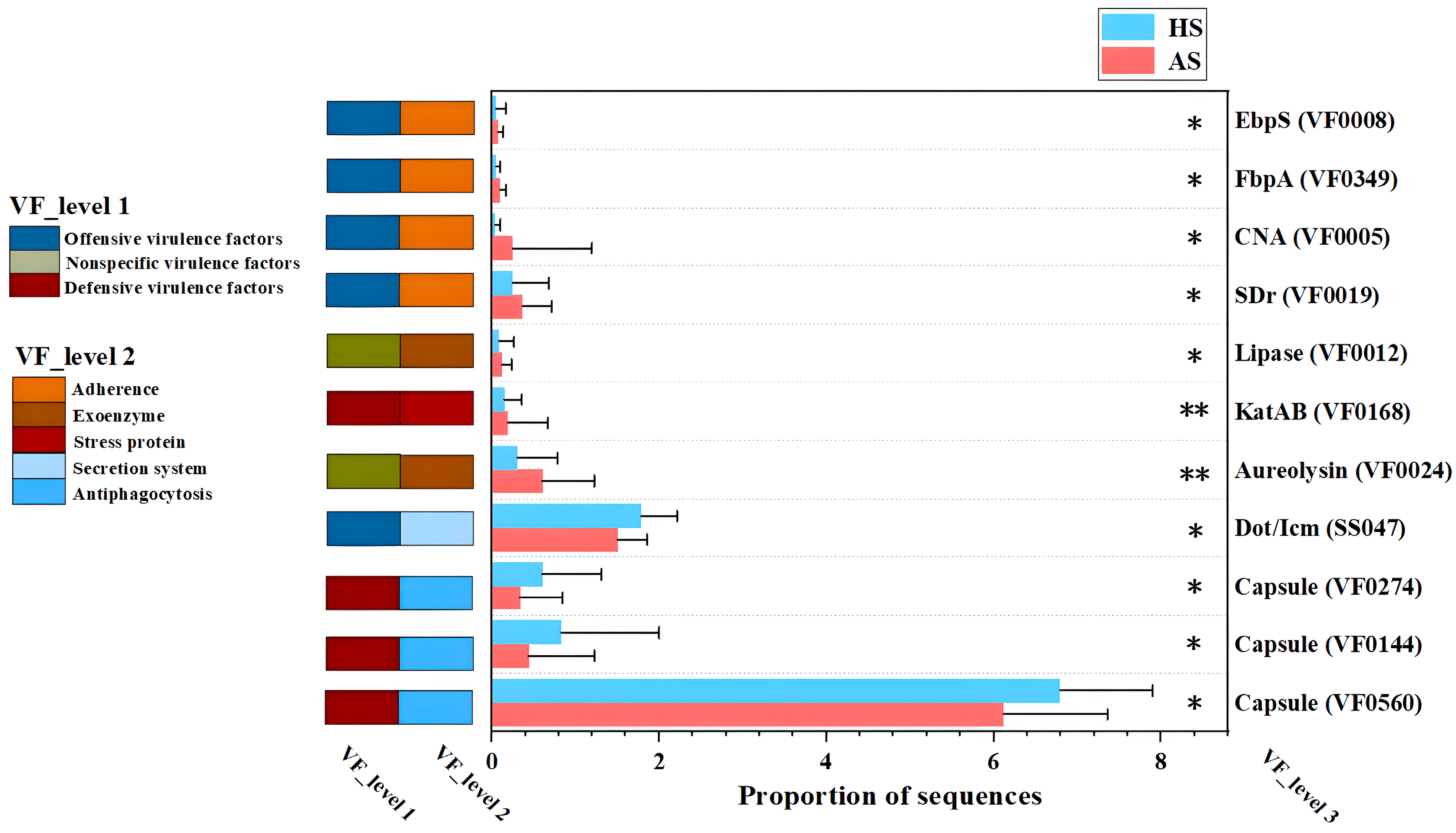
3.6. Analysis of Virulence Factors and Drug Resistance Genes in the Facial Microbiota of the Acne and Healthy Groups
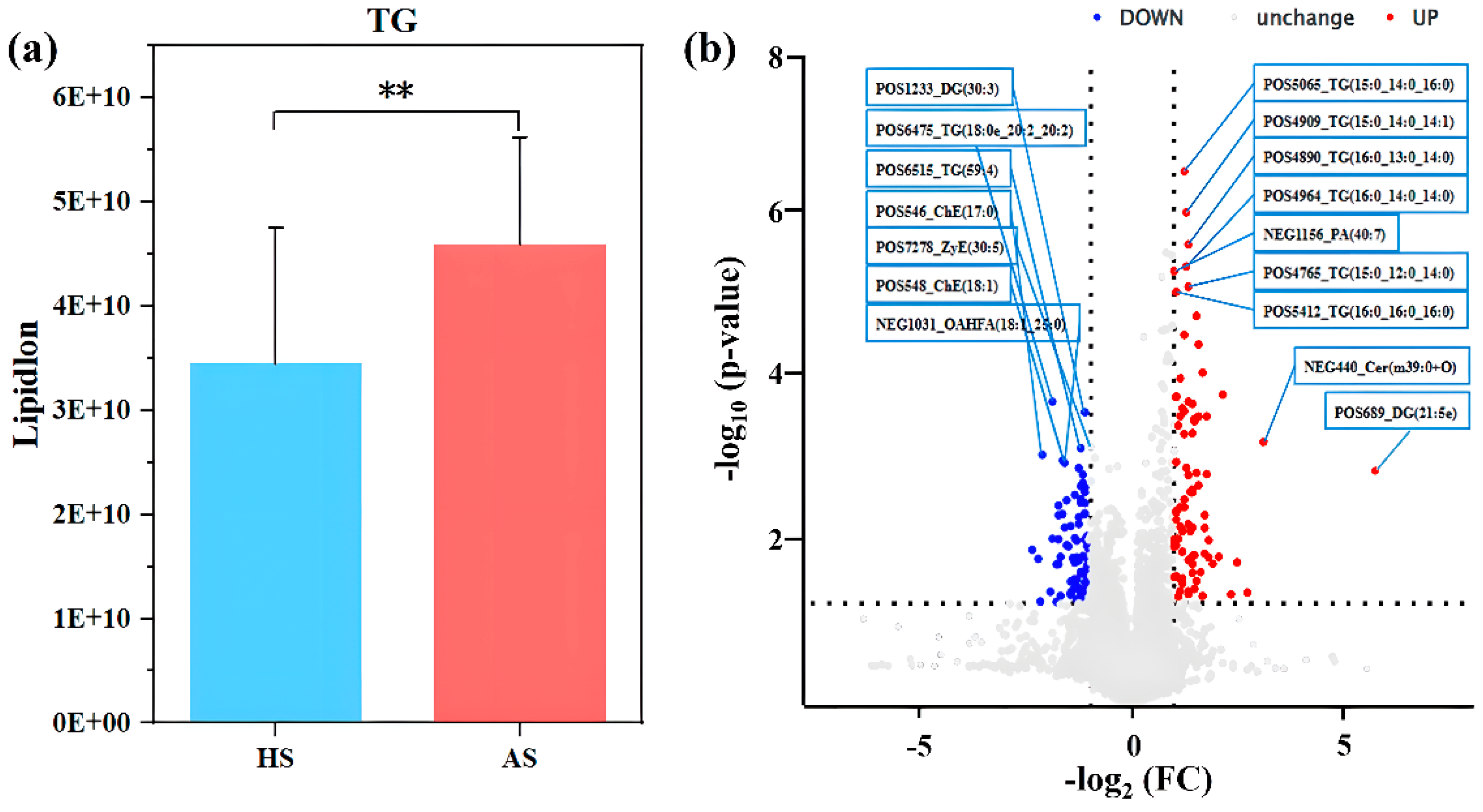
3.7. Differences in Facial Skin Lipids Between the Acne and Healthy Groups
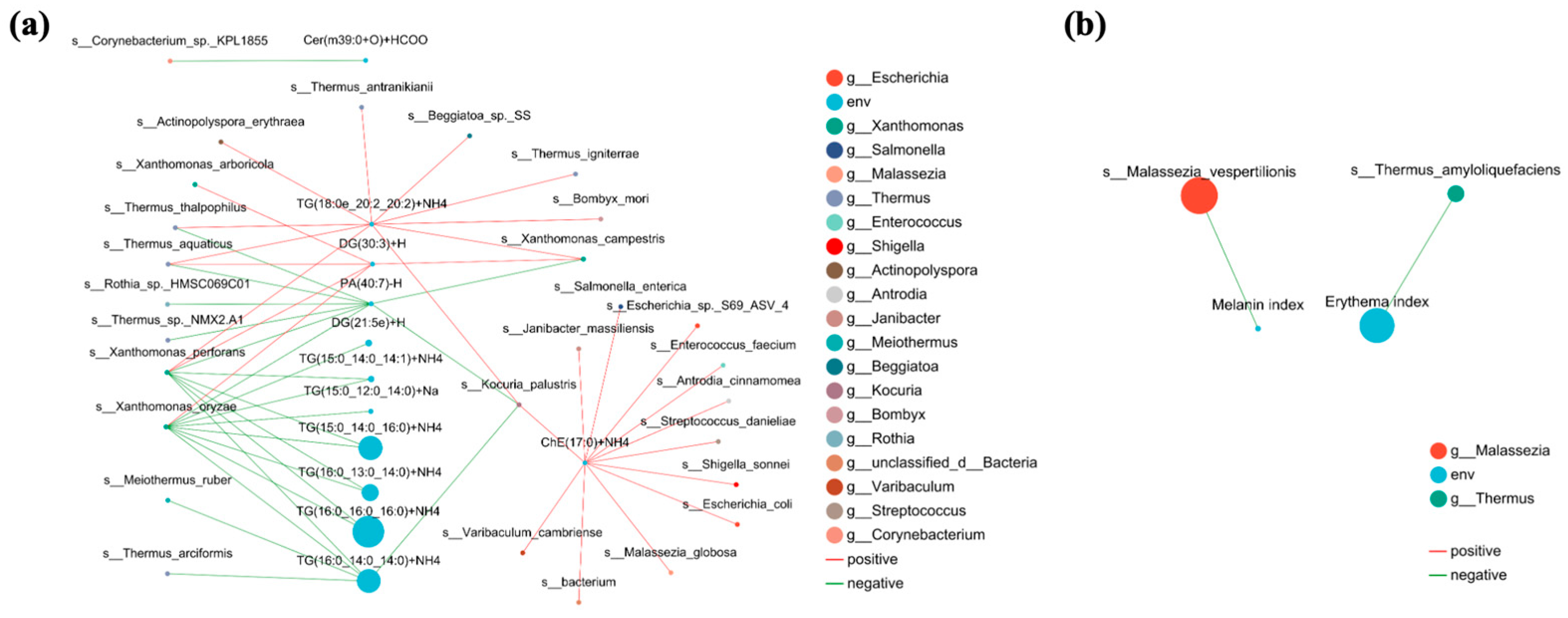
3.8. Relationship Between Facial Skin Microbiome, Differential Skin Lipids, Clinical Factors in Acne and Healthy Groups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, H.; Zhang, T.C.; Yin, X.L.; Man, J.Y.; Yang, X.R.; Lu, M. Magnitude and temporal trend of acne vulgaris burden in 204 countries and territories from 1990 to 2019: An analysis from the Global Burden of Disease Study 2019. Br. J. Dermatol. 2022, 186, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Fan, M.; Fan, X.; Chen, J.; Xiang, L.F.; Ma, Y. Progress on Multiomics Research on Acne Vulgaris: A Literature Review. J. Investig. Dermatol. 2025, 145, 2162–2169. [Google Scholar] [CrossRef] [PubMed]
- Dréno, B.; Pécastaings, S.; Corvec, S.; Veraldi, S.; Khammari, A.; Roques, C. Cutibacterium acnes (Propionibacterium acnes) and acne vulgaris: A brief look at the latest updates. J. Eur. Acad. Dermatol. Venereol. 2018, 32 (Suppl. S2), 5–14. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. TLR signaling. Semin. Immunol. 2007, 19, 24–32. [Google Scholar]
- Sanford, J.A.; O’Neill, A.M.; Zouboulis, C.C.; Gallo, R.L. Short-Chain Fatty Acids from Cutibacterium acnes Activate Both a Canonical and Epigenetic Inflammatory Response in Human Sebocytes. J. Immunol. 2019, 202, 1767–1776. [Google Scholar] [CrossRef] [PubMed]
- Mias, C.; Mengeaud, V.; Bessou-Touya, S.; Duplan, H. Recent advances in understanding inflammatory acne: Deciphering the relationship between Cutibacterium acnes and Th17 inflammatory pathway. J. Eur. Acad. Dermatol. Venereol. 2023, 37 (Suppl. S2), 3–11. [Google Scholar] [CrossRef] [PubMed]
- Paugam, C.; Corvec, S.; Saint-Jean, M.; Le Moigne, M.; Khammari, A.; Boisrobert, A.; Nguyen, J.; Gaultier, A.; Dréno, B. Propionibacterium acnes phylotypes and acne severity: An observational prospective study. J. Eur. Acad. Dermatol. Venereol. 2017, 31, e398–e399. [Google Scholar] [CrossRef]
- Borrel, V.; Gannesen, A.V.; Barreau, M.; Gaviard, C.; Duclairoir-Poc, C.; Hardouin, J.; Konto-Ghiorghi, Y.; Lefeuvre, L.; Feuilloley, M.G.J. Adaptation of acneic and non acneic strains of Cutibacterium acnes to sebum-like environment. Microbiologyopen 2019, 8, e00841. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; O’Neill, A.M.; Williams, M.R.; Cau, L.; Nakatsuji, T.; Horswill, A.R.; Gallo, R.L. Short chain fatty acids produced by Cutibacterium acnes inhibit biofilm formation by Staphylococcus epidermidis. Sci. Rep. 2020, 10, 21237. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dreno, B.; Dekio, I.; Baldwin, H.; Demessant, A.L.; Dagnelie, M.A.; Khammari, A.; Corvec, S. Acne microbiome: From phyla to phylotypes. J. Eur. Acad. Dermatol. Venereol. 2024, 38, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Fournière, M.; Latire, T.; Souak, D.; Feuilloley, M.G.J.; Bedoux, G. Staphylococcus epidermidis and Cutibacterium acnes: Two major sentinels of skin microbiota and the influence of cosmetics. Microorganisms 2020, 8, 1752. [Google Scholar] [CrossRef]
- Yu, T.; Xu, X.; Liu, Y.; Wang, X.; Wu, S.; Qiu, Z.; Liu, X.; Pan, X.; Gu, C.; Wang, S.; et al. Multi-omics signatures reveal genomic and functional heterogeneity of Cutibacterium acnes in normal and diseased skin. Cell Host Microbe 2024, 32, 1129–1146.e8. [Google Scholar] [CrossRef]
- Camera, E.; Ludovici, M.; Tortorella, S.; Sinagra, J.L.; Capitanio, B.; Goracci, L.; Picardo, M. Use of lipidomics to investigate sebum dysfunction in juvenile acne. J. Lipid Res. 2016, 57, 1051–1058. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Sun, Q.; Gao, J.; Liu, Q.; Tian, H.; Ding, H.; Qiao, J.; Chen, H. Quantitative lipidomics profiling of skin surface lipids and skin barrier function evaluation in patients with acne vulgaris. Arch. Dermatol. Res. 2025, 317, 349. [Google Scholar] [CrossRef] [PubMed]
- Pocock, S.J. Clinical Trials: A Practical Approach; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Xiao, Y. High—Throughput Sequencing Analysis of the Facial Microbiota Before and After 2% Supramolecular Salicylic Acid Treatment in Patients with Moderate Acne Vulgaris. Master’s Thesis, Southwest Medical University, Luzhou, China, 2022. [Google Scholar]
- Mayer, W.; Weibel, M.; De Luca, C.; Ibragimova, G.; Trakhtman, I.; Kharaeva, Z.; Chandler, D.L.; Korkina, L. Biomolecules of Fermented Tropical Fruits and Fermenting Microbes as Regulators of Human Hair Loss, Hair Quality, and Scalp Microbiota. Biomolecules 2023, 13, 699. [Google Scholar] [CrossRef]
- Yuan, Y.; Xu, F.; Jin, M.; Wang, X.; Hu, X.; Zhao, M.; Cheng, X.; Luo, J.; Jiao, L.; Betancor, M.B.; et al. Untargeted lipidomics reveals metabolic responses to different dietary n-3 PUFA in juvenile swimming crab (Portunus trituberculatus). Food Chem. 2021, 354, 129570. [Google Scholar] [CrossRef]
- Wei, J.; Dai, W.; Pan, X.; Zhong, Y.; Xu, N.; Ye, P.; Wang, J.; Li, J.; Yang, F.; Luo, J.; et al. Identifying the Novel Gut Microbial Metabolite Contributing to Metabolic Syndrome in Children Based on Integrative Analyses of Microbiome-Metabolome Signatures. Microbiol. Spectr. 2023, 11, e0377122. [Google Scholar] [CrossRef]
- Akhtar, A.A.; Turner, D.P. The role of bacterial ATP-binding cassette (ABC) transporters in pathogenesis and virulence: Therapeutic and vaccine potential. Microb. Pathog. 2022, 171, 105734. [Google Scholar] [CrossRef] [PubMed]
- Vuong, C.; Otto, M. Staphylococcus epidermidis infections. Microbes Infect. 2002, 4, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.J.; Geoghe-gan, J.A.; Ganesh, V.K.; Höök, M. Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 2014, 12, 49–62. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Patti, J.M.; Bremell, T.; Krajewska-Pietrasik, D.; Abdelnour, A.; Tarkowski, A.; Rydén, C.; Höök, M. The Staphylococcus aureus collagen adhesin is a virulence determinant in experimental septic arthritis. Infect. Immun. 1994, 62, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Corrigan, R.M.; Rigby, D.; Handley, P.; Foster, T.J. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 2007, 153 Pt 8, 2435–2446. [Google Scholar] [CrossRef] [PubMed]
- Rollof, J.; Braconier, J.H.; Söderström, C.; Nilsson-Ehle, P. Interference of Staphylococcus aureus lipase with human granulocyte function. Eur. J. Clin. Microbiol. Infect. Dis. 1988, 7, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Sieprawska-Lupa, M.; Mydel, P.; Krawczyk, K.; Wójcik, K.; Puklo, M.; Lupa, B.; Suder, P.; Silber-ring, J.; Reed, M.; Pohl, J.; et al. Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob. Agents Chemother. 2004, 48, 4673–4679. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sanz, R.; Marı, N.I.; Ruiz-Santa-Quiteria, J.A.; Orden, J.A.; Cid, D.; Diez, R.M.; Silhadi, K.S.; Amils, R.; de la Fuente, R. Catalase deficiency in Staphylococcus aureus subsp. anaerobius is associated with natural loss-of-function mutations within the structural gene. Microbiology 2000, 146 Pt 2, 465–475. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Panmanee, W.; Hassett, D.J. Differential roles of OxyR-controlled antioxidant enzymes alkyl hydroperoxide reductase (AhpCF) and catalase (KatB) in the protection of Pseudomonas aeruginosa against hydrogen peroxide in biofilm vs. planktonic culture. FEMS Microbiol. Lett. 2009, 295, 238–244. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, B.; Park, S.; Thompson, C.D.; Li, X.; Lee, J.C. Antibodies to Staphylococcus aureus capsular polysaccharides 5 and 8 perform similarly in vitro but are functionally distinct in vivo. Virulence 2017, 8, 859–874. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Luo, Z.Q.; Isberg, R.R. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. USA 2004, 101, 841–846. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- De Pessemier, B.; López, C.D.; Taelman, S.; Verdonck, M.; Chen, Y.; Stockman, A.; Lambert, J.; Van de Wiele, T.; Callewaert, C. Comparative Whole Metagenome Analysis in Lesional and Nonlesional Scalp Areas of Patients with Psoriasis Capitis and Healthy Individuals. J. Investig. Dermatol. 2025, 145, 605–617.e14. [Google Scholar] [CrossRef] [PubMed]
- De La Hoz-Romo, M.C.; Díaz, L.; Gómez-León, J.; Quintero, M.; Villamil, L. Marine actinobacteria metabolites: Unlocking new treatments for acne vulgaris. Front. Microbiol. 2025, 15, 1501951. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Harder, J.; Tsuruta, D.; Murakami, M.; Kurokawa, I. What is the role of antimicrobial peptides (AMP) in acne vulgaris? Exp. Dermatol. 2013, 22, 386–391. [Google Scholar] [CrossRef]
- Li, X.; He, C.; Chen, Z.; Zhou, C.; Gan, Y.; Jia, Y. A review of the role of sebum in the mechanism of acne pathogenesis. J. Cosmet. Dermatol. 2017, 16, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Callewaert, C.; Nakatsuji, T.; Knight, R.; Kosciolek, T.; Vrbanac, A.; Kotol, P.; Ardeleanu, M.; Hultsch, T.; Guttman-Yassky, E.; Bissonnette, R.; et al. IL-4Rα Blockade by Dupilumab Decreases Staphylococcus aureus Colonization and Increases Microbial Diversity in Atopic Dermatitis. J. Investig. Dermatol. 2020, 140, 191–202.e7. [Google Scholar] [CrossRef] [PubMed]
- Vieira-Silva, S.; Sabino, J.; Valles-Colomer, M.; Falony, G.; Kathagen, G.; Caenepeel, C.; Cleynen, I.; van der Merwe, S.; Vermeire, S.; Raes, J. Quantitative microbiome profiling disentangles inflammation- and bile duct obstruction-associated microbiota alterations across PSC/IBD diagnoses. Nat. Microbiol. 2019, 4, 1826–1831. [Google Scholar] [CrossRef]
- Liang, M.C.; Li, J.Q.; Wu, X.Y.; Mo, X.H.; Ju, Q. Analysis of hair follicle microbiota in non-lesional areas of patients with moderate-to-severe acne vulgaris: A single-center cross-sectional study. Shanghai Jiaotong Univ. Med. Sci. 2024, 44, 1094–1103. [Google Scholar]
- Guo, Z.; Yang, Y.; Wu, Q.; Liu, M.; Zhou, L.; Zhang, L.; Dong, D. New insights into the characteristic skin microorganisms in different grades of acne and different acne sites. Front. Microbiol. 2023, 14, 1167923. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Manurung, T.H.P.; Sitohang, I.B.S.; Agustin, T. Staphylococcus caprae and Staphylococcus epidermidis define the skin microbiome among different grades of acne vulgaris. Arch. Dermatol. Res. 2024, 317, 156. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Loss, M.; Thompson, K.G.; Agostinho-Hunt, A.; James, G.A.; Mongodin, E.F.; Rosenthal, I.; Cheng, N.; Leung, S.; Chien, A.L.; Kang, S. Noninflammatory comedones have greater diversity in microbiome and are more prone to biofilm formation than inflammatory lesions of acne vulgaris. Int. J. Dermatol. 2021, 60, 589–596. [Google Scholar] [CrossRef]
- Wang, W.; Dernst, A.; Martin, B.; Lorenzi, L.; Cadefau-Fabregat, M.; Phulphagar, K.; Wagener, A.; Budden, C.; Stair, N.; Wagner, T.; et al. Butyrate and propionate are microbial danger signals that activate the NLRP3 inflammasome in human macrophages upon TLR stimulation. Cell Rep. 2024, 43, 114736. [Google Scholar] [CrossRef] [PubMed]
- Almoughrabie, S.; Cau, L.; Cavagnero, K.; O’Neill, A.M.; Li, F.; Roso-Mares, A.; Mainzer, C.; Closs, B.; Kolar, M.J.; Williams, K.J.; et al. Commensal Cutibacterium acnes induce epidermal lipid synthesis important for skin barrier function. Sci. Adv. 2023, 9, eadg6262. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- He, Q.; Shu, H.; Peng, Y.; Xu, Y.; Liu, L.; Zhou, J.; Zhao, J.; Xiong, X.; Li, C. Untargeted metabolomics analysis of plasma metabolic characteristics in patients with acne and insulin resistance. Amino Acids 2023, 55, 1417–1428. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- O’Neill, A.M.; Gallo, R.L. Host-microbiome interactions and recent progress into understanding the biology of acne vulgaris. Microbiome 2018, 6, 177. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Varin-Simon, J.; Colin, M.; Velard, F.; Tang-Fichaux, M.; Ohl, X.; Mongaret, C.; Gangloff, S.C.; Reffuveille, F. Cutibacterium acnes biofilm formation is influenced by bone microenvironment, implant surfaces and bacterial internalization. BMC Microbiol. 2024, 24, 270, Erratum in BMC Microbiol. 2025, 25, 48. https://doi.org/10.1186/s12866-025-03777-z. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dagnelie, M.A.; Corvec, S.; Timon-David, E.; Khammari, A.; Dréno, B. Cutibacterium acnes and Staphylococcus epidermidis: The unmissable modulators of skin inflammatory response. Exp. Dermatol. 2022, 31, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Okoro, O.E.; Adenle, A.; Ludovici, M.; Truglio, M.; Marini, F.; Camera, E. Lipidomics of facial sebum in the comparison between acne and non-acne adolescents with dark skin. Sci. Rep. 2021, 11, 16591, Erratum in Sci. Rep. 2021, 11, 17974. [Google Scholar]
- Christensen, G.J.; Scholz, C.F.; Enghild, J.; Rohde, H.; Kilian, M.; Thürmer, A.; Brzuszkiewicz, E.; Lomholt, H.B.; Brüggemann, H. Antagonism between Staphylococcus epidermidis and Propionibacterium acnes and its genomic basis. BMC Genom. 2016, 17, 152. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hamann, T.; Brüggemann, H.; Feidenhansl, C.; Rruci, E.; Gallinger, J.; Gallinat, S.; Hüpeden, J. Distinct Intraspecies Variation of Cutibacterium acnes and Staphylococcus epidermidis in Acne Vulgaris and Healthy Skin. Microorganisms 2025, 13, 299. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, Y.; Kuo, S.; Shu, M.; Yu, J.; Huang, S.; Dai, A.; Two, A.; Gallo, R.L.; Huang, C.M. Staphylococcus epidermidis in the human skin microbiome mediates fermentation to inhibit the growth of Propionibacterium acnes: Implications of probiotics in acne vulgaris. Appl. Microbiol. Biotechnol. 2014, 98, 411–424. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ochlich, D.; Rademacher, F.; Drerup, K.A.; Gläser, R.; Harder, J. The influence of the commensal skin bacterium Staphylococcus epidermidis on the epidermal barrier and inflammation: Implications for atopic dermatitis. Exp. Dermatol. 2023, 32, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Han, H.S.; Shin, S.H.; Choi, B.Y.; Koo, N.; Lim, S.; Son, D.; Chung, M.J.; Park, K.Y.; Sul, W.J. A split face study on the effect of an anti-acne product containing fermentation products of Enterococcus faecalis CBT SL-5 on skin microbiome modification and acne improvement. J. Microbiol. 2022, 60, 488–495, Erratum in J. Microbiol. 2022, 60, 766. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef]
- Noguchi, H.; Park, J.; Takagi, T. MetaGene: Prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006, 34, 5623–5630. [Google Scholar] [CrossRef]

| Physicochemical Indexes | HS (Mean ± SD) | AS (Mean ± SD) | p-Value |
|---|---|---|---|
| Moisture content | 79.49 ± 14.34 | 74.62 ± 18.11 | 0.24 |
| TWEL | 16.80 ± 3.77 | 25.81 ± 7.20 | <0.001 |
| Oil content | 17.30 ± 11.95 | 35.27 ± 16.67 | <0.001 |
| Melanin index | 20.71 ± 22.20 | 57.20 ± 34.57 | <0.001 |
| Erythema index | 227.34 ± 51.77 | 353.84 ± 67.50 | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, X.; Han, Z.; Meng, J.; Zhao, H.; Zhou, M.; Wang, M.; Kang, S.; Shi, Q.; Li, H.; Lu, F.; et al. Integrated Metagenomic and Lipidomic Profiling Reveals Dysregulation of Facial Skin Microbiome in Moderate Acne Vulgaris. Microorganisms 2025, 13, 2674. https://doi.org/10.3390/microorganisms13122674
Qi X, Han Z, Meng J, Zhao H, Zhou M, Wang M, Kang S, Shi Q, Li H, Lu F, et al. Integrated Metagenomic and Lipidomic Profiling Reveals Dysregulation of Facial Skin Microbiome in Moderate Acne Vulgaris. Microorganisms. 2025; 13(12):2674. https://doi.org/10.3390/microorganisms13122674
Chicago/Turabian StyleQi, Xiaoye, Zhaoying Han, Jie Meng, Hongrui Zhao, Maoyuan Zhou, Meichao Wang, Shengze Kang, Qingying Shi, Hongyan Li, Fuping Lu, and et al. 2025. "Integrated Metagenomic and Lipidomic Profiling Reveals Dysregulation of Facial Skin Microbiome in Moderate Acne Vulgaris" Microorganisms 13, no. 12: 2674. https://doi.org/10.3390/microorganisms13122674
APA StyleQi, X., Han, Z., Meng, J., Zhao, H., Zhou, M., Wang, M., Kang, S., Shi, Q., Li, H., Lu, F., & Zhao, H. (2025). Integrated Metagenomic and Lipidomic Profiling Reveals Dysregulation of Facial Skin Microbiome in Moderate Acne Vulgaris. Microorganisms, 13(12), 2674. https://doi.org/10.3390/microorganisms13122674






