Abstract
Malolactic fermentation (MLF) is a bioprocess driven by lactic acid bacteria (LAB), which is desired in red and highly acidic white wines. Among all LAB, Oenococcus oeni is the main species in wine, followed by Lactiplantibacillus plantarum. The harsh conditions found in wine—not only due to the low nutrient concentration but also the presence of antimicrobial compounds such as ethanol, high acidity, SO2, and polyphenols—can compromise MLF performance. In recent years, the use of certain non-Saccharomyces yeasts, such as Torulaspora delbrueckii or Metschnikowia pulcherrima, as starter cultures for alcoholic fermentation, has emerged as a promising strategy to improve MLF. In this study, we evaluated the effect of four different fractions from a T. delbrueckii strain on MLF performance. First, the positive impact of this strain as a starter culture on O. oeni growth was confirmed; then, yeast-derived compounds were tested in different wines. Two fractions showed the most promising results in reducing MLF duration: the inactivated yeast fraction and the autolysate fraction. Those enhanced bacterial viability and promoted mannoprotein consumption. These findings highlight the potential of T. delbrueckii-derived compounds as enological tools to support MLF under restrictive wine conditions.
1. Introduction
Malolactic fermentation (MLF) is a biotransformation driven by lactic acid bacteria (LAB) in wine. It consists of the decarboxylation of L-malic acid into L-lactic acid [1,2] that generally occurs after alcoholic fermentation (AF). As MLF reduces acidity, it is highly recommended in red winemaking and also for high-acidity white wines. Additionally, microbial stability is increased, and the organoleptic profile is modified [3]. During AF, wine yeasts consume nutrients and produce toxic metabolites, such as ethanol and medium-chain fatty acids; therefore, they leave a harsh environment for microbial growth. Among the enological yeasts, Saccharomyces cerevisiae is the dominant species from the middle to final fermentative stages [4]. It has been domesticated to the wine fermentative process, and it is part of the resident microbiota of cellars [5]. Nevertheless, during the first fermentative stages, a vast diversity of other yeasts, known as non-Saccharomyces, are predominant, which come from the grape skins and are subsequently found in grape must [6].
Also, some LAB have been progressively domesticated to this enological niche, being the most important the species Oenococcus oeni [7,8,9]. This bacterium has developed specific mechanisms to resist the wine environment, characterized by a low pH and moderate to high ethanol concentration, and to enable bacterial growth and survival in a very poor nutrient medium [10]. In this sense, L-malic acid degradation is one of those specific mechanisms for survival in wine found in LAB, which enables energy acquisition from L-malic acid decarboxylation.
After the completion of AF, wine yeasts undergo an autolytic process [11,12], in which they release some compounds to the wine, such as nitrogen compounds (amino acids, peptides, and proteins), vitamins, or mannoproteins, among others [13,14,15]. Thus, enriching wine with nutrients that can eventually be used by LAB. That is why, generally, MLF occurs after AF, when wine is enriched with those yeast-derived compounds. At this stage, yeast cells are no longer metabolically active, which reduces competition, and together with the release of yeast-derived compounds, creates favorable conditions for LAB growth and the initiation of MLF.
The harsh conditions found in wine can be somewhat counteracted by inoculating non-Saccharomyces for AF [16]. Their initial low population in grape must and their low ethanol resistance, together with the usual inoculation of S. cerevisiae in must, make it difficult to observe any changes in wine composition. In addition, as it is carried out for S. cerevisiae, these yeasts can also be inoculated at the beginning of AF to intentionally modulate wine characteristics [17,18,19,20,21,22,23]. Among the non-Saccharomyces yeasts, some strains of Torulaspora delbrueckii, Metschnikowia pulcherrima, and Lachancea thermotolerans are already available as commercial starter cultures [20]. These yeasts not only modulate wine organoleptic profile but also modulate other compounds that directly impact O. oeni, and consequently, impact the MLF performance [16]. Indeed, non-Saccharomyces can be inoculated as immobilized cultures [24,25], or even LAB that can help to counteract and protect the cultures from the harsh environment [26], or even yeasts and LAB together [27]. There are several authors who reported positive effects on MLF duration reduction using non-Saccharomyces in AF from different winemaking, mainly with strains belonging to T. delbrueckii [28,29,30] and M. pulcherrima [31,32] species. To this regard, some positive outcomes from the chemical modulation of wine composition by these two species are as follows: (i) the decrease in medium chain fatty acids, (ii) the increase in the pH value, (iii) the reduction in the ethanol content, (iv) the enhancement of mannoprotein release, and (v) the decrease in sulfur dioxide concentration, among other [33,34,35].
Interestingly, fractions from different enological yeast species present specific modulation on MLF duration. For instance, Balmaseda et al. [36] reported a general enhanced MLF when performing with yeasts’ lees of T. delbrueckii, but slowed down when fermenting in the presence of some S. cerevisiae yeast lees. In addition, the general outcome when supplementing wines with lees or yeast fractions is a positive reduction in MLF duration. In this sense, to our knowledge, there are no commercial non-Saccharomyces fractions for this purpose; they are all derived from S. cerevisiae.
Altogether, the aim of this work was to evaluate the potential use of some fractions obtained from a specific T. delbrueckii strain by different treatments on the MLF performance of O. oeni in some selected harsh red and white wines, characterized by low pH, high ethanol content, and high polyphenolic composition.
2. Materials and Methods
2.1. Wines, Yeast Fractions, and Microbial Strains
First, during vintage 2023, about 120 kg of Tempranillo grapes from the experimental cellar of the Universitat Rovira i Virgili were destemmed, and 60 kg were transferred each to two 100 kg steel tanks. Grape must general parameters were density 1091.8 g/L, primary amino nitrogen 57 mgN/L, ammonia 34 mgN/L, and pH 3.44. Sulfur dioxide in the form of K2S2O5 (Fisher Scientific, Madrid, Spain) was added to each tank at a final concentration of 20 g/hL. After one hour, tanks were inoculated: one with S. cerevisiae LMD38 (Lallemand SAS, Montreal, QC, Canada), and the other with T. delbrueckii LMD84 (Lallemand SAS, Montreal, QC, Canada), following rehydration instructions. Fermenting must was manually punched down every day, and samples were taken to monitor AF progression. After 48 h, NUTRIENT VITTM (Lallemand SAS, Montreal, QC, Canada) was added (20 g/hL). Then, S. cerevisiae was inoculated into the tank previously inoculated with T. delbrueckii. Once AF was finished (<2 g/L of residual sugars), wines were settled for 5 days at 4 °C.
Second, four wines from different cultivars were used in this study. These wines were provided by Lallemand SAS during vintage 2024, obtained after AF not heat-treated but stored at 4 °C before the experiment, representing harsh wines where MLF was performed. There were three red wines, including two Merlot wines (wine A and wine B) and a Tempranillo wine (wine C), and a single white Chardonnay wine (wine D). The initial composition of these wines is summarized in Table 1.

Table 1.
Wines used in this study with some oenological parameters. * PAN—Primary Amino Nitrogen.
The fractions used in this study were also provided by Lallemand SAS. They were obtained with different treatments applied to the biomass of a specific strain of T. delbrueckii LMD84. The fractions were named as: inactivated yeast extract (Tdi) resulting of a heat-treated cream (yeast cream at 70 °C for 15 min) then dried, autolysate extract (A) obtained after 20 h keeping the biomass at 55 °C followed by drying, yeast extract (YE), and cell wall extract (CW) were respective soluble and insoluble fractions resulting from the autolysis followed by a separation by centrifugation and then dried.
To perform MLF, commercial freeze-dried O. oeni strains (Lallemand SAS) were used following the manufacturer’s instructions, inoculating 1 g/hL of the product into each wine.
For the vinification of vintage 2023, LAA1, LAB6, LAB9, LAB2013, LAC20, and LAA4 O. oeni strains (Lallemand SAS) were used. For the subsequent trials, O. oeni LAB6 was used; then, other strains were used to confirm the observed effects in wine D: LAA1, LAB2013, LAA4, LAC20, and LAB9.
2.2. Experimental Fermentations
2.2.1. Effect of T. delbrueckii as Starter Culture
S. cerevisiae control and T. delbrueckii racked wines were aliquoted in 24 different 50 mL-tubes and inoculated with the six O. oeni strains (1 g/hL), following the manufacturer’s instructions, in triplicate. Three tubes were left without inoculation. Approximately twice a week, samples were taken, and L-malic acid concentration was quantified by enzymatic methods (Analyzer Y15, Biosystems, Barcelona, Spain). When [L-malic acid] < 0.10 g/L, MLF was considered as finished, and the main oenological parameters were determined. Bacterial viability was monitored periodically on MRSmf plates (55 g/L MRS broth (BD™ Difco™, Fisher Scientific, Madrid, Spain), 4 g/L DL-malic acid, 5 g/L fructose, 100 mg/L nystatin and 20 g/L of agar ((Panreac Química, Castellar del Valles, Spain), pH 5.00).
2.2.2. Screening of Different T. delbrueckii Fractions in Harsh Wines
Five aliquots of 50 mL of each wine (wines A, B, C, and D) were prepared and supplemented with 20 g/hL of each fraction (Td, A, YE, and CW). The remaining aliquot without supplementation was used as a control. After 24 h, 10 mL of wine were transferred to a 10 mL syringe (Luer-Lok™, BD, Madrid, Spain) coupled to a needle (0.8 × 40 mm, B. Braun, Melsungen, Germany) as described in Balmaseda et al. (2024) [37]. This syringe was used as a control for spontaneous fermentation. Then, the remaining volume was inoculated with 1 g/hL of O. oeni LAB6. Once inoculated, two additional syringes were filled with the inoculated wines, and each fermentation was performed in duplicate. The fermentations were incubated statically at 20 °C. Approximately twice a week, samples were taken, and L-malic acid concentration was quantified by enzymatic methods (Analyzer Y15, Biosystems). When [L-malic acid] < 0.10 g/L, MLF was considered as finished, and some oenological parameters were determined. After 65 days of inoculation, the unfinished MLF wines were considered stuck.
2.2.3. Confirmation of Results in Harsh Wines
From the four studied wines in Section 2.2.1, wine C and wine D were selected to further characterize the effect of the fractions. The fractions Tdi and A were also chosen for this second trial, supporting the shortest MLF durations. Fermentations were carried out similarly to the previous section, inoculating LAB6 into wines C and D, supplemented with extracts Tdi and A. In this case, bacterial viability was determined before inoculation (t0), two, seven, and 14 days after inoculation by plating on MRSmf plates.
Similarly, the observed effects were confirmed in tubes containing 10 mL of wine D inoculated with LAA1, LAB2013, LAA4, LAC20, and LAB9. Wine D was supplemented with 20 g/hL of Tdi fraction. A control wine D without supplementation was also inoculated. Samples were taken weekly to measure L-malic acid concentration and bacterial viability. MLF was considered finished when <0.1 g/L of L-malic acid was determined. After 70 days, wines with the MLF not completed were considered as stuck fermentations. Fermentations were performed in duplicate.
2.3. Wine Characterization
pH was determined using a Crison micro pH 2002 pH-meter (Barcelona, Spain). D-lactic acid, L-lactic acid, acetic acid, citric acid, glycerol, ammonium, and PAN concentrations were determined with the analyzer Y15 (Biosystems, Barcelona, Spain).
Mannoprotein concentration was estimated by enzymatic measurement of mannose coming from a hydrolysis of the total precipitated polysaccharides with ethanol, following the method of Balmaseda et al. [38]. Briefly, 5 mL of wine was mixed with 5 volumes of 95% (v/v) ethanol and precipitated overnight at 4 °C. After washing the pellet twice with 10 mL of 95% ethanol, pellets were dried at 30 °C for 30′ in vacuum. Then, the pellet was resuspended in 1 mL of 5 M H2SO4, incubated at 90 °C for 1 h, and neutralized with 1 mL of 10 M NaOH. Finally, the sample was centrifuged (8500 rpm, 5′) and the supernatant was used to determine the free sugar concentration (D-glucose, D-fructose, and D-mannose) using the K-MANGL kit (Megazyme, Wicklow, Ireland). Concentration is expressed as mg mannose equivalents/L [35].
Amino acid quantification was performed as previously described in Balmaseda et al. [36]. In short, filtered samples were derivatized in borate buffer and methanol with the addition of L-2-aminoadipic acid as an internal standard, and incubated at 80 °C. The HPLC (Agilent 1100, Agilent Technologies, Waldbronn, Germany) was equipped with a DAD ultraviolet (Agilent Technologies, Waldbronn, Germany), and separation was performed on a Hypersil ODS C18 column (Agilent Technologies, Waldbronn, Germany) with a particle size of 5 μm (250 mm × 4.6 mm) at 20 °C, with sodium acetate buffer (pH 5.80) and an acetonitrile/methanol mixture as mobile phases. Amino acid concentrations were calculated against external calibration curves and normalized to the internal standard.
2.4. α-Mannosidase Activity
The determination of α-mannosidase activity was based on a protocol to measure β-glucosidase activity in O. oeni [39] with the modifications proposed by Toraño et al. (2024) [40]. Briefly, the mannosidase activity of all O. oeni strains was characterized in a wine-like medium. Two-milliliter samples were centrifuged, and the pellet was retained (corresponding to about 2 × 107 CFU/mL) for analysis. The pellets were resuspended in 1 mL of 0.1 M sodium acetate buffer, pH 5.10. 25 μL of the resuspended pellet was added to 75 μL of substrate (25 mM p-nitrophenyl-α-D-mannopyranoside [p-NMan, (Sigma-Aldrich, Barcelona, Spain)] diluted in 0.1 M sodium acetate buffer, pH 5.10) and incubated for 30 min at 37 °C. Next, 100 μL of 1 M Na2CO3 was added. The reaction tubes were centrifuged, and the assay was read against the blank at 400 nm using a Spectro Star Nano (BMG Labtech, Ortenberg, Germany). From this measurement, the concentration of liberated p-nitrophenol (p-NP) was determined from a calibration curve taken from standard p-NP, where one unit of α-mannosidase activity (U) corresponded to 1 μmol of p-nitrophenol released per minute. The enzymatic activity (U) data were normalized to 1 L of culture. All the samples were collected to obtain the same population of 2 × 107 CFU/mL, considering the growth curve (OD 600 nm vs. CFU/mL) of each strain. The viability of the samples was confirmed via CFU counting on MRSmf plates.
2.5. Statistical Analysis
All the statistical analyses of the results were performed using the statistics software XLSTAT version 2020.2.3 (Addinsoft, Paris, France). The analysis of variance was carried out by ANOVA with a subsequent Tukey HSD test to determine the significant differences between the samples: the confidence interval used was 95% and the statistical level of significance was set at p ≤ 0.05. The considered ANOVA factors were the supplementation with fractions of wine after MLF and the inoculated O. oeni strain; variables were the individual analytical results obtained in the experiment.
3. Results
3.1. Validating T. delbrueckii as a Malolactic Fermentation Enhancing Starter Culture
The Tempranillo wines produced during the vintage 2023 with S. cerevisiae or sequential fermentation with T. delbrueckii and S. cerevisiae finished after 10 and 16 days, respectively (Table 2). The AF duration is usually extended when using sequential or coinoculated AF strategies with non-Saccharomyces and S. cerevisiae strains [29]. Also, the potential reduction in MLF when inoculating T. delbrueckii was observed. In all inoculated wines, no matter the inoculated O. oeni strain, MLF was completed before it finished in the control wine, inoculated with S. cerevisiae only (Table 2).

Table 2.
Alcoholic (AF) and malolactic fermentation (MLF) duration (days) in Tempranillo wines from vintage 2023 produced by the inoculation of S. cerevisiae (Sc) or a sequential fermentation of T. delbrueckii and S. cerevisiae (Td + Sc). Oo: O. oeni, Sp: spontaneous MLF without LAB inoculation. Fermentations were carried out in triplicate.
The sequentially inoculated wine presented significantly lower ethanol content of about 13.6% (v/v) with respect to the control S. cerevisiae with about 14.1% (v/v). Slightly higher pH was measured in the sequentially inoculated wine (3.45 on average), compared to the control wine (3.41 on average), as observed by other authors [41]. Although these parameters are not definitive indicators of the potential benefits of T. delbrueckii on MLF, some of them have been reported to be closely associated with a positive effect on O. oeni [33].
The obtained wines were then inoculated with six different O. oeni strains. The MLF duration in the T. delbrueckii wines was significantly reduced by half in all O. oeni inoculated wines (Table 2). The observed results are related to the enhanced bacterial population found at the end of AF in those wines: about 105 CFU/mL with respect to the 102 CFU/mL of the control wine. This has been observed by several other authors when inoculating T. delbrueckii as a starter [42,43]. Under these conditions, the inoculation of commercial O. oeni strains did not notably accelerate MLF, possibly because the wines already harbored a substantial indigenous LAB population (around 7 × 105 CFU/mL) prior to inoculation. Thus, it is possible that MLF in the T. delbrueckii wines was driven mainly by the native microbiota, although the inoculated O. oeni strains may also have contributed.
On the other hand, the bacterial population in the S. cerevisiae control wine was <102 CFU/mL. In this wine, there was a significant reduction in the MLF duration when inoculating the six O. oeni strains.
The conditions found for this vinification were not as harsh as those of other wines with high acidity, high ethanol content, or high polyphenolic content [30]. The modulation caused by inoculating T. delbrueckii that enhanced O. oeni growth up to concentrations able to perform spontaneous MLF in a short time was confirmed. Based on these observations, the study proceeded to assess the potential benefits of supplementation with extracts derived from the same T. delbrueckii strain under more harsh conditions for O. oeni development, using wines produced exclusively with S. cerevisiae as the sole starter culture.
3.2. Screening of Different T. delbrueckii Fractions in Harsh Wines
In this second trial, we explored the effect of all four fractions from T. delbrueckii in four wines (Table 1), representing different matrices to perform MLF in harsh conditions (low pH, high polyphenolic index, or the combination of them, even low L-malic acid concentration). The initial bacterial population was <10 CFU/mL in wines B, C, and D. The population in wine A was about 5 × 103 CFU/mL before inoculation.
What was first observed was that wines supplemented or not with the different fractions were not able to start L-malic consumption without the inoculation of selected O. oeni strains (Figure 1). This confirms the harsh MLF scenario, in which a spontaneous MLF was not possible. In the inoculated wines, O. oeni started the L-malic acid consumption in all four tested wines, regardless of the supplementation. However, in all wines but wine A, the control fermentation with no fraction was stuck (Figure 1).
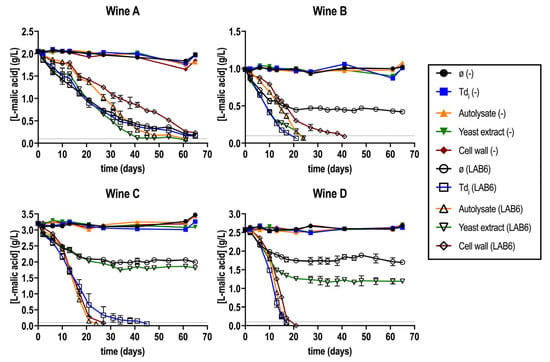
Figure 1.
Malolactic fermentation (MLF) performance in wines (A–D) supplemented or not (ø) with all the T. delbrueckii fractions (Tdi, A, YE, CW), inoculated with O. oeni (LAB6) or with no inoculation (−). The gray horizontal line represents [L-malic acid] = 0.1 g/L, considered as finished MLF. Fermentations were carried out in duplicate.
In general, there was a positive effect on the reduction in MLF duration in all wines when supplementing with the fractions (Figure 1) [40]. The only exception was wine A, where all the conditions behaved like the control with no addition. In this regard, wine A exhibited the lowest ethanol content and lower color intensity compared to the other two red wines (Table 1). Supplementation with the YE fraction resulted in stuck MLF in wines C and D, similarly to the control. In the case of the CW fraction, it prolonged MLF duration specifically in wine D compared to the control. This suggests that, although yeast-derived fractions can release compounds that potentially benefit MLF, not all fractions are equally effective in enhancing bacterial performance. In particular, the YE fraction exhibited a lower stimulatory effect on MLF progression compared to the other tested fractions. This observation is consistent with previous reports indicating that some yeast lees can, in certain cases, extend the duration of MLF beyond that of the control [36].
The general chemical characterization of the obtained wines after MLF was similar in all cases, except for the L-malic and L-lactic acid concentrations that depended on the completion of MLF (Table A1). As expected, wines in which MLF was completed exhibited higher pH values, regardless of fraction addition. This trend was also reflected in L-malic and L-lactic acid concentrations, which are directly related to MLF progression and, consequently, influence the pH [2]. Higher D-lactic acid concentrations were also observed in wines where MLF had been completed, independently of fraction supplementation. In addition, in two wines in particular, wine A and wine D, where MLF finished, showed lower citric acid concentrations and higher acetic acid levels. This is likely associated with increased metabolic activity of O. oeni, which commonly consumes citric acid during fermentation [1]. No clear trends were observed for glycerol. Regarding PAN and ammonium, interpretation is challenging because O. oeni can release these compounds through its protease and peptidase activity [44,45]. Nevertheless, wines supplemented with fractions tended to show higher levels of these compounds, independently of whether MLF was completed or not.
From all four wines tested, two were selected for further characterization: wine C and D, representing a harsh red (the one with the highest ethanol content) and a white wine, respectively. Fractions Tdi and A were also selected for the next experimentation since they showed the best MLF duration reduction (Figure 1).
3.3. Confirmation of the Potential of T. delbrueckii Fractions
Fermentations in wine C and D, supplemented with fractions Tdi and A, were repeated similarly to the previous section. In this case, fermentations were maintained for up to 40 days, obtaining similar results (Figure 2).
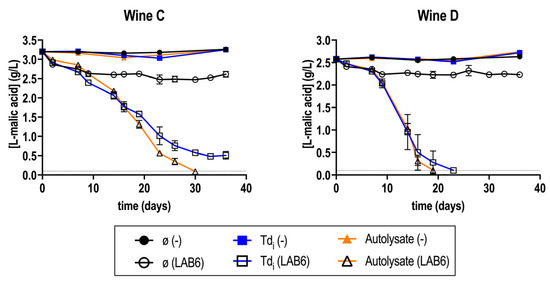
Figure 2.
Malolactic fermentation (MLF) performance in wine C and D supplemented or not (ø) with T. delbrueckii fractions (Tdi, and A), inoculated with O. oeni (LAB6) or with no inoculation (−). The gray horizontal line represents [L-malic acid] = 0.1 g/L, considered as finished MLF. Fermentations were carried out in duplicate.
MLF in these wines was again difficult to finish. Spontaneous MLF did not occur, and fractions were necessary for MLF completion (Figure 2). This time, the supplementation with Tdi in wine C was stopped at [L-malic acid] = 0.5 g/L.
For a better understanding of the observed MLF dynamics, viability was determined after inoculation (t0), and after two, seven, and 14 days (Table 3). The initial viability was about 6 log CFU/mL in all wines and treatments. Then, O. oeni population increased during the first week, mainly in wine C (Table 3), with no statistical differences between the supplemented or non-supplemented wines. The differences came after two weeks of fermentation, when the wines supplemented with Tdi and A fractions showed an increased population of about 1 log compared to the control (Table 3). Hence, the addition of these two fractions improved the survival of O. oeni in wine conditions. This effect has also been observed when preparing O. oeni cells in the presence of mannoproteins prior to inoculation in synthetic wine [46].

Table 3.
Viability of LAB population on MRSmf plates (log CFU/mL) in wine C and D, supplemented or not (ø) with T. delbrueckii fractions (Tdi and A), inoculated with O. oeni (LAB6) at different sampling points (t = 2, 7, and 14 days after inoculation). Fermentations were carried out in duplicate.
Mannoproteins are glycoproteins present in the yeast cell walls that are released during AF and subsequent yeast autolysis [47]. They consist of 80–90% mannose polysaccharides and 5–10% protein [48].
The presence of mannoproteins has been associated with enhanced O. oeni survival, and this is related to late MLF stages [38,46,49,50]. These macromolecules constitute one of the potential key compounds in wine lees [47,51] for O. oeni [16]. The amino acid fraction or even the glycolytic one seems to have a positive impact on O. oeni. In this sense, it has been demonstrated that O. oeni has specific glycoside and peptidase activities that enable the release of sugars and amino acids [44,52,53], which could be used by the bacterium [52,54]. On the other hand, other authors have shown that mannoproteins could adsorb medium-chain fatty acids, thus detoxifying wine for O. oeni [55,56].
The ability of each strain to degrade mannoproteins was evaluated in WLM for all O. oeni strains. All strains exhibited α-mannosidase activity under wine-like conditions (Figure 3), even without the addition of mannoprotein fractions, which are known to enhance this enzymatic activity [40]. This suggests that α-mannosidase activity is likely constitutive in all tested strains. Notably, O. oeni strains LAB6 and LAC20 showed the highest α-mannosidase activity (Figure 3).
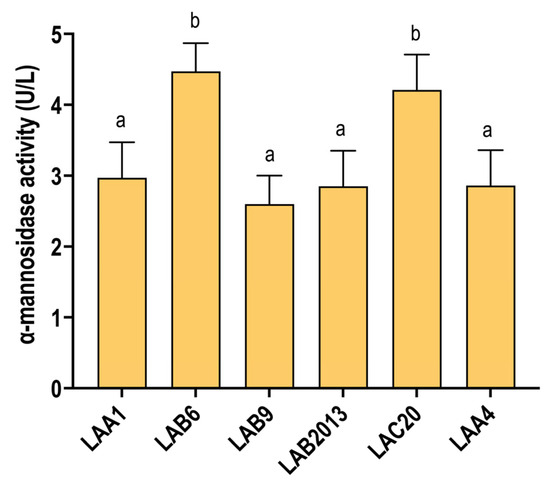
Figure 3.
Mannosidase activity in the middle of MLF of the different O. oeni strains. Data are the mean values of triplicate assays expressed in units of enzymatic activity per liter (U/L). Letters indicate a significant difference between values of each condition using the Tukey (HSD) test at p < 0.05. Fermentations were carried out in duplicate.
The mannoprotein concentration in these wines, C and D, was determined (Figure 4). The supplementation of fractions Tdi and A increased mannoprotein concentration in wines without O. oeni inoculation in both wines. Interestingly, a decrease in mannoprotein concentration was observed in those supplemented wines, but not in the control wine. This could be related to the low viability (Table 3) and metabolic activity observed in the MLF performance (Figure 2). Mannoprotein consumption has been reported to be enhanced with higher concentrations [38] under wine conditions, as it was observed in this experiment (Figure 4). In addition, mannoprotein concentration was higher in wine D compared to wine C, as it was the mannoprotein consumption.
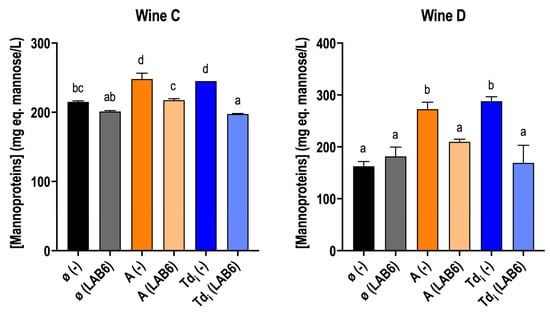
Figure 4.
Mannoprotein concentration (mg mannose eq./L) in wine C and D supplemented or not (ø) with T. delbrueckii fractions (Tdi and A), inoculated with O. oeni (LAB6) or with no inoculation (−). Measurements are from wines after MLF, in case they finished with the addition of the extracts, or after 40 days. (−) wines were taken as a reference wine without inoculation to compare the potential mannoprotein consumption by O. oeni. Letters mean that values are significant at p < 0.05. Fermentations were carried out in duplicate.
Amino acid concentration was also determined. Nitrogen compounds are not abundant in wine after AF. Indeed, there is a complex dynamic since amino acids can be consumed by LAB, while they are again released from peptides or proteins [54,57,58,59]. The free amino acid concentration is usually maintained after MLF, compared to the concentration found before this fermentation [57,58]. In this experiment, the results align with the literature, as no significant changes were observed in these wines regardless of whether O. oeni was inoculated or not (Figure 5). Even so, a trend can be observed, mainly in wine C, where wines after MLF showed a slight increase in free amino acids, which could indicate hydrolysis from peptides [44,54] prior to their consumption.
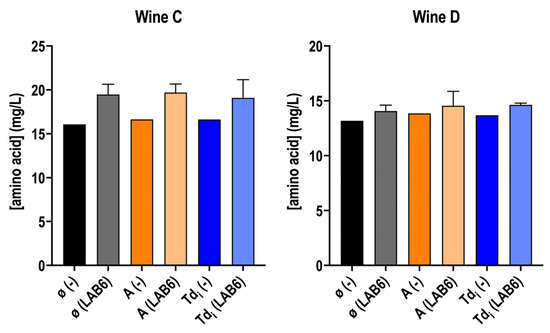
Figure 5.
Total free amino acid concentration in wine C and D supplemented or not (ø) with T. delbrueckii fractions (Tdi, and A), inoculated with O. oeni (LAB6) or with no inoculation (−). Measurements are from wines after MLF, in case they finished—with the addition of the fractions—or after 40 days. (−) wines were taken as a reference wine without inoculation to compare the potential mannoprotein consumption by O. oeni. Fermentations were carried out in duplicate.
Considering the obtained results, the Chardonnay wine, wine D, was selected for testing the other four O. oeni strains with the Tdi extract. In this trial, the fraction also showed its potential for reducing MLF duration with all five tested strains (Table 4). Indeed, the control wine did not finish MLF (Table 4). When adding the Tdi fraction, all strains could finish the MLF. Similar durations, about 20 days, were obtained with LAA4, LAC20, and LAB9 strains. LAA1 and LAB2013 showed more extended durations. Once again, confirming the potential of non-Saccharomyces yeasts for promoting O. oeni growth and MLF, not only as a living starter culture but also as an activator extract for MLF [36].

Table 4.
Malolactic fermentation duration and residual average L-malic concentration after 40 days of fermentation of wine D inoculated with O. oeni LAA1, LAB2013, LAA4, LAC20, LAB9, and supplemented or not (ø) with the Tdi fraction. Spontaneous MLF is not included since it did not occur. -: uncompleted MLF, n.d.: not detected. Fermentations were carried out in duplicate.
Indeed, the effect on maintaining or even promoting bacterial viability was also observed in this confirmation test. All tested strains showed increased viable population in wines supplemented with the Tdi extract after 13 and 20 days of fermentation (Table 4), aligned with the previous observed results of LAB6 in wines C and D (Table 3). The observed increase in viability was up to 1 log in most strains and at most sampling points, except for day 6 of fermentation. This is consistent with previous findings that reported a positive effect associated with prolonged fermentation durations (Table 3). Nevertheless, no significant decrease in viability was observed in relation to the supplementation with the Tdi fraction.
Altogether, it was confirmed that the supplementation with T. delbrueckii fractions improved MLF performance by reducing the fermentative duration (Figure 1 and Figure 2). This was consistent for different types of wine and for different O. oeni strains (Figure 5). From all four extracts, the best results were obtained with the Tdi and A extracts (Figure 1). The monitoring of bacterial viability during the first two weeks of fermentation showed that viability was maintained in those supplemented wines (Table 3), which could suggest a survival enhancement for O. oeni [46]. In this regard, an increase in mannoprotein consumption was observed in those supplemented wines (Figure 4), which is also related to better adaptation to wine conditions.
The study provides insights into how specific fractions can influence bacterial behavior, offering potential technological applications for promoting MLF in harsh conditions. Future work should address sensory and aroma evaluations, test these fractions at pilot and industrial scale, to evaluate the feasibility and applicability in winemaking.
4. Conclusions
In this study, different fractions of T. delbrueckii were used to evaluate their potential to improve MLF in wine. The red and white wines selected for the experiment represented harsh conditions for MLF, as no spontaneous MLF occurred in any of the wines. However, supplementation with the four tested fractions—particularly Tdi and A—enabled MLF when a commercial O. oeni strain was inoculated. Under these conditions, the commercial strain stuck in the wines without Tdi fraction supplementation, leaving more than half of the initial L-malic acid unconsumed. This outcome was confirmed in three independent experiments. Notably, the viability of O. oeni improved two weeks after inoculation in wines supplemented with the extracts. In addition, mannoprotein consumption was detected in the supplemented wines, suggesting a potential detoxifying role of the extracts and a possible enhancement of O. oeni survival mechanisms. These results support the potential application of T. delbrueckii-derived fractions, and not only as a living starter culture, mainly Tdi and A fractions, as enological tools, which can promote and facilitate MLF under harsh wine conditions, thus saving time and ensuring the completion of the process. Further research will focus on larger-scale trials and the evaluation of sensory, technological, and practical outcomes to better understand the potential of these fractions in winemaking.
Author Contributions
Conceptualization, A.B., N.R., M.D.-B. and C.R.; methodology, A.B., N.R., B.L., M.D.-B. and C.R.; formal analysis, A.B. and C.R.; investigation, A.B., P.T. and B.L.; resources, J.M.H., S.K.-W. and M.D.-B.; data curation, A.B.; writing—original draft preparation, A.B.; writing—review and editing, A.B., N.R., M.D.-B. and C.R.; visualization, A.B.; supervision, C.R.; project administration, C.R.; funding acquisition, N.R. and C.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded through contract T23140S between the Fundació Universitat Rovira i Virgili (FURV) and Lallemand SAS.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
Four of the authors, José M. Heras, Sibylle Krieger-Weber, Benjamin Leroux, and Magali Deleris-Bou, work for the company Lallemand SAS. The other authors declare no conflicts of interest.
Appendix A

Table A1.
Duration of MLF in wines A, B, C, and D inoculated with O. oeni LAB6 and the chemical characterization of the obtained wines after MLF. Ø: no fraction. Tdi, A, YE, and CW mean inactivated yeast extract (Tdi), autolysate (A), yeast extract (YE), and cell wall extract (CW).
Table A1.
Duration of MLF in wines A, B, C, and D inoculated with O. oeni LAB6 and the chemical characterization of the obtained wines after MLF. Ø: no fraction. Tdi, A, YE, and CW mean inactivated yeast extract (Tdi), autolysate (A), yeast extract (YE), and cell wall extract (CW).
| Duration (d) | L-Malic Acid (g/L) | pH | L-Lactic Acid (g/L) | D-Lactic Acid (g/L) | Citric Acid (mg/L) | Acetic Acid (g/L) | Glycerol (g/L) | PAN (mg N/L) | NH4 + (mg N/L) | |
|---|---|---|---|---|---|---|---|---|---|---|
| A-Ø | ~0.20 b | 3.19 ± 0.01 a | 1.13 ± 0.01 a | 0.14 ± 0.01 ab | 77 ± 1 | 0.29 ± 0.02 a | 7.30 ± 0.57 ab | 8 ± 2 ab | 22 ± 2 b | |
| A-Tdi | ~0.16 a | 3.19 ± 0.01 a | 1.12 ± 0.03 a | 0.13 ± 0.01 a | 77 ± 8 | 0.28 ± 0.02 a | 7.10 ± 0.01 a | 8 ± 1 a | 26 ± 6 b | |
| A-A | 61 | n.d. | 3.31 ± 0.01 b | 1.27 ± 0.05 b | 0.15 ± 0.01 bc | 56 ± 1 | 0.40 ± 0.02 b | 8.45 ± 0.07 ab | 12 ± 2 ab | 15 ± 1 ab |
| A-YE | 61 | n.d. | 3.30 ± 0.01 b | 1.18 ± 0.04 ab | 0.15 ± 0.01 c | 61 ± 20 | 0.37 ± 0.01 b | 8.50 ± 0.28 b | 14 ± 1 b | 9 ± 1 a |
| A-CW | ~0.24 c | 3.18 ± 0.01 a | 1.11 ± 0.01 a | 0.13 ± 0.01 a | 79 ± 1 | 0.28 ± 0.01 a | 7.40 ± 0.42 ab | 14 ± 1 b | 9 ± 2 a | |
| B-Ø | ~0.42 | 3.32 ± 0.01 a | 0.36 ± 0.01 a | 0.12 ± 0.04 | 192 ± 11 | 0.46 ± 0.01 | 8.85 ± 0.07 a | 65 ± 1 a | 57 ± 3 a | |
| B-Tdi | 21 | n.d. | 3.42 ± 0.04 b | 0.58 ± 0.03 c | 0.12 ± 0.01 | 209 ± 1 | 0.52 ± 0.01 | 9.40 ± 0.14 ab | 70 ± 1 a | 68 ± 2 b |
| B-A | 24 | n.d. | 3.44 ± 0.02 b | 0.54 ± 0.02 bc | 0.11 ± 0.01 | 201 ± 5 | 0.51 ± 0.04 | 9.45 ± 0.07 ab | 72 ± 1 ab | 61 ± 1 ab |
| B-YE | 24 | n.d. | 3.45 ± 0.01 b | 0.50 ± 0.02 b | 0.11 ± 0.01 | 199 ± 11 | 0.50 ± 0.01 | 9.45 ± 0.07 ab | 86 ± 5 c | 63 ± 10 ab |
| B-CW | 41 | n.d. | 3.45 ± 0.02 b | 0.54 ± 0.01 bc | 0.11 ± 0.01 | 199 ± 23 | 0.51 ± 0.03 | 9.70 ± 0.28 b | 83 ± 4 bc | 68 ± 2 ab |
| C-Ø | ~2.00 b | 3.70 ± 0.01 | 0.77 ± 0.01 a | 0.25 ± 0.02 a | 280 ± 11 | 0.34 ± 0.01 a | 8.90 ± 0.01 | 43 ± 1 a | 22 ± 5 | |
| C-Tdi | 45 | n.d. | 3.82 ± 0.11 | 2.07 ± 0.11 b | 0.38 ± 0.02 abc | 210 ± 6 | 0.42 ± 0.01 b | 9.35 ± 0.21 | 49 ± 1 ab | 44 ± 4 |
| C-A | 24 | n.d. | 3.80 ± 0.14 | 2.14 ± 0.07 b | 0.42 ± 0.03 c | 251 ± 3 | 0.41 ± 0.01 b | 9.50 ± 0.01 | 52 ± 3 ab | 34 ± 8 |
| C-YE | ~1.81 a | 3.72 ± 0.01 | 0.91 ± 0.01 a | 0.26 ± 0.05 ab | 283 ± 39 | 0.35 ± 0.01 a | 8.80 ± 0.14 | 57 ± 3 b | 25 ± 1 | |
| C-CW | 27 | n.d. | 3.84 ± 0.08 | 2.05 ± 0.15 b | 0.42 ± 0.06 bc | 242 ± 8 | 0.41 ± 0.02 b | 9.50 ± 0.57 | 58 ± 4 b | 27 ± 3 |
| D-Ø | ~1.70 b | 3.29 ± 0.01 a | 0.58 ± 0.05 a | 0.11 ± 0.01 a | 182 ± 14 c | 0.49 ± 0.01 | 9.55 ± 0.07 | 35 ± 1 | n.d. | |
| D-Tdi | 17 | n.d. | 3.46 ± 0.01 b | 1.63 ± 0.10 c | 0.13 ± 0.01 ab | 149 ± 11 abc | 0.56 ± 0.01 | 9.35 ± 0.21 | 31 ± 6 | 7 ± 3 |
| D-A | 17 | n.d. | 3.47 ± 0.01 b | 1.67 ± 0.16 c | 0.13 ± 0.01 ab | 145 ± 4 ab | 0.57 ± 0.05 | 9.70 ± 0.14 | 36 ± 8 | n.d. |
| D-YE | ~1.19 a | 3.30 ± 0.01 a | 0.96 ± 0.04 b | 0.12 ± 0.01 a | 178 ± 1 bc | 0.55 ± 0.04 | 9.65 ± 0.35 | 41 ± 1 | n.d. | |
| D-CW | 21 | n.d. | 3.48 ± 0.01 b | 1.67 ± 0.01 c | 0.15 ± 0.01 b | 131 ± 5 a | 0.58 ± 0.01 | 9.65 ± 0.35 | 37 ± 1 | 8 ± 1 |
Superscripts mean that values are significant at p < 0.05. No letter means no significant difference. Statistics were conducted for each wine independently.
References
- Lerm, E.; Engelbrecht, L.; du Toit, M. Malolactic Fermentation: The ABC’s of MLF. S. Afr. J. Enol. Vitic. 2010, 31, 186–212. [Google Scholar] [CrossRef]
- Davis, C.R.; Wibowo, D.; Eschenbruch, R.; Lee, T.H.; Fleet, G.H. Practical Implications of Malolactic Fermentation: A Review. Am. J. Enol. Vitic. 1985, 36, 290–301. [Google Scholar] [CrossRef]
- Liu, S.Q. Malolactic Fermentation in Wine—Beyond Deacidification. J. Appl. Microbiol. 2002, 92, 589–601. [Google Scholar] [CrossRef]
- Albergaria, H.; Arneborg, N. Dominance of Saccharomyces cerevisiae in Alcoholic Fermentation Processes: Role of Physiological Fitness and Microbial Interactions. Appl. Microbial. Biotechnol. 2016, 100, 2035–2046. [Google Scholar] [CrossRef] [PubMed]
- Börlin, M.; Venet, P.; Claisse, O.; Salin, F.; Legras, J.L.; Masneuf-Pomarede, I. Cellar-Associated Saccharomyces cerevisiae Population Structure Revealed High-Level Diversity and Perennial Persistence at Sauternes Wine Estates. Appl. Environ. Microbiol. 2016, 82, 2909–2918. [Google Scholar] [CrossRef]
- Padilla, B.; García-Fernández, D.; González, B.; Izidoro, I.; Esteve-Zarzoso, B.; Beltran, G.; Mas, A. Yeast Biodiversity from DOQ Priorat Uninoculated Fermentations. Front. Microbiol. 2016, 7, 930. [Google Scholar] [CrossRef]
- Balmaseda, A.; Lorentzen, M.; Dutilh, L.; Bauduin, R.; Guichard, H.; Ollivier, S.; Miot-Sertier, C.; Lucas, P.M. Alcoholic Fermentation Drives the Selection of Oenococcus oeni Strains in Wine but Not in Cider. Int. J. Food Microbiol. 2023, 400, 110276. [Google Scholar] [CrossRef]
- Lorentzen, M.P.; Campbell-Sills, H.; Jorgensen, T.S.; Nielsen, T.K.; Coton, M.; Coton, E.; Hansen, L.; Lucas, P.M. Expanding the Biodiversity of Oenococcus oeni through Comparative Genomics of Apple Cider and Kombucha Strains. BMC Genom. 2019, 20, 300. [Google Scholar] [CrossRef]
- Campbell-Sills, H.; Khoury, M.E.; Gammacurta, M.; Miot-sertier, C.; Claisse, O.; Spano, G.; Revel, G.D.; Lucas, P. Two Different Oenococcus oeni Lineages Are Associated to Either Red or White Wines in Burgundy: Genomics and Metabolomics Insights. OENO One 2017, 51, 309–322. [Google Scholar] [CrossRef]
- Bech-Terkilsen, S.; Westman, J.O.; Swiegers, J.H.; Siegumfeldt, H. Oenococcus Oeni, a Species Born and Moulded in Wine: A Critical Review of the Stress Impacts of Wine and the Physiological Responses. Aust. J. Grape Wine Res. 2020, 26, 188–206. [Google Scholar] [CrossRef]
- Alexandre, H.; Guilloux-Benatier, M. Yeast Autolysis in Sparkling Wine—A Review. Aust. J. Grape Wine Res. 2006, 12, 119–127. [Google Scholar] [CrossRef]
- Sawyer, S.; Longo, R.; Solomon, M.; Nicolotti, L.; Westmore, H.; Merry, A.; Gnoinski, G.; Ylia, A.; Dambergs, R.; Kerslake, F. Autolysis and the Duration of Ageing on Lees Independently Influence the Aroma Composition of Traditional Method Sparkling Wine. Aust. J. Grape Wine Res. 2022, 28, 146–159. [Google Scholar] [CrossRef]
- Charpentier, C.; Aussenac, J.; Charpentier, M.; Prome, J.C.; Duteurtre, B.; Feuillat, M. Release of Nucleotides and Nucleosides during Yeast Autolysis: Kinetics and Potential Impact on Flavor. J. Agric. Food Chem. 2005, 53, 3000–3007. [Google Scholar] [CrossRef]
- Martínez-Rodriguez, A.J.; Carrascosa, A.V.; Polo, M.C. Release of Nitrogen Compounds to the Extracellular Medium by Three Strains of Saccharomyces cerevisiae During Induced Autolysis in a Model Wine System. Int. J. Food Microbiol. 2001, 68, 155–160. [Google Scholar] [CrossRef]
- Palomero, F.; Morata, A.; Benito, S.; González, M.C.; Suárez-Lepe, J.A. Conventional and Enzyme-Assisted Autolysis during Ageing over Lees in Red Wines: Influence on the Release of Polysaccharides from Yeast Cell Walls and on Wine Monomeric Anthocyanin Content. Food Chem. 2007, 105, 838–846. [Google Scholar] [CrossRef]
- Balmaseda, A.; Bordons, A.; Reguant, C.; Bautista-Gallego, J. Non-Saccharomyces in Wine: Effect upon Oenococcus oeni and Malolactic Fermentation. Front. Microbiol. 2018, 9, 534. [Google Scholar] [CrossRef]
- Jolly, N.P.; Augustyn, O.P.H.; Pretorius, I.S. The Role and Use of Non-Saccharomyces Yeasts in Wine Production. S. Afr. J. Enol. Vitic. 2006, 27, 15–39. [Google Scholar] [CrossRef]
- Padilla, B.; Gil, J.V.; Manzanares, P. Past and Future of Non-Saccharomyces Yeasts: From Spoilage Microorganisms to Biotechnological Tools for Improving Wine Aroma Complexity. Front. Microbiol. 2016, 7, 411. [Google Scholar] [CrossRef]
- Escribano-Viana, R.; Portu, J.; Garijo, P.; López, R.; Santamaría, P.; López-Alfaro, I.; Gutiérrez, A.R.; González-Arenzana, L. Effect of the Sequential Inoculation of Non-Saccharomyces/Saccharomyces on the Anthocyans and Stilbenes Composition of Tempranillo Wines. Front. Microbiol. 2019, 10, 773. [Google Scholar] [CrossRef] [PubMed]
- Vejarano, R.; Gil-Calderón, A. Commercially Available Non-Saccharomyces Yeasts for Winemaking: Current Market, Advantages over Saccharomyces, Biocompatibility, and Safety. Fermentation 2021, 7, 171. [Google Scholar] [CrossRef]
- Canonico, L.; Solomon, M.; Comitini, F.; Ciani, M.; Varela, C. Volatile Profile of Reduced Alcohol Wines Fermented with Selected Non-Saccharomyces Yeasts Under Different Aeration Conditions. Food Microbiol. 2019, 84, 103247. [Google Scholar] [CrossRef]
- Ivit, N.N.; Kemp, B. The Impact of Non-Saccharomyces Yeast on Traditional Method Sparkling The Impact of Non-Saccharomyces Yeast on Traditional Method Sparkling Wine. Fermentation 2018, 4, 73. [Google Scholar] [CrossRef]
- Zhang, B.; Tang, C.; Yang, D.; Liu, H.; Xue, J.; Duan, C.; Yan, G. Effects of Three Indigenous Non-Saccharomyces Yeasts and Their Pairwise Combinations in Co-Fermentation with Saccharomyces Cerevisiae on Volatile Compounds of Petit Manseng Wines. Food Chem. 2022, 368, 130807. [Google Scholar] [CrossRef] [PubMed]
- Canonico, L.; Comitini, F.; Oro, L.; Ciani, M. Sequential Fermentation with Selected Immobilized Non-Saccharomyces Yeast for Reduction of Ethanol Content in Wine. Front. Microbiol. 2016, 7, 278. [Google Scholar] [CrossRef]
- Moreno-García, J.; García-Martínez, T.; Mauricio, J.C.; Moreno, J. Yeast Immobilization Systems for Alcoholic Wine Fermentations: Actual Trends and Future Perspectives. Front. Microbiol. 2018, 9, 241. [Google Scholar] [CrossRef] [PubMed]
- Guzzon, R.; Carturan, G.; Krieger-Weber, S.; Cavazza, A. Use of Organo-Silica Immobilized Bacteria Produced in a Pilot Scale Plant to Induce Malolactic Fermentation in Wines That Contain Lysozyme. Ann. Microbiol. 2012, 62, 381–390. [Google Scholar] [CrossRef]
- Nikolaou, A.; Sgouros, G.; Mitropoulou, G.; Santarmaki, V.; Kourkoutas, Y. Freeze-Dried Immobilized Kefir Culture in Low Alcohol Winemaking. Foods 2020, 9, 115. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-de-Villa, C.; Gombau, J.; Poblet, M.; Bordons, A.; Canals, J.M.; Zamora, F.; Reguant, C.; Rozès, N. Sequential Inoculation of Torulaspora delbrueckii and Saccharomyces cerevisiae in Rosé Wines Enhances Malolactic Fermentation and Potentially Improves Colour Stability. LWT 2023, 190, 115540. [Google Scholar] [CrossRef]
- Ruiz-de-Villa, C.; Poblet, M.; Bordons, A.; Reguant, C.; Rozès, N. Comparative Study of Inoculation Strategies of Torulaspora delbrueckii and Saccharomyces cerevisiae on the Performance of Alcoholic and Malolactic Fermentations in an Optimized Synthetic Grape Must. Int. J. Food Microbiol. 2023, 404, 110367. [Google Scholar] [CrossRef]
- Balmaseda, A.; Rozès, N.; Bordons, A.; Reguant, C. Torulaspora delbrueckii Promotes Malolactic Fermentation in High Polyphenolic Red Wines. LWT 2021, 148, 111777. [Google Scholar] [CrossRef]
- Balmaseda, A.; Rozès, N.; Bordons, A.; Reguant, C. Molecular Adaptation Response of Oenococcus oeni in Non-Saccharomyces Fermented Wines: A Comparative Multi-Omics Approach. Int. J. Food Microbiol. 2022, 362, 109490. [Google Scholar] [CrossRef] [PubMed]
- Balmaseda, A.; Rozès, N.; Leal, M.Á.; Bordons, A.; Reguant, C. Impact of Changes in Wine Composition Produced by Non-Saccharomyces on Malolactic Fermentation. Int. J. Food Microbiol. 2021, 337, 108954. [Google Scholar] [CrossRef] [PubMed]
- Balmaseda, A.; Rozès, N.; Bordons, A.; Reguant, C. The Use of Torulaspora delbrueckii to Improve Malolactic Fermentation. Microb. Biotechnol. 2024, 17, e14302. [Google Scholar] [CrossRef] [PubMed]
- Morata, A.; Loira, I.; Escott, C.; Manuel, J.; Su, A. Applications of Metschnikowia pulcherrima in Wine Biotechnology. Fermentation 2019, 5, 63. [Google Scholar] [CrossRef]
- Contreras, A.; Hidalgo, C.; Schmidt, S.; Henschke, P.A.; Curtin, C.; Varela, C. The Application of Non-Saccharomyces Yeast in Fermentations with Limited Aeration as a Strategy for the Production of Wine with Reduced Alcohol Content. Int. J. Food Microbiol. 2015, 205, 7–15. [Google Scholar] [CrossRef]
- Balmaseda, A.; Rozès, N.; Bordons, A.; Reguant, C. Simulated Lees of Different Yeast Species Modify the Performance of Malolactic Fermentation by Oenococcus oeni in Wine-like Medium. Food Microbiol. 2021, 99, 103839. [Google Scholar] [CrossRef]
- Balmaseda, A.; Rozès, N.; Bordons, A.; Reguant, C. Characterization of Malolactic Fermentation by Lactiplantibacillus plantarum in Red Grape Must. LWT 2024, 199, 116070. [Google Scholar] [CrossRef]
- Balmaseda, A.; Aniballi, L.; Rozès, N.; Bordons, A. Use of Yeast Mannoproteins by Oenococcus oeni during Malolactic Fermentation under Different Oenological Conditions. Foods 2021, 10, 1540. [Google Scholar] [CrossRef] [PubMed]
- Olguín, N.; Alegret, J.O.; Bordons, A.; Reguant, C. β-Glucosidase Activity and Bgl Gene Expression of Oenococcus oeni Strains in Model Media and Cabernet Sauvignon Wine. Am. J. Enol. Vitic. 2011, 62, 99–105. [Google Scholar] [CrossRef]
- Toraño, P.; Martín-García, A.; Bordons, A.; Rozès, N.; Reguant, C. Enhancing Wine Malolactic Fermentation: Variable Effect of Yeast Mannoproteins on Oenococcus oeni Strains. Food Microbiol. 2024, 127, 104689. [Google Scholar] [CrossRef]
- Martín-García, A.; Balmaseda, A.; Bordons, A.; Reguant, C. Effect of the Inoculation Strategy of Non-Saccharomyces Yeasts on Wine Malolactic Fermentation. OENO One 2020, 54, 101–108. [Google Scholar] [CrossRef]
- Balmaseda, A.; Rozès, N.; Bordons, A.; Reguant, C. Modulation of a Defined Community of Oenococcus oeni Strains by Torulaspora delbrueckii and Its Impact on Malolactic Fermentation. Aust. J. Grape Wine Res. 2022, 28, 374–382. [Google Scholar] [CrossRef]
- Ramírez, M.; Velázquez, R.; Maqueda, M.; Zamora, E.; López-Piñeiro, A.; Hernández, L.M. Influence of the Dominance of Must Fermentation by Torulaspora delbrueckii on the Malolactic Fermentation and Organoleptic Quality of Red Table Wine. Int. J. Food Microbiol. 2016, 238, 311–319. [Google Scholar] [CrossRef]
- Ritt, J.; Remize, F.; Grandvalet, C.; Guzzo, J.; Atlan, D.; Alexandre, H. Peptidases Specific for Proline-Containing Peptides and Their Unusual Peptide-Dependent Regulation in Oenococcus oeni. J. Appl. Microbiol. 2009, 106, 801–813. [Google Scholar] [CrossRef]
- Alcaide-Hidalgo, J.M.; Moreno-Arribas, M.V.; Polo, M.C.; Pueyo, E. Partial Characterization of Peptides from Red Wines. Changes During Malolactic Fermentation and Ageing with Lees. Food Chem. 2008, 107, 622–630. [Google Scholar] [CrossRef]
- Toraño, P.; Gombau, J.; Mejías, I.; Bordons, A.; Rozès, N.; Reguant, C. Evaluation of the Addition of Yeast Mannoprotein to Oenococcus oeni Starter Cultures to Improve Wine Malolactic Fermentation. Fermentation 2024, 10, 52. [Google Scholar] [CrossRef]
- Juega, M.; Carrascosa, A.V.; Martinez-Rodriguez, A.J. Effect of Short Ageing on Lees on the Mannoprotein Content, Aromatic Profile, and Sensorial Character of White Wines. J. Food Sci. 2015, 80, M384–M388. [Google Scholar] [CrossRef] [PubMed]
- Lipke, P.N.; Ovalle, R. Cell Wall Architecture in Yeast: New Structure and New Challenge. J. Bacteriol. 1998, 180, 3735–3740. [Google Scholar] [CrossRef] [PubMed]
- Patynowsky, R.J.; Jiranek, V.; Markides, A.J. Yeast Viability During Fermentation and Sur Lie Ageing of a Defined Medium and Subsequent Growth of Oenococcus oeni. Aust. J. Grape Wine Res. 2002, 8, 62–69. [Google Scholar] [CrossRef]
- Diez, L.; Guadalupe, Z.; Ayestarán, B.; Ruiz-Larrea, F. Effect of Yeast Mannoproteins and Grape Polysaccharides on the Growth of Wine Lactic Acid and Acetic Acid Bacteria. J. Agric. Food Chem. 2010, 58, 7731–7739. [Google Scholar] [CrossRef]
- Dupin, I.V.S.; McKinnon, B.M.; Ryan, C.; Boulay, M.; Markides, A.J.; Jones, G.P.; Williams, P.J.; Waters, E.J. Saccharomyces cerevisiae Mannoproteins That Protect Wine from Protein Haze: Their Release during Fermentation and Lees Contact and a Proposal for Their Mechanism of Action. J. Agric. Food Chem. 2000, 48, 3098–3105. [Google Scholar] [CrossRef]
- Jamal, Z.; Miot-Sertier, C.; Thibau, F.; Dutilh, L.; Lonvaud-Funel, A.; Ballestra, P.; Le Marrec, C.; Dols-Lafargue, M. Distribution and Functions of Phosphotransferase System Genes in the Genome of the Lactic Acid Bacterium Oenococcus oeni. Appl. Environ. Microbiol. 2013, 79, 3371–3379. [Google Scholar] [CrossRef] [PubMed]
- Michlmayr, H.; Nauer, S.; Brandes, W.; Schümann, C.; Kulbe, K.D.; Andrés, M.; Eder, R. Release of Wine Monoterpenes from Natural Precursors by Glycosidases from Oenococcus oeni. Food Chem. 2012, 135, 80–87. [Google Scholar] [CrossRef]
- Balmaseda, A.; Rozès, N.; Bordons, A.; Alexandre, H.; Reguant, C. Evaluating the Impact of Torulaspora delbrueckii and Amino Acid Concentration on the Nitrogen Metabolism of Oenococcus oeni. LWT 2024, 210, 116838. [Google Scholar] [CrossRef]
- Guilloux-Benatier, M.; Guerreau, J.; Feuillat, M. Influence of Initial Colloid Content on Yeast Macromolecule Production and on the Metabolism of Wine Microorganisms. Am. J. Enol. Vitic. 1995, 46, 486–492. [Google Scholar] [CrossRef]
- Guilloux-Benatier, M.; Chassagne, D. Comparison of Components Released by Fermented or Active Dried Yeasts After Aging on Lees in a Model Wine. J. Agric. Food Chem. 2003, 51, 746–751. [Google Scholar] [CrossRef]
- Guadalupe, Z.; Ayestara, B.; Herna, Z. Amino Acids and Biogenic Amines in Red Varietal Wines: The Role of Grape Variety, Malolactic Fermentation and Vintage. Eur. Food Res. Technol. 2013, 237, 887–895. [Google Scholar] [CrossRef]
- Pozo-Bayón, M.A.; G-Alegría, E.; Polo, M.C.; Tenorio, C.; Martín-Álvarez, P.J.; Calvo de la Banda, M.T.; Ruiz-Larrea, F.; Moreno-Arribas, M.V. Wine Volatile and Amino Acid Composition after Malolactic Fermentation: Effect of Oenococcus oeni and Lactobacillus plantarum Starter Cultures. J. Agric. Food Chem. 2005, 53, 8729–8735. [Google Scholar] [CrossRef] [PubMed]
- Alcaide-Hidalgo, J.M.; Moreno-Arribas, M.V.; Martín-Álvarez, P.J.; Polo, M.C. Influence of Malolactic Fermentation, Postfermentative Treatments and Ageing with Lees on Nitrogen Compounds of Red Wines. Food Chem. 2007, 103, 572–581. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).