Metabolic Engineering of Bacillus licheniformis for High-Yield L-Lactic Acid and Galactooligosaccharide Retention in Complementary Synbiotics Production
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Vectors, Primers, and Chemicals
2.2. Gene Deletion and Construction of Recombinant Bacteria
2.3. Flask Fermentation Experiments
2.4. Bioreactor Experiments
2.5. Analytical Methods
2.6. Metabolic Network Analysis of B. licheniformis
2.7. Data Statistics and Analysis
3. Results
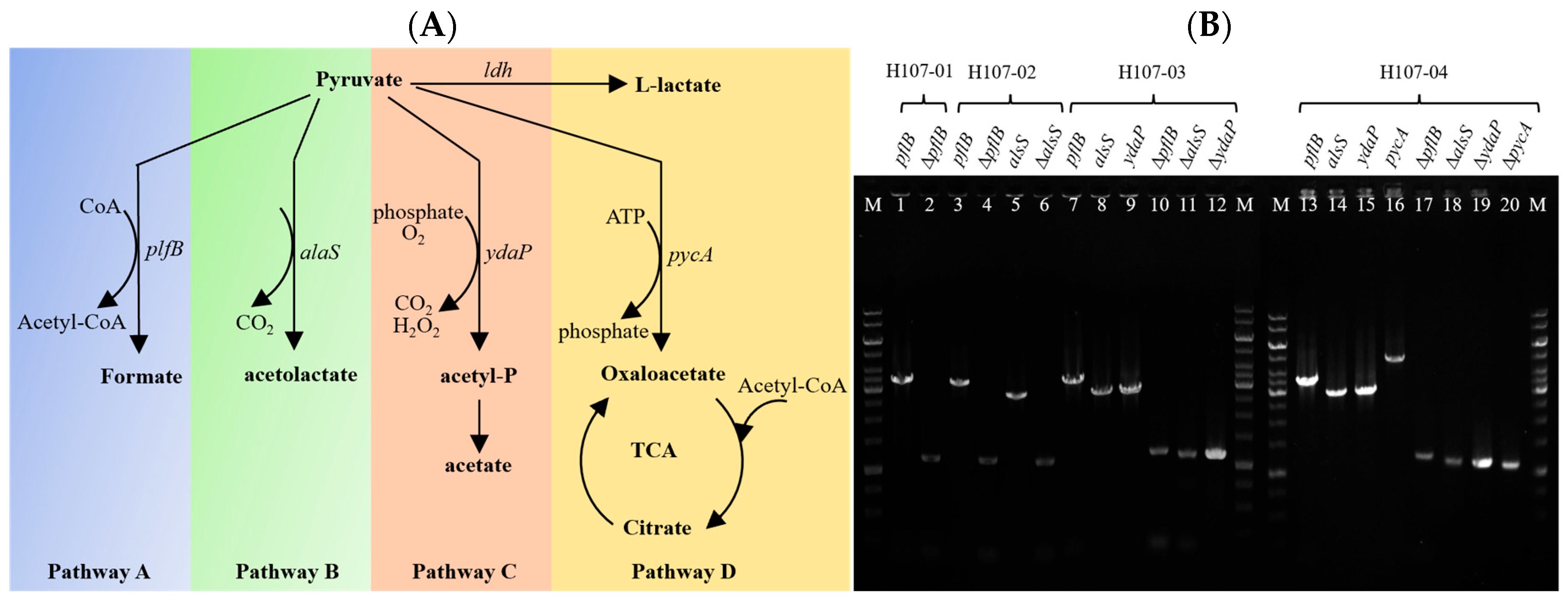
3.1. Metabolic Network Analysis of Pyruvate in B. licheniformis
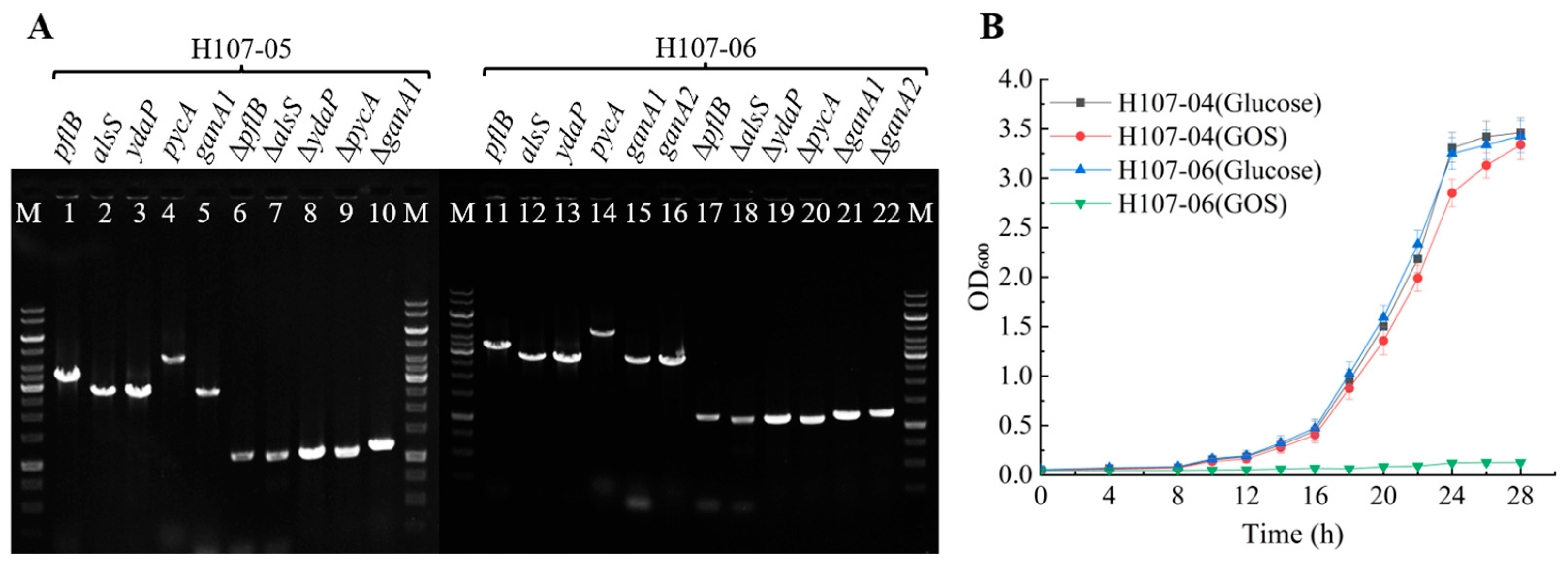
3.2. Selection of Lactose and GOS Non-Metabolizing Mutants
3.3. Enhancement of L-Lactic Acid Synthesis in B. licheniformis
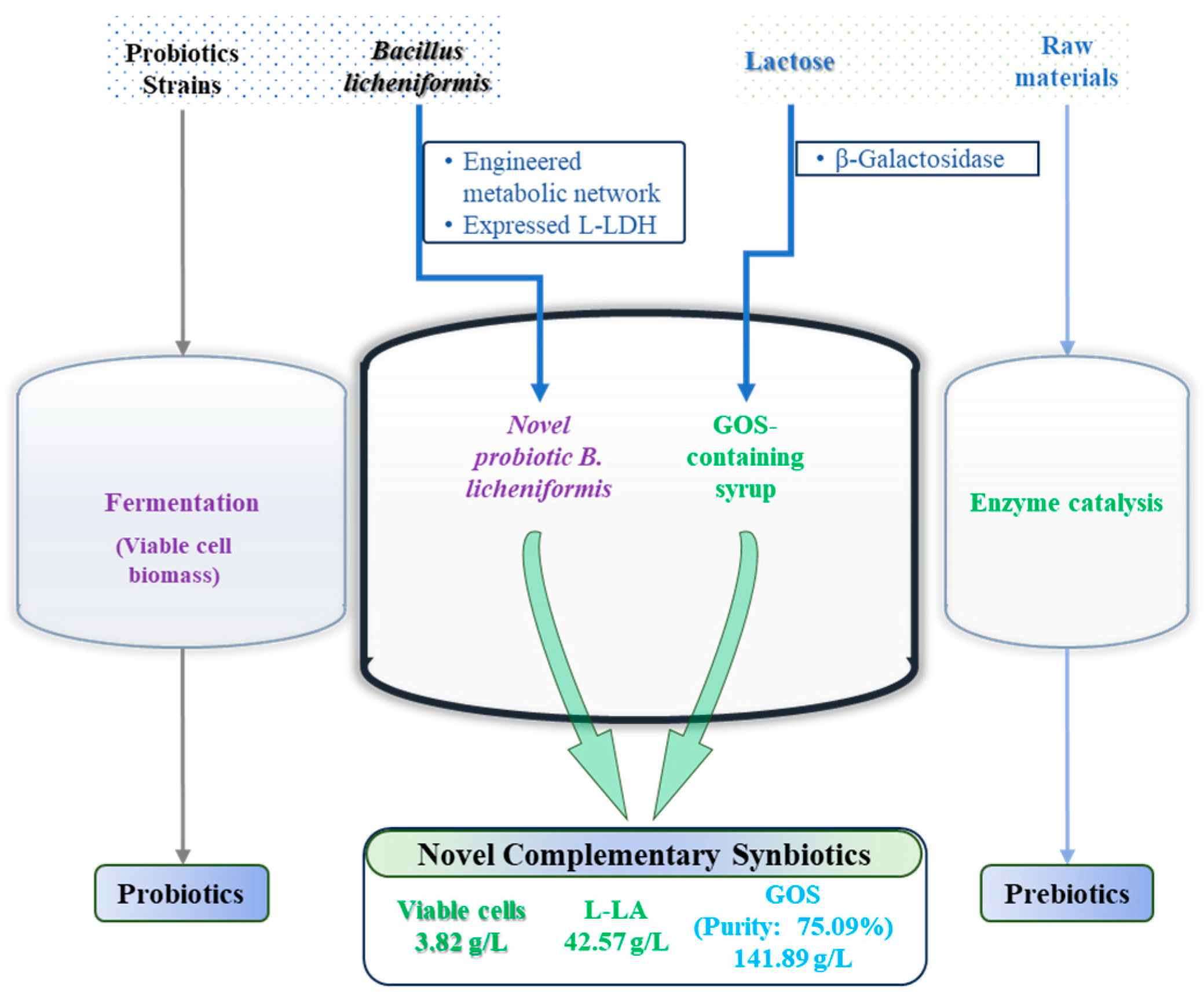
3.4. Establishment and Optimization of a Complementary Synbiotic Production Process
4. Discussion
5. Study Limitations and Future Perspectives
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mounir, M.; Ibijbijen, A.; Farih, K.; Rabetafika, H.N.; Razafindralambo, H.L. Synbiotics and their antioxidant properties, mechanisms, and benefits on human and animal health: A narrative review. Biomolecules 2022, 12, 1443. [Google Scholar] [CrossRef]
- Luzzi, A.; Briata, I.M.; Di Napoli, I.; Giugliano, S.; Di Sabatino, A.; Rescigno, M.; Cena, H. Prebiotics, probiotics, synbiotics and postbiotics to adolescents in metabolic syndrome. Clin. Nutr. 2024, 43, 1433–1446. [Google Scholar] [CrossRef] [PubMed]
- Khursheed, R.; Gulati, M.; Wadhwa, S.; Vishwas, S.; Sharma, D.S.; Corrie, L.; Alam, A.; Alnasser, S.M.; Aba Alkhayl, F.F.; Parveen, Z.; et al. Multifaceted role of synbiotics as nutraceuticals, therapeutics and carrier for drug delivery. Chem. Biol. Interact. 2022, 368, 110223. [Google Scholar] [CrossRef] [PubMed]
- Gomez Quintero, D.F.; Kok, C.R.; Hutkins, R. The future of synbiotics: Rational formulation and design. Front. Microbiol. 2022, 13, 919725. [Google Scholar] [CrossRef]
- Lee, S.; Choi, S.P.; Choi, H.J.; Jeong, H.; Park, Y.S. A comprehensive review of synbiotics: An emerging paradigm in health promotion and disease management. World J. Microbiol. Biotechnol. 2024, 40, 280. [Google Scholar] [CrossRef]
- Fukaya, M.; Yokoyama, Y.; Usui, H.; Fujieda, H.; Sakatoku, Y.; Takahashi, T.; Miyata, K.; Niikura, M.; Sugimoto, T.; Asahara, T.; et al. Impact of synbiotics treatment on bacteremia induced during neoadjuvant chemotherapy for esophageal cancer: A randomised controlled trial. Clin. Nutr. 2021, 40, 5781–5791. [Google Scholar] [CrossRef]
- Kanazawa, A.; Aida, M.; Yoshida, Y.; Kaga, H.; Katahira, T.; Suzuki, L.; Tamaki, S.; Sato, J.; Goto, H.; Azuma, K.; et al. Effects of synbiotic supplemen tation on chronic inflammation and the gut microbiota in obese patients with type 2 diabetes mellitus: A randomized controlled study. Nutrients 2021, 13, 558. [Google Scholar] [CrossRef]
- Swanson, K.S.; Gibson, G.R.; Hutkins, R.; Reimer, R.A.; Reid, G.; Verbeke, K.; Scott, K.P.; Holscher, H.D.; Azad, M.B.; Delzenne, N.M.; et al. The international scientific association for probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 687–701. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Niu, D.; Liu, J.; Jin, Z.; Mchunu, N.P.; Singh, S.; Wang, Z. Enhancing β-galactosidase performance for galactooligosaccharides preparation via strategic glucose re-tunneling. Int. J. Mol. Sci. 2024, 25, 12316. [Google Scholar] [CrossRef]
- Muras, A.; Romero, M.; Mayer, C.; Otero, A. Biotechnological applications of Bacillus licheniformis. Crit. Rev. Biotechnol. 2021, 41, 609–627. [Google Scholar] [CrossRef]
- He, H.; Yu, Q.; Ding, Z.; Zhang, L.; Shi, G.; Li, Y. Biotechnological and food synthetic biology potential of platform strain: Bacillus licheniformis. Synth. Syst. Biotechnol. 2023, 8, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Ma, L.; Xu, Z.; Soteyome, T.; Yuan, L.; Yang, Z.; Jiao, X.A. Invited review: Role of Bacillus licheniformis in the dairy industry-Friends or foes? J. Dairy Sci. 2024, 107, 7520–7532. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; He, P.; Hu, S.; Yu, Y.; Wang, X.; Ishaq, A.R.; Chen, S. Promoting cell growth for biochemicals production via boosting the synthesis of L/D-alanine and D-alanyl-D-alanine in Bacillus licheniformis. World J. Microbiol. Biotechnol. 2023, 39, 115. [Google Scholar] [CrossRef]
- Wang, C.; Niu, D.; Zhou, Y.; Liu, H.; Mchunu, N.P.; Zhang, M.; Singh, S.; Wang, Z. Synergistic engineering of the twin-arginine translocation (Tat) pathway and membrane capacity enhances extracellular production of amylosucrase in Bacillus licheniformis. Microorganisms 2025, 13, 1179. [Google Scholar] [CrossRef]
- Hofvendahl, K.; Hahn-Hägerdal, B. Factors affecting the fermentative lactic acid production from renewable resources. Enzyme Microb. Technol. 2000, 26, 87–107. [Google Scholar] [CrossRef]
- Sun, S.; Li, H.; Chen, J.; Qian, Q. Lactic Acid: No Longer an inert and end-product of glycolysis. Physiology 2017, 32, 453–463. [Google Scholar] [CrossRef]
- Bilal, M.; Niu, D.; Wang, Z. Novel enzyme-fermentation process for bioconversion of restaurant food waste into isomaltooligosaccharide-and L-lactic acid-enriched animal feed. Front. Sustain. Food Syst. 2024, 8, 1326304. [Google Scholar] [CrossRef]
- Elsayed, E.A.; Danial, E.N.; Wadaan, M.A.; El-Enshasy, H.A. Production of β-galactosidase in shake-flask and stirred tank bioreactor cultivations by a newly isolated Bacillus licheniformis strain. Biocatal. Agric. Biotechnol. 2019, 20, 101231. [Google Scholar] [CrossRef]
- Ren, Z.Q.; Liu, L.H.; Tang, T.X.; Huang, K.; Huang, Z. Effectively increase the L(+)-isomer proportion of ethyl lactate in Baijiu by isolating and applying L(+)-lactic acid-producing bacteria. Food Biosci. 2025, 63, 105615. [Google Scholar] [CrossRef]
- Li, B.; Cai, D.; Chen, S. Metabolic engineering of central carbon metabolism of Bacillus licheniformis for enhanced production of poly-γ-glutamic acid. Appl. Biochem. Biotechnol. 2021, 193, 3540–3552. [Google Scholar] [CrossRef]
- Shen, P.; Niu, D.; Liu, X.; Tian, K.; Permaul, K.; Singh, S.; McHunu, N.P.; Wang, Z. High-efficiency chromosomal integrativeamplification strategy for overexpressing alpha-amylase in Bacillus licheniformis. J. Ind. Microbiol. Biotechnol. 2022, 49, kuac009. [Google Scholar] [CrossRef]
- Wang, C.; Niu, D.; McHunu, N.P.; Zhang, M.; Singh, S.; Wang, Z. Secretory expression of amylosucrase in Bacillus licheniformis through twin-arginine translocation pathway. J. Ind. Microbiol. Biotechnol. 2024, 51, kuae004. [Google Scholar] [CrossRef] [PubMed]
- Tian, K.; Shi, G.; Lu, F.; Singh, S.; Wang, Z. High-efficiency L-lactate production from glycerol by metabolically engineered Escherichia coli. Chin. J. Biotechnol. 2013, 29, 1268–1277. [Google Scholar]
- Sambrook, J.; Russel, D. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Press: Woodbury, NY, USA, 2001. [Google Scholar]
- Xu, M.; Ma, J.; Wang, Z. Effect of high osmolarity on electro-transformation efficiency of bacteria. J. Wuxi Uni. Light Ind. 2004, 23, 98–100. [Google Scholar]
- Zhou, L.; Zuo, Z.; Chen, X.; Niu, D.; Tian, K.; Prior, B.A.; Shen, W.; Shi, G.; Singh, S.; Wang, Z. Evaluation of genetic manipulation strategies on D-lactate production by Escherichia coli. Curr. Microbiol. 2011, 62, 981–989. [Google Scholar] [CrossRef]
- Zhou, L.; Niu, D.; Tian, K.; Chen, X.; Prior, B.A.; Shen, W.; Shi, G.; Singh, S.; Wang, Z. Genetically switched D-lactate production in Escherichia coli. Metab. Eng. 2012, 14, 560–568. [Google Scholar] [CrossRef]
- Niu, D.; Tian, K.; Prior, B.A.; Wang, M.; Wang, Z.; Lu, F.; Singh, S. Highly efficient L-lactate production using engineered Escherichia coli with dissimilar temperature optima for L-lactate formation and cell growth. Microb. Cell Fact. 2014, 13, 78. [Google Scholar] [CrossRef]
- Wang, M.; Niu, D.; Gao, M.; Wang, A.; Zhao, W.; Permaul, K.; Singh, S.; Wang, Z. Decoupling sucrose utilization from oxygen-responsive regulation for high-efficiency L-lactic acid production in Escherichia coli. Biotechnol. Biofuels Bioprod. 2025, 18, 101. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Prakash, G.; Lali, A.M. 2,3-Butanediol production using soy-based nitrogen source and fermentation process evaluation by a novel isolate of Bacillus licheniformis BL1. Prep. Biochem. Biotechnol. 2021, 51, 1046–1055. [Google Scholar] [CrossRef]
- Niu, D.; Wang, Z. Development of a pair of bifunctional expression vectors for Escherichia coli and Bacillus licheniformis. J. Ind. Microbiol. Biotechnol. 2007, 34, 357–362. [Google Scholar] [CrossRef]
- Jin, Q.; Jin, G.; Ju, J.; Xu, L.; Tang, L.; Fu, Y.; Hou, R.; Atala, A.; Zhao, W. Bioprinting small-diameter vascular vessel with endothelium and smooth muscle by the approach of two-step crosslinking process. Biotechnol. Bioeng. 2022, 119, 1679–1690. [Google Scholar] [CrossRef]
- Abedin, M.M.; Chourasia, R.; Phukon, L.C.; Sarkar, P.; Ray, R.C.; Singh, S.P.; Rai, A.K. Lactic acid bacteria in the functional food industry: Biotechnological properties and potential applications. Crit. Rev. Food Sci. Nutr. 2024, 64, 10730–10748. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Yu, Z.; Liu, W.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Probiotic characteristics of Bacillus coagulans and associated implications for human health and diseases. J. Funct. Foods 2020, 64, 103643. [Google Scholar] [CrossRef]
- Wang, X.; Pan, J.; Wu, J.; Chen, X.; Gao, C.; Song, W.; Wei, W.; Liu, J.; Liu, L. Regulation of intracellular level of ATP and NADH in Escherichia coli to promote succinic acid production. Chin. J. Biotechnol. 2023, 39, 3236–3252. [Google Scholar] [CrossRef]
- Yin, X.; Li, J.; Shin, H.D.; Du, G.; Liu, L.; Chen, J. Metabolic engineering in the biotechnological production of organic acids in the tricarboxylic acid cycle of microorganisms: Advances and prospects. Biotechnol. Adv. 2015, 33, 830–841. [Google Scholar] [CrossRef]
- Zhang, S.; Wakai, S.; Sasakura, N.; Tsutsumi, H.; Hata, Y.; Ogino, C.; Kondo, A. Pyruvate metabolism redirection for biological production of commodity chemicals in aerobic fungus Aspergillus oryzae. Metab. Eng. 2020, 61, 225–237. [Google Scholar] [CrossRef]
- Chang, D.E.; Jung, H.C.; Rhee, J.S.; Pan, J.G. Homofermentative production of D- or L-lactate in metabolically engineered Escherichia coli RR1. Appl. Environ. Microbiol. 1999, 65, 1384–1389. [Google Scholar] [CrossRef]
- Xu, X.; Wang, H.; Chen, X.; Wu, J.; Gao, C.; Song, W.; Wei, W.; Liu, J.; Liu, Y.; Liu, L. Metabolic engineering of the substrate utilization pathway in Escherichia coli increases L-lysine production. Chin. J. Biotechnol. 2024, 40, 2513–2527. [Google Scholar] [CrossRef]
- Xu, J.Z.; Yu, H.B.; Han, M.; Liu, L.M.; Zhang, W.G. Metabolic engineering of glucose uptake systems in corynebacterium glutamicum for improving the efficiency of L-lysine production. J. Ind. Microbiol. Biotechnol. 2019, 46, 937–949. [Google Scholar] [CrossRef]
| Strains/Vectors | Characteristics | Source |
|---|---|---|
| Strains | ||
| E. coli JM109 | endA1, recA1, gyrA96, thi, hsdR17, relA1, supE44, λ-, Δ(lac-proAB), [F’, traD36, proAB, laqIqZΔM15] | Lab stock |
| B. licheniformis H107 | Wild-type B. licheniformis | Lab stock |
| B. licheniformis H107-01 | ∆pflB | This study |
| B. licheniformis H107-02 | ∆pflB, ∆alsS | This study |
| B. licheniformis H107-03 | ∆pflB, ∆alsS, ∆ydaP | This study |
| B. licheniformis H107-04 | ∆pflB, ∆alsS, ∆ydaP, ∆pycA, | This study |
| B. licheniformis H107-04A | ∆pflB, ∆alsS, ∆ydaP, ∆pycA, pHY-P43-BcoaLDH | This study |
| B. licheniformis H107-05 | ∆pflB, ∆alsS, ∆ydaP, ∆pycA, ∆ganA2 | This study |
| B. licheniformis H107-06 | ∆pflB, ∆alsS, ∆ydaP, ∆pycA, ∆ganA2, ∆ganA1 | This study |
| B. licheniformis H107-06A | ∆pflB, ∆alsS, ∆ydaP, ∆pycA, ∆ganA2, ∆ganA1, pHY-P43-BcoaLDH | This study |
| Vectors | ||
| pUB-EX | KmR, Derived from T2(2)-ori, E. coli-Bacillus shµttle vector, thermosensitive replication | [21,22] |
| pUB-EX-pflB | KmR, thermosensitive plasmid, used for deletion of pflB | This study |
| pUB-EX-alsS | KmR, thermosensitive plasmid, used for deletion of alsS | This study |
| pUB-EX-ydaP | KmR, thermosensitive plasmid, used for deletion of ydaP | This study |
| pUB-EX-pycA | KmR, thermosensitive plasmid, used for deletion of pycA | This study |
| pUB-EX-ganA2 | KmR, thermosensitive plasmid, used for deletion of ganA2 | Lab stock |
| pUB-EX-ganA1 | KmR, thermosensitive plasmid, used for deletion of ganA1 | Lab stock |
| pHY-P43-BcoaLDH | TetR, AmpR, P43, BcoaLDH | [23] |
| Strains | L-Lactic Acid (g·L−1) | Acetic Acid (g·L−1) | Formic Acid (g·L−1) | Succinic Acid (g·L−1) | Biomass (g·L−1) |
|---|---|---|---|---|---|
| WT | 0.29 ± 0.03 | 0.54 ± 0.03 | 0.63 ± 0.12 | 3.75 ± 0.28 | 1.52 ± 0.11 |
| H107-01 | 0.52 ± 0.04 * | 0.40 ± 0.02 * | ND | 2.85 ± 0.21 * | 1.43 ± 0.09 |
| H107-02 | 0.59 ± 0.04 * | 0.91 ± 0.06 ** | ND | 2.66 ± 0.19 * | 1.38 ± 0.10 |
| H107-03 | 0.58 ± 0.03 ** | 0.85 ± 0.06 ** | ND | 2.84 ± 0.21 * | 1.35 ± 0.08 |
| H107-04 | 1.06 ± 0.07 ** | 0.70 ± 0.05 ** | ND | 3.01 ± 0.22 * | 1.31 ± 0.09 |
| Cultivation Conditions | L-Lactic Acid (g·L−1) | Glucose Consumption (g·L−1) | Conversion Rate (%) | Biomass (g·L−1) |
|---|---|---|---|---|
| Aerobic | 2.52 ± 0.23 | 8.64 ± 0.46 | 29.15 ± 1.23 | 2.13 ± 0.15 |
| Microaerobic | 3.24 ± 0.31 | 9.43 ± 0.49 | 34.37 ± 1.67 | 1.82 ± 0.12 |
| Anaerobic | 4.45 ± 0.38 | 9.90 ± 0.52 | 44.93 ± 1.94 | 1.57 ± 0.11 |
| Indicator | Wild Type | H107-06A | Fold Increase |
|---|---|---|---|
| L-Lactic acid (g·L−1) | 2.89 ± 0.45 | 42.56 ± 1.85 *** | 14.7 |
| Glucose conversion rate (%) | 25.38 ± 1.86 | 85.12 ± 2.35 *** | 4.4 |
| Biomass (g·L−1) | 2.55 ± 0.18 | 3.82 ± 0.15 *** | 1.5 |
| Relative purity of GOS (%) | 53.67 ± 2.12 | 75.09 ± 1.85 *** | 1.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, J.; Mu, T.; Niu, D.; Ding, Z.; Mchunu, N.P.; Zhang, M.; Singh, S.; Wang, Z. Metabolic Engineering of Bacillus licheniformis for High-Yield L-Lactic Acid and Galactooligosaccharide Retention in Complementary Synbiotics Production. Microorganisms 2025, 13, 2530. https://doi.org/10.3390/microorganisms13112530
Zhao J, Mu T, Niu D, Ding Z, Mchunu NP, Zhang M, Singh S, Wang Z. Metabolic Engineering of Bacillus licheniformis for High-Yield L-Lactic Acid and Galactooligosaccharide Retention in Complementary Synbiotics Production. Microorganisms. 2025; 13(11):2530. https://doi.org/10.3390/microorganisms13112530
Chicago/Turabian StyleZhao, Jihua, Teng Mu, Dandan Niu, Zhongzhen Ding, Nokuthula Peace Mchunu, Meng Zhang, Suren Singh, and Zhengxiang Wang. 2025. "Metabolic Engineering of Bacillus licheniformis for High-Yield L-Lactic Acid and Galactooligosaccharide Retention in Complementary Synbiotics Production" Microorganisms 13, no. 11: 2530. https://doi.org/10.3390/microorganisms13112530
APA StyleZhao, J., Mu, T., Niu, D., Ding, Z., Mchunu, N. P., Zhang, M., Singh, S., & Wang, Z. (2025). Metabolic Engineering of Bacillus licheniformis for High-Yield L-Lactic Acid and Galactooligosaccharide Retention in Complementary Synbiotics Production. Microorganisms, 13(11), 2530. https://doi.org/10.3390/microorganisms13112530





