Enhanced Tolerance to Antifungals as a General Feature of Rho− Mutants in Yeast Species: Implications to Positive Selection of Respiratory Deficiency
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Media Used
2.2. Sensitivity Testing
2.3. Replica Plating
2.4. Spot Tests
2.5. Mode of Inhibition Assays
2.6. Data Analysis
3. Results
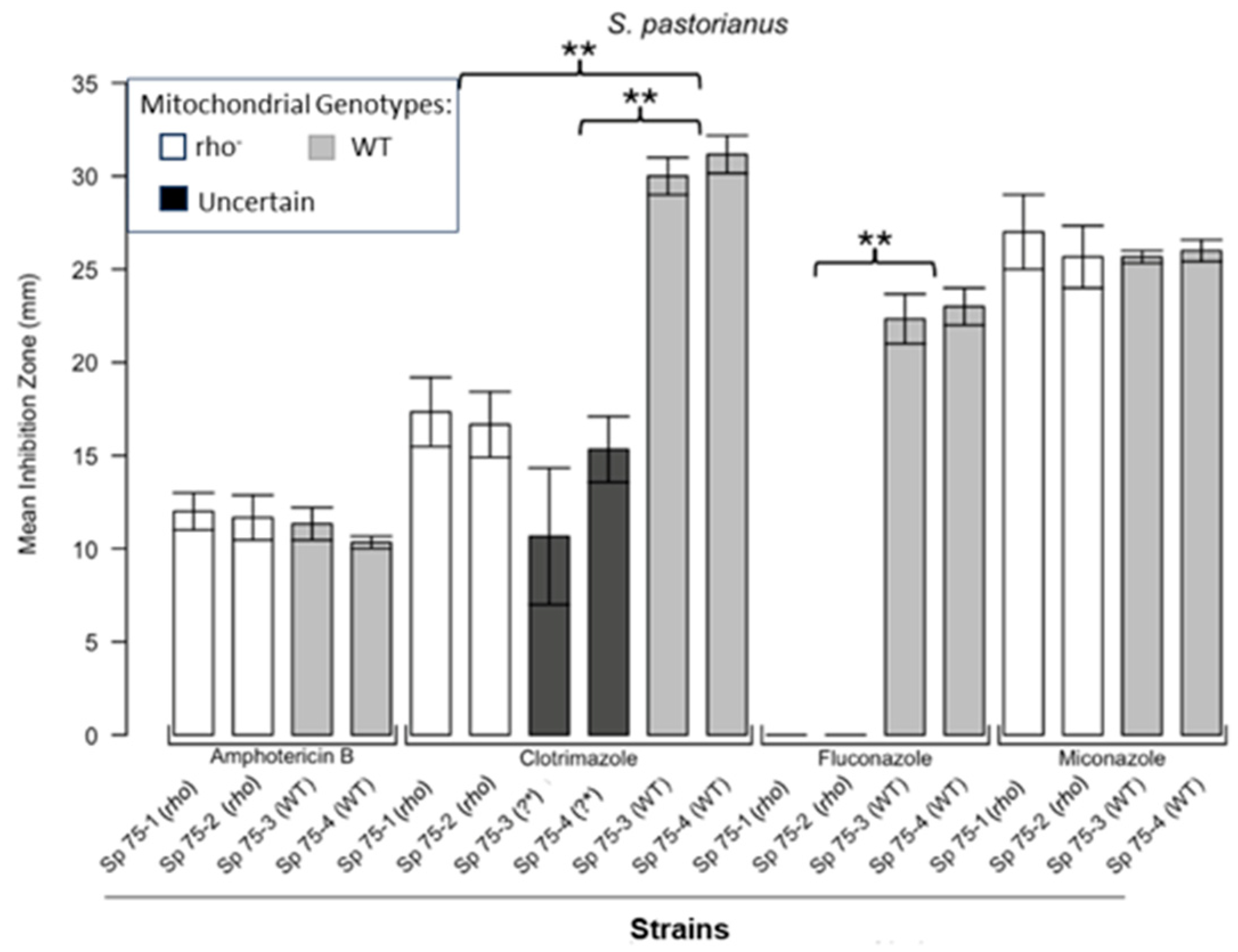
3.1. Respiratory Deficient Rho− Mutants of Petite-Positive Yeast Species Are More Tolerant to Some Antifungals than Their WT Counterparts
3.2. Variations of ZOI Patterns Are Strain- and Drug-Dependent
3.3. Incomplete Inhibition Outcomes Predominantly from Spontaneous Generation of Respiratory-Deficient Cells with Enhanced Tolerance to Antifungals
3.4. Modelling Experiments Towards Positive Selection of Respiratory-Deficient Mutants
3.5. Modes of Inhibition of Yeast Cells by Antifungals
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DMSO | Dimethyl sulfoxide |
| MATa | Mating type a |
| MATalpha | Mating type alpha |
| MoI | Mode of inhibition |
| mtDNA | Mitochondrial DNA |
| NCYC | National Collection of Yeast Cultures |
| OXPHOS | Oxidative phosphorylation |
| PIF1 | [from: Petite Integration Frequency] A nuclear gene encoding DNA helicase functioning in nucleus and mitochondria |
| RAD5 | [from: RADiation sensitive] A nuclear gene encoding a DNA-dependent ATPase and DNA helicase |
| SEM | Standard error of the mean |
| W303 | A genetic background of a yeast model, derived from K6001 background |
| WT | Wild type |
| YEPD | Yeast extract peptone dextrose medium (dextrose = glucose) |
| YEPG | Yeast extract peptone glycerol medium |
| ZOI | Zone of inhibition |
| ZOIs | Zones of inhibition |
References
- Williamson, D. Timeline—The Curious History of Yeast Mitochondrial DNA. Nat. Rev. Genet. 2002, 3, 475–481. [Google Scholar] [CrossRef]
- Burger, G.; Gray, M.; Lang, B. Mitochondrial Genomes: Anything Goes. Trends Genet. 2003, 19, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Gualberto, J.; Newton, K. Plant Mitochondrial Genomes: Dynamics and Mechanisms of Mutation. Annu. Rev. Plant Biol. 2017, 68, 225–252. [Google Scholar] [CrossRef] [PubMed]
- Malina, C.; Larsson, C.; Nielsen, J. Yeast Mitochondria: An Overview of Mitochondrial Biology and the Potential of Mitochondrial Systems Biology. FEMS Yeast Res. 2018, 18, foy040. [Google Scholar] [CrossRef] [PubMed]
- Macreadie, I. My Early Years of Yeast Mitochondrial Genetics. Microorganisms 2024, 12, 2077. [Google Scholar] [CrossRef]
- Gorman, G.; Chinnery, P.; DiMauro, S.; Hirano, M.; Koga, Y.; McFarland, R.; Suomalainen, A.; Thorburn, D.; Zeviani, M.; Turnbull, D. Mitochondrial diseases. Nat. Rev. Dis. Primers 2016, 2, 16080. [Google Scholar] [CrossRef] [PubMed]
- Alston, C.; Rocha, M.; Lax, N.; Turnbull, D.; Taylor, R. The Genetics and Pathology of Mitochondrial Disease. J. Pathol. 2017, 241, 236–250. [Google Scholar] [CrossRef]
- Kopinski, P.; Singh, L.; Zhang, S.; Lott, M.; Wallace, D. Mitochondrial DNA Variation and Cancer. Nat. Rev. Cancer 2021, 21, 431–445. [Google Scholar] [CrossRef] [PubMed]
- Fekete, V.; Cierna, M.; Poláková, S.; Piskur, J.; Sulo, P. Transition of the Ability to Generate Petites in the Saccharomyces Kluyveromyces Complex. FEMS Yeast Res. 2007, 7, 1237–1247. [Google Scholar] [CrossRef] [PubMed]
- Bulder, C.J.E.A. Induction of Petite Mutation and Inhibition of Synthesis of Respiratory Enzymes in Various Yeasts. Antonie Van Leeuwenhoek 1964, 30, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.; Labedzka-Dmoch, K. Retrogreat Signaling: The Lessons We Learn from Yeast. Iubmb Life 2024, 76, 26–37. [Google Scholar] [CrossRef]
- Hsu, P.-C.; Lu, T.-C.; Hung, P.-H.; Leu, J.-Y. Protein Moonlighting by a Target Gene Dominates Phenotypic Divergence of the Sef1 Transcriptional Regulatory Network in Yeasts. Nucleic Acids Res. 2024, 52, 13914–13930. [Google Scholar] [CrossRef] [PubMed]
- Grant, C.M.; MacIver, F.H.; Dawes, I.W. Mitochondrial Function Is Required for Resistance to Oxidative Stress in the Yeast Saccharomyces cerevisiae. FEBS Lett. 1997, 410, 219–222. [Google Scholar] [CrossRef]
- Rasmussen, A.; Chatterjee, A.; Rasmussen, L.; Singh, K. Mitochondria-Mediated Nuclear Mutator Phenotype in Saccharomyces cerevisiae. Nucleic Acids Res. 2003, 31, 3909–3917. [Google Scholar] [CrossRef]
- Gao, Q.; Liou, L.-C.; Ren, Q.; Bao, X.; Zhang, Z. Salt Stress Causes Cell Wall Damage in Yeast Cells Lacking Mitochondrial DNA. Microb. Cell 2014, 1, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Zubko, E.I.; Zubko, M.K. Deficiencies in Mitochondrial DNA Compromise the Survival of Yeast Cells at Critically High Temperatures. Microbiol. Res. 2014, 169, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Cal, M.; Matyjaszczyk, I.; Filik, K.; Ogórek, R.; Ko, Y.; Ulaszewski, S. Mitochondrial Function Are Disturbed in the Presence of the Anticancer Drug, 3-Bromopyruvate. Int. J. Mol. Sci. 2021, 22, 6640. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Peris, D.; Hittinger, C.; Sia, E.; Fay, J. Mitochondria-Encoded Genes Contribute to Evolution of Heat and Cold Tolerance in Yeast. Sci. Adv. 2019, 5, eaav1848. [Google Scholar] [CrossRef]
- Huang, C.; Lu, M.; Chang, Y.; Li, W. Experimental Evolution of Yeast for High-Temperature Tolerance. Mol. Biol. Evol. 2018, 35, 1823–1839. [Google Scholar] [CrossRef] [PubMed]
- Bouchara, J.; Zouhair, R.; Le Boudouil, S.; Renier, G.; Filmon, R.; Chabasse, D.; Hallet, J.; Defontaine, A. In-Vivo Selection of an Azole-Resistant Petite Mutant of Candida glabrata. J. Med. Microbiol. 2000, 49, 977–984. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Sanguinetti, M.; De Bernardis, F.; Torelli, R.; Posteraro, B.; Vandeputte, P.; Sanglard, D. Loss of Mitochondrial Functions Associated with Azole Resistance in Candida glabrata Results in Enhanced Virulence in Mice. Antimicrob. Agents Chemother. 2011, 55, 1852–1860. [Google Scholar] [CrossRef]
- Siscar-Lewin, S.; Gabaldón, T.; Aldejohann, A.; Kurzai, O.; Hube, B.; Brunke, S. Transient Mitochondria Dysfunction Confers Fungal Cross-Resistance against Phagocytic Killing and Fluconazole. mBio 2021, 12, e0112821. [Google Scholar] [CrossRef] [PubMed]
- Badrane, H.; Cheng, S.; Dupont, C.; Hao, B.; Driscoll, E.; Morder, K.; Liu, G.; Newbrough, A.; Fleres, G.; Kaul, D.; et al. Genotypic Diversity and Unrecognized Antifungal Resistance Among Populations of Candida glabrata from Positive Blood Cultures. Nat. Commun. 2023, 14, 5918. [Google Scholar] [CrossRef]
- Kontoyiannis, D. Modulation of Fluconazole Sensitivity by the Interaction Of Mitochondria and Erg3p in Saccharomyces cerevisiae. J. Antimicrob. Chemother. 2000, 46, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Adams, A.; Gottschling, D.E.; Kaiser, C.A.; Stearns, T. Methods in Yeast Genetics, 1997th ed.; CSHL Press: Cold Spring Harbor, NY, USA, 1997; xiv+177pp. [Google Scholar]
- Lederberg, J.; Lederberg, E.M. Replica Plating and Indirect Selection of Bacterial Mutants. J. Bacteriol. 1952, 63, 399–406. [Google Scholar] [CrossRef]
- Platania, V.; Douglas, T.E.L.; Zubko, M.K.; Ward, D.; Pietryga, K.; Chatzinikolaidou, M. Phloroglucinol-Enhanced Whey Protein Isolate Hydrogels with Antimicrobial Activity for Tissue Engineering. Mater. Sci. Eng. C-Mater. Biol. Appl. 2021, 129, 112412. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Daneshnia, F.; Hovhannisyan, H.; Fuentes, D.; Cabrera, N.; Quinteros, C.; Ilkit, M.; Ünal, N.; Hilmioglu-Polat, S.; Jabeen, K.; et al. Overlooked Candida glabrata Petites Are Echinocandin Tolerant, Induce Host Inflammatory Responses, and Display Poor In Vivo Fitness. mBio 2023, 14, e0118023. [Google Scholar] [CrossRef]
- Katsipoulaki, M.; Stappers, M.; Malavia-Jones, D.; Brunke, S.; Hube, B.; Gow, N. Candida albicans and Candida glabrata: Global Priority Pathogens. Microbiol. Mol. Biol. Rev. 2024, 88, e0002123. [Google Scholar] [CrossRef]
- Lee, Y.; Robbins, N.; Cowen, L.E. Molecular Mechanisms Governing Antifungal Drug Resistance. Npj Antimicrob. Resist. 2023, 1, 5. [Google Scholar] [CrossRef]
- Kontoyiannis, D.P.; Lewis, R.E. Antifungal Drug Resistance of Pathogenic Fungi. Lancet 2002, 359, 1135–1144. [Google Scholar] [CrossRef] [PubMed]
- Muellner, J.; Schmidt, K. Yeast Genome Maintenance by the Multifunctional PIF1 DNA Helicase Family. Genes 2020, 11, 224. [Google Scholar] [CrossRef]
- Haffter, P.; Fox, T. Nuclear Mutations in the Petite-Negative Yeast Schizosaccharomyces-Pombe Allow Growth of Cells Lacking Mitochondrial-Dna. Genetics 1992, 131, 255–260. [Google Scholar] [CrossRef]
- Slonimski, P.P.; Perrodin, G.; Croft, J.H. Ethidium Bromide Induced Mutation of Yeast Mitochondria: Complete Transformation of Cells into Respiratory Deficient Non-Chromosomal “Petites”. Biochem. Biophys. Res. Commun. 1968, 30, 232–239. [Google Scholar] [CrossRef]
- Goldring, E.; Grossman, L.; Krupnick, D.; Cryer, D.; Marmur, J. Petite Mutation in Yeast—Loss of Mitochondrial Deoxyribonucleic Acid During Induction of Petites with Ethidium Bromide. J. Mol. Biol. 1970, 52, 323–325. [Google Scholar] [CrossRef] [PubMed]
- De Chiara, M.; Friedrich, A.; Barré, B.; Breitenbach, M.; Schacherer, J.; Liti, G. Discordant Evolution of Mitochondrial and Nuclear Yeast Genomes at Population Level. BMC Biol. 2020, 18, 49. [Google Scholar] [CrossRef]
- Paliwal, S.; Fiumera, A.C.; Fiumera, H.L. Mitochondrial-Nuclear Epistasis Contributes to Phenotypic Variation and Coadaptation in Natural Isolates of Saccharomyces cerevisiae. Genetics 2014, 198, 1251–1265. [Google Scholar] [CrossRef] [PubMed]
- Heluane, H.; Spencer, J.F.; Spencer, D.; Defigueroa, L.; Callieri, D.A.S. Characterization of Hybrids Obtained by Protoplast Fusion, between Pachysolen-Tannophilus and Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 1993, 40, 98–100. [Google Scholar] [CrossRef]
- Curran, B.P.; Bugeja, V.C. Protoplast Fusion in Saccharomyces Cerevisiae. Methods Mol. Biol. 1996, 53, 45–49. [Google Scholar]
- Choi, G.W.; Um, H.J.; Kang, H.W.; Kim, Y.; Kim, M.; Kim, Y.H. Bioethanol Production by a Flocculent Hybrid, Chfy0321 Obtained by Protoplast Fusion between Saccharomyces cerevisiae and Saccharomyces bayanus. Biomass Bioenergy 2010, 34, 1232–1242. [Google Scholar] [CrossRef]
- Steensels, J.; Snoek, T.; Meersman, E.; Nicolino, M.P.; Voordeckers, K.; Verstrepen, K.J. Improving Industrial Yeast Strains: Exploiting Natural and Artificial Diversity. FEMS Microbiol. Rev. 2014, 38, 947–995. [Google Scholar] [CrossRef]
- Horn, P.; Wilkie, D. Selective advantage of the cytoplasmic respiratory mutant of Saccharomyces cerevisiae in a cobalt medium. Heredity 1966, 21, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, P.; Ni, J.; Wong, T. Synergistic and antagonistic interactions of triclosan with various antibiotics in bacteria. J. Environ. Sci. Health Part C-Toxicol. Carcinog. 2020, 38, 187–203. [Google Scholar] [CrossRef] [PubMed]
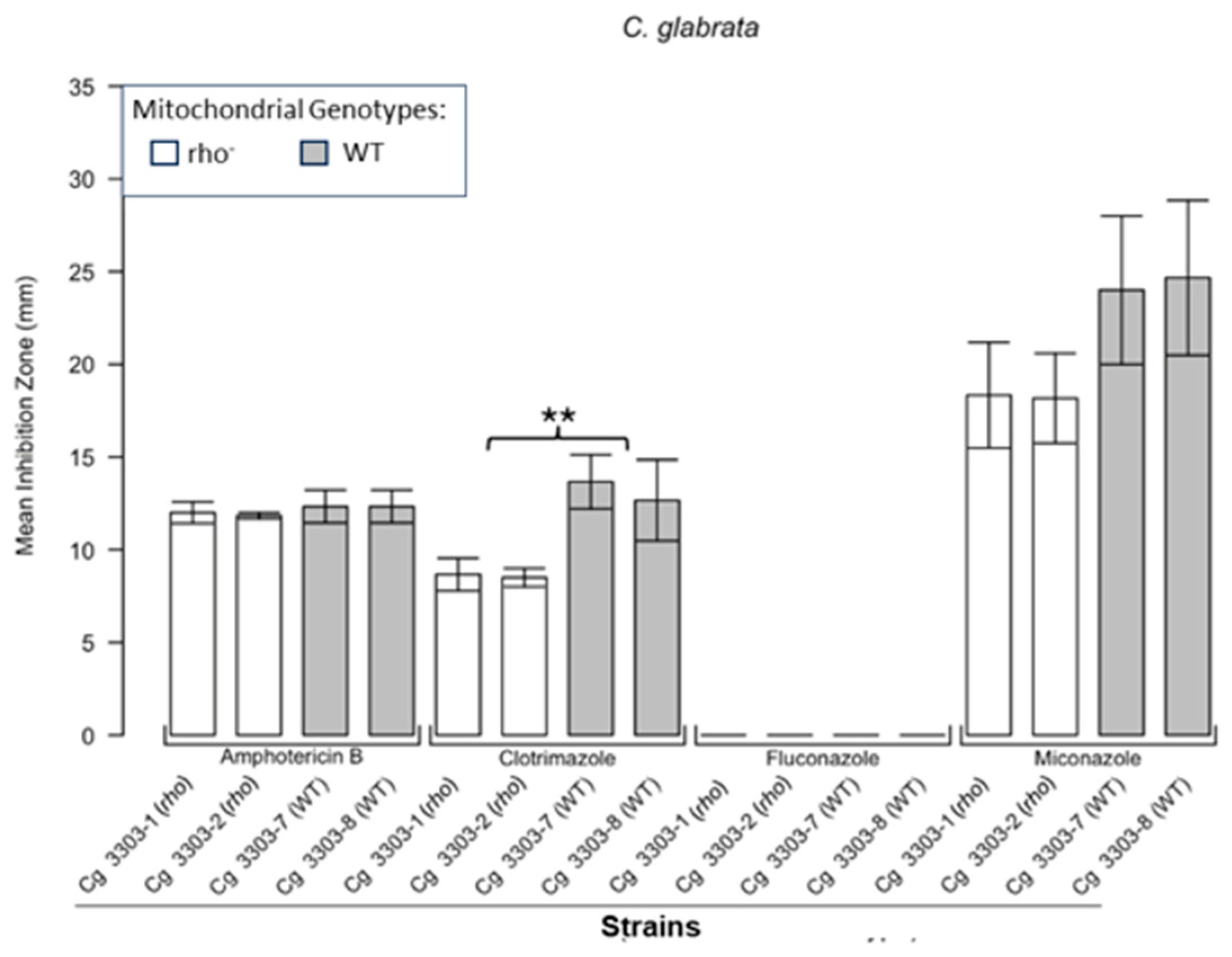
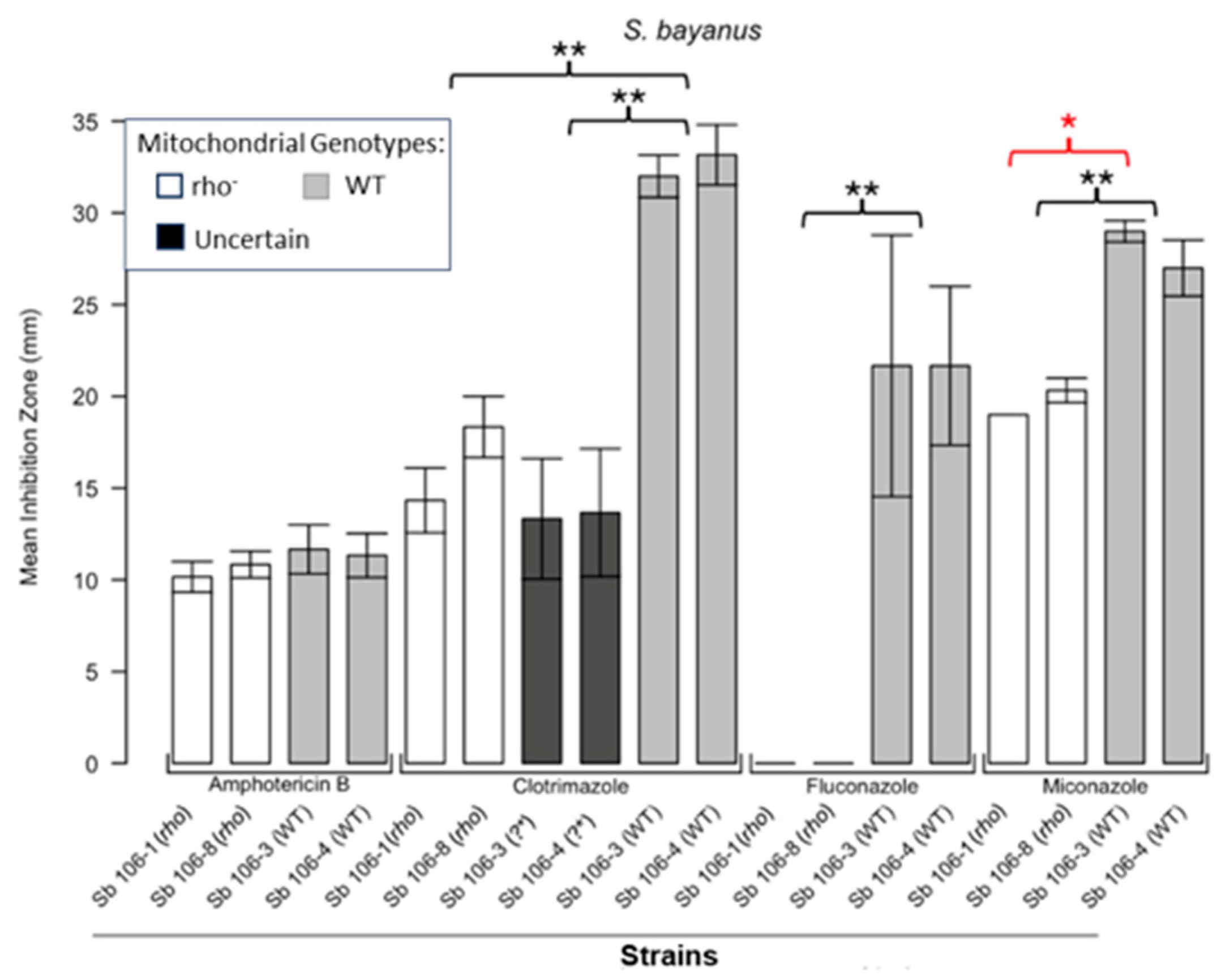
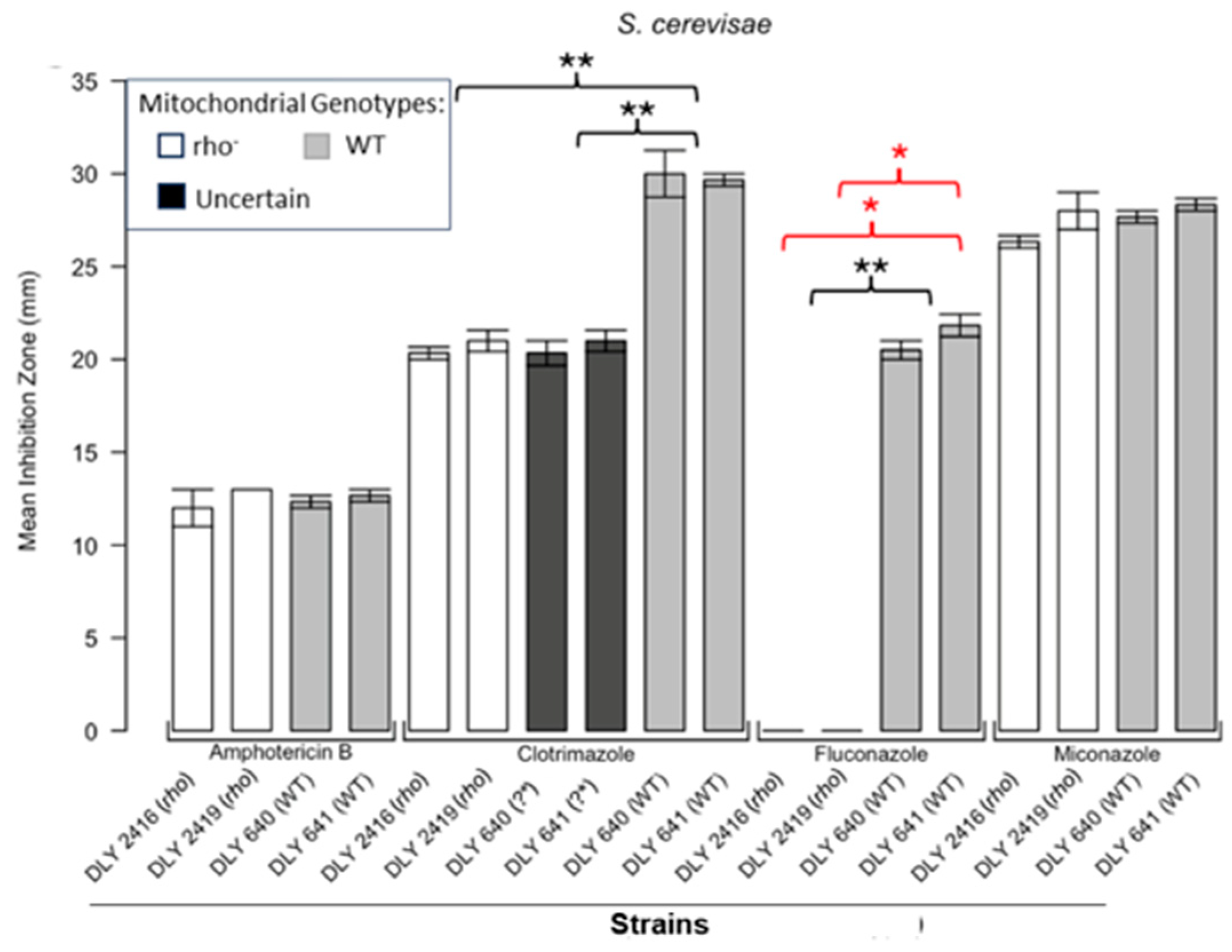
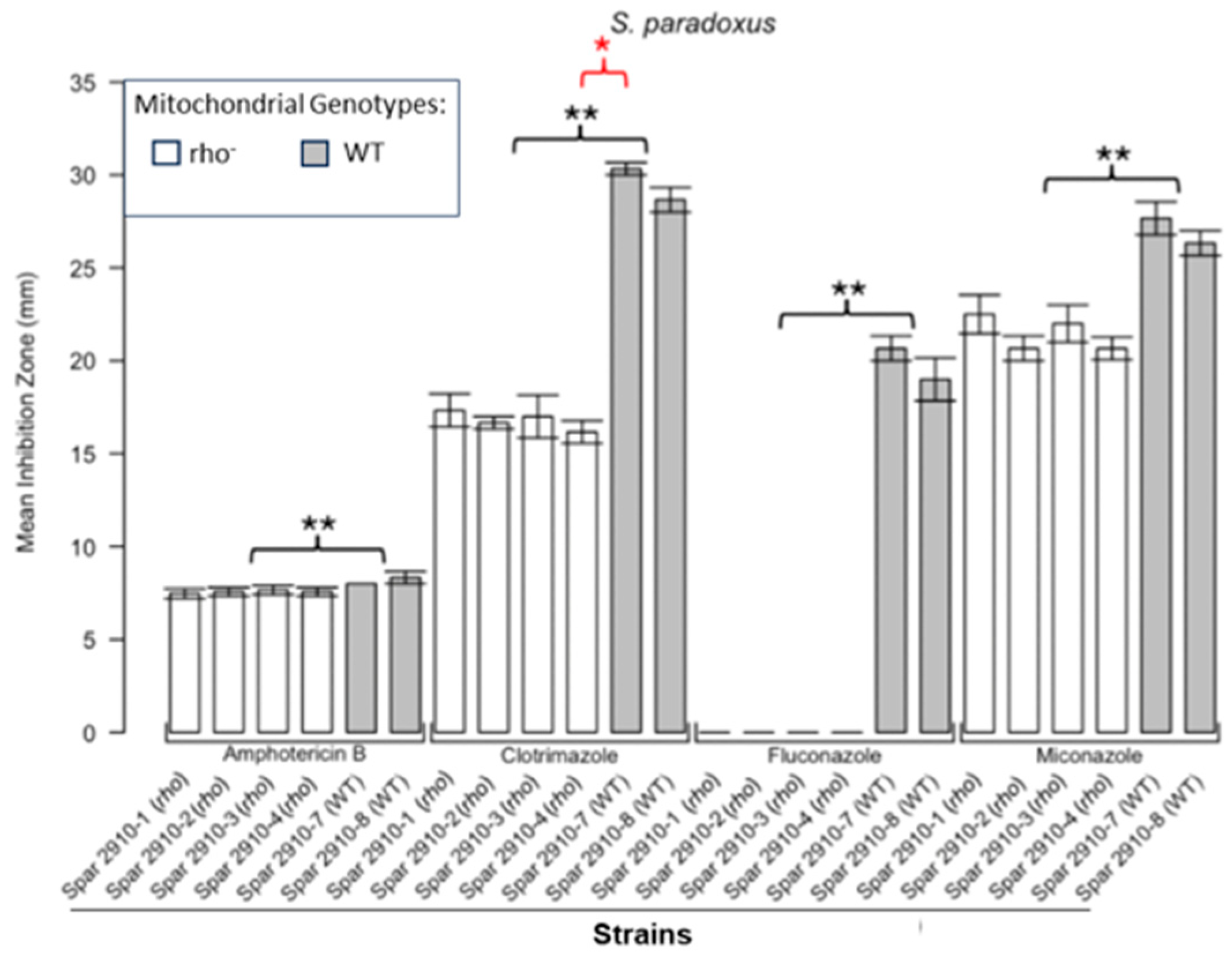



| Strain | Description/Genotype | Origin |
|---|---|---|
| Candida glabrata | ||
| NCYC 3303 | the strain deposited in 2005 by C. Bond (NCYC) {91} # | NCYC * |
| Cg 3303-2 | rho mutant induced with ethidium bromide {92} | ** |
| Cg 3303-7 | an individual wild-type clone (respiratory proficient) {97} | ** |
| Cg 3303-8 | an individual wild-type clone (respiratory proficient) {98} | ** |
| Saccharomyces bayanus | ||
| NCYC 106 | the strain deposited in 1920 by A. Klocker (Carlsberg Laboratories, Denmark) as Saccharomyces willianus | NCYC |
| Sb106-1 | spontaneous rho mutant of NCYC 106 {21} | ** |
| Sb106-3 | an individual wild-type clone of NCYC 106 (respiratory proficient) {29} | ** |
| Sb106-4 | an individual wild-type clone of NCYC 106 (respiratory proficient) {30} | ** |
| Sb106-8 | rho mutant induced with ethidium bromide {27} | ** |
| S. cerevisiae | ||
| DLY 640 | MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 GAL + psi+ ssd1-d2 RAD5 (wild type, W303 background, from R. Rothstein) {42} | D. Lydall |
| DLY 641 | MATalpha ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 GAL+psi+ ssd1-d2 RAD5 (wild type, W303 background, from R. Rothstein) {40} | D. Lydall |
| DLY 2416 | respiratory-deficient mutant generated by treatment of DLY 640 with ethidium bromide and proved to be a rho mutant {43} | ** |
| DLY 2419 | respiratory-deficient mutant generated by the treatment of DLY 641 with ethidium bromide and proved to be a rho mutant {41} | ** |
| S. paradoxus | ||
| NCYC 2910 | the strain deposited in 1999 by Ed Louis (University of Oxford, UK) | |
| Spar 2910-1 | rho mutant induced with ethidium bromide {115} | ** |
| Spar 2910-2 | rho mutant induced with ethidium bromide {116} | ** |
| Spar 2910-3 | rho mutant induced with ethidium bromide {117} | ** |
| Spar 2910-4 | rho mutant induced with ethidium bromide {118} | ** |
| Spar 2910-7 | an individual wild-type clone (respiratory proficient) {121} | ** |
| Spar 2910-8 | an individual wild-type clone (respiratory proficient) {122} | ** |
| S. pastorianus | ||
| NCYC 75 | the strain deposited in 1920 by A. Klocker (Carlsberg Laboratories, Denmark) as Saccharomyces carlsbergensis | NCYC |
| Sp75-1 | spontaneous rho mutant of NCYC 75 {1} | ** |
| Sp75-2 | spontaneous rho mutant of NCYC 75 {2} | ** |
| Sp75-3 | an individual wild-type clone of NCYC 75 (respiratory proficient) {7} | ** |
| Sp75-4 | an individual wild-type clone of NCYC 75 (respiratory proficient) {8} | ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Johnson, Z.; Nadim, F.; Zubko, M.K. Enhanced Tolerance to Antifungals as a General Feature of Rho− Mutants in Yeast Species: Implications to Positive Selection of Respiratory Deficiency. Microorganisms 2025, 13, 99. https://doi.org/10.3390/microorganisms13010099
Johnson Z, Nadim F, Zubko MK. Enhanced Tolerance to Antifungals as a General Feature of Rho− Mutants in Yeast Species: Implications to Positive Selection of Respiratory Deficiency. Microorganisms. 2025; 13(1):99. https://doi.org/10.3390/microorganisms13010099
Chicago/Turabian StyleJohnson, Zachary, Farhan Nadim, and Mikhajlo K. Zubko. 2025. "Enhanced Tolerance to Antifungals as a General Feature of Rho− Mutants in Yeast Species: Implications to Positive Selection of Respiratory Deficiency" Microorganisms 13, no. 1: 99. https://doi.org/10.3390/microorganisms13010099
APA StyleJohnson, Z., Nadim, F., & Zubko, M. K. (2025). Enhanced Tolerance to Antifungals as a General Feature of Rho− Mutants in Yeast Species: Implications to Positive Selection of Respiratory Deficiency. Microorganisms, 13(1), 99. https://doi.org/10.3390/microorganisms13010099






