Mycobacteria Exploit Host GPR84 to Dampen Pro-Inflammatory Responses and Promote Infection in Macrophages
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Blood Samples from TB Patients
2.2. Bacteria Strain
2.3. Cell Culture
2.4. Infection of Macrophages
2.5. qRT-PCR Detection of GPR84 mRNA Expression
2.6. Animals
2.7. Mouse Infected with Mm-Wasabi, Histopathological Analysis, Nile-Red Staining, and Acid-Fast Staining
2.7.1. Mouse Infected with Mm-Wasabi
2.7.2. Histopathological Analysis
2.7.3. Nile Red Staining
2.7.4. Immunofluorescence Assay
2.8. Immunofluorescence Microscopy Imaging
2.9. Flow Cytometry
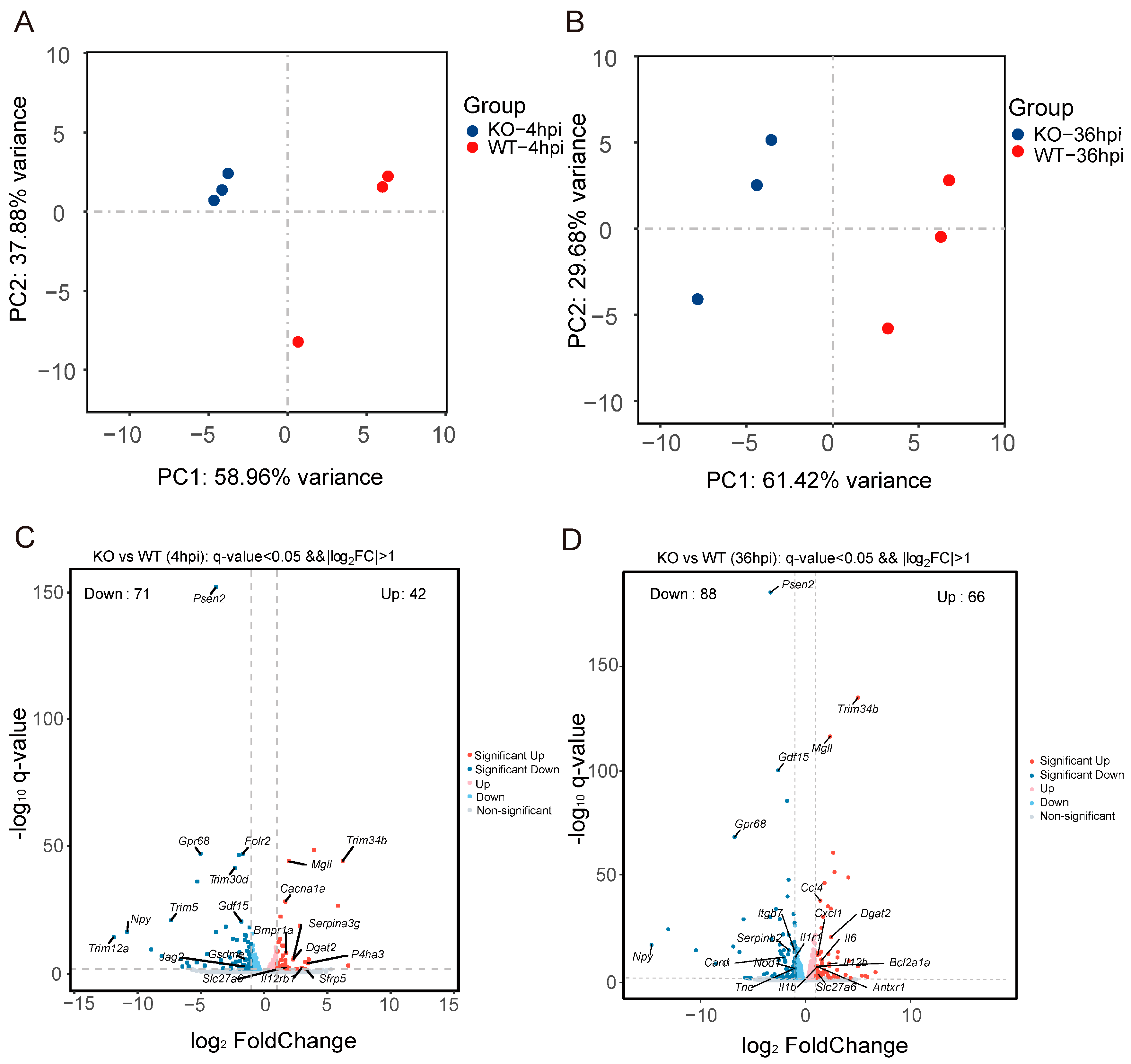
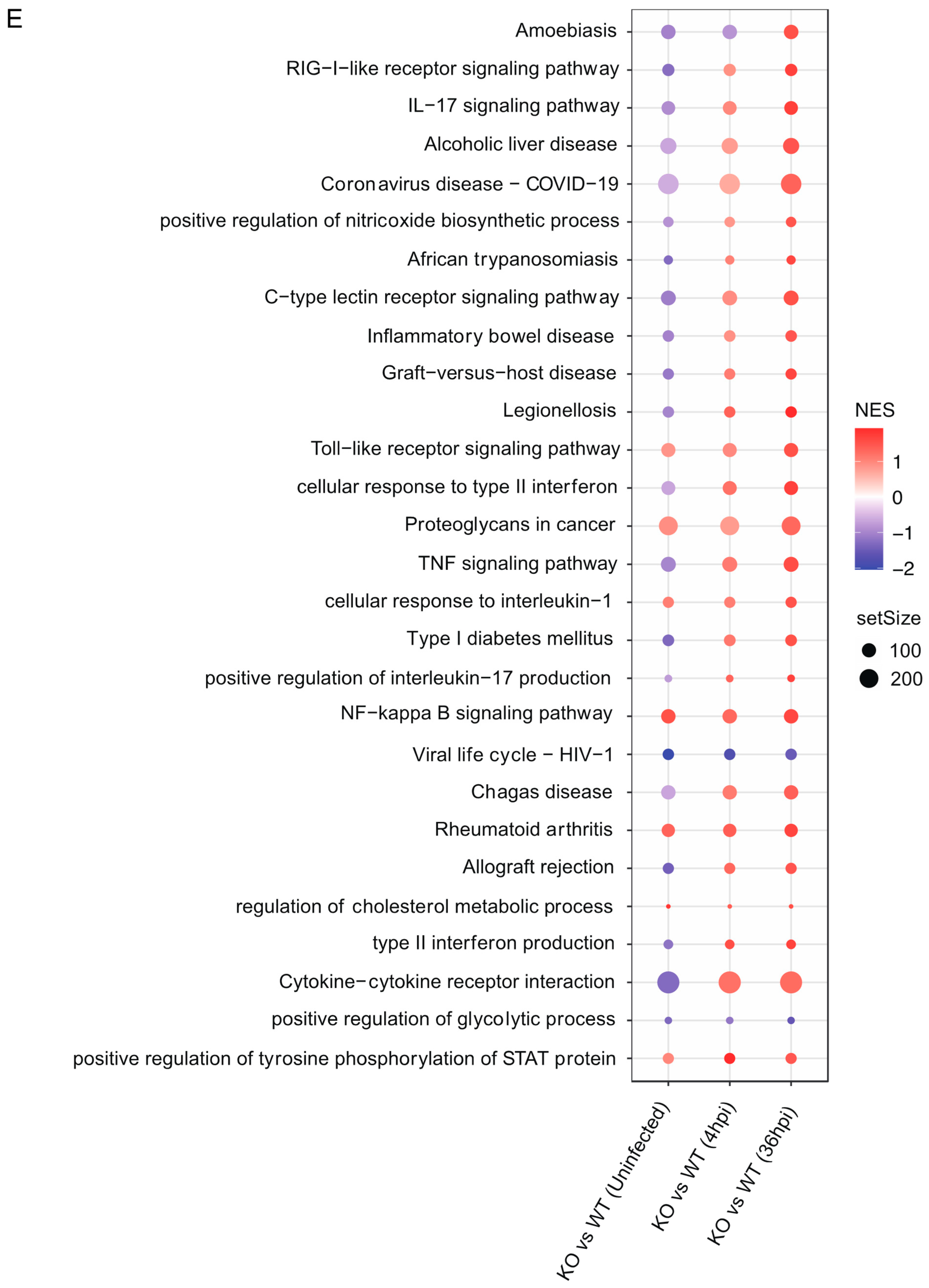
2.10. RNA Sequencing and Transcriptome Analysis
2.11. Data Analysis
3. Results
3.1. Mycobacterial Infection Induces Upregulation of GPR84 Expression
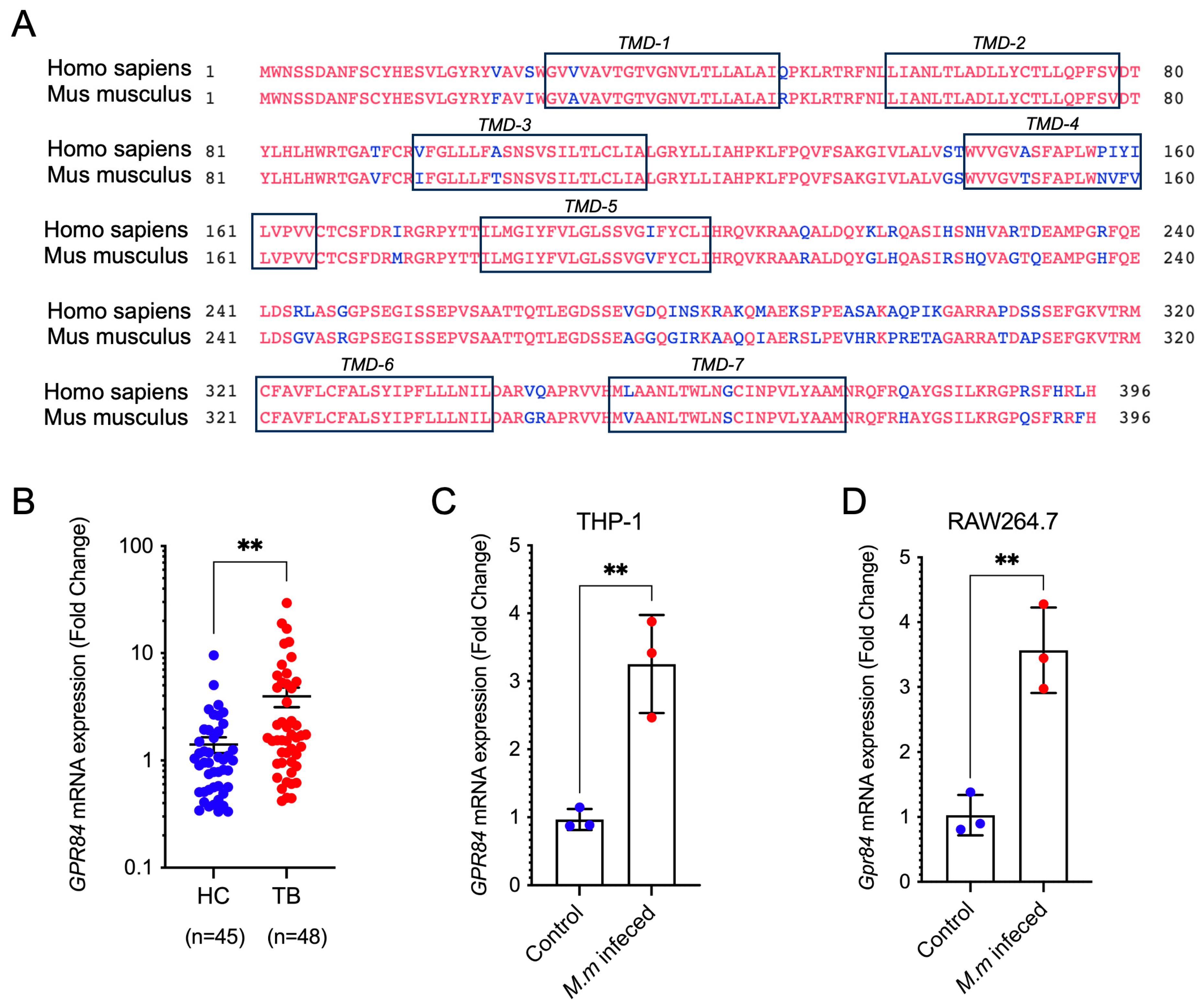
3.2. GPR84 Aggravates Mycobacterium-Induced Tissue Damage
3.3. GPR84 Promotes Intracellular Proliferation of Mm-Wasabi and LD Accumulation
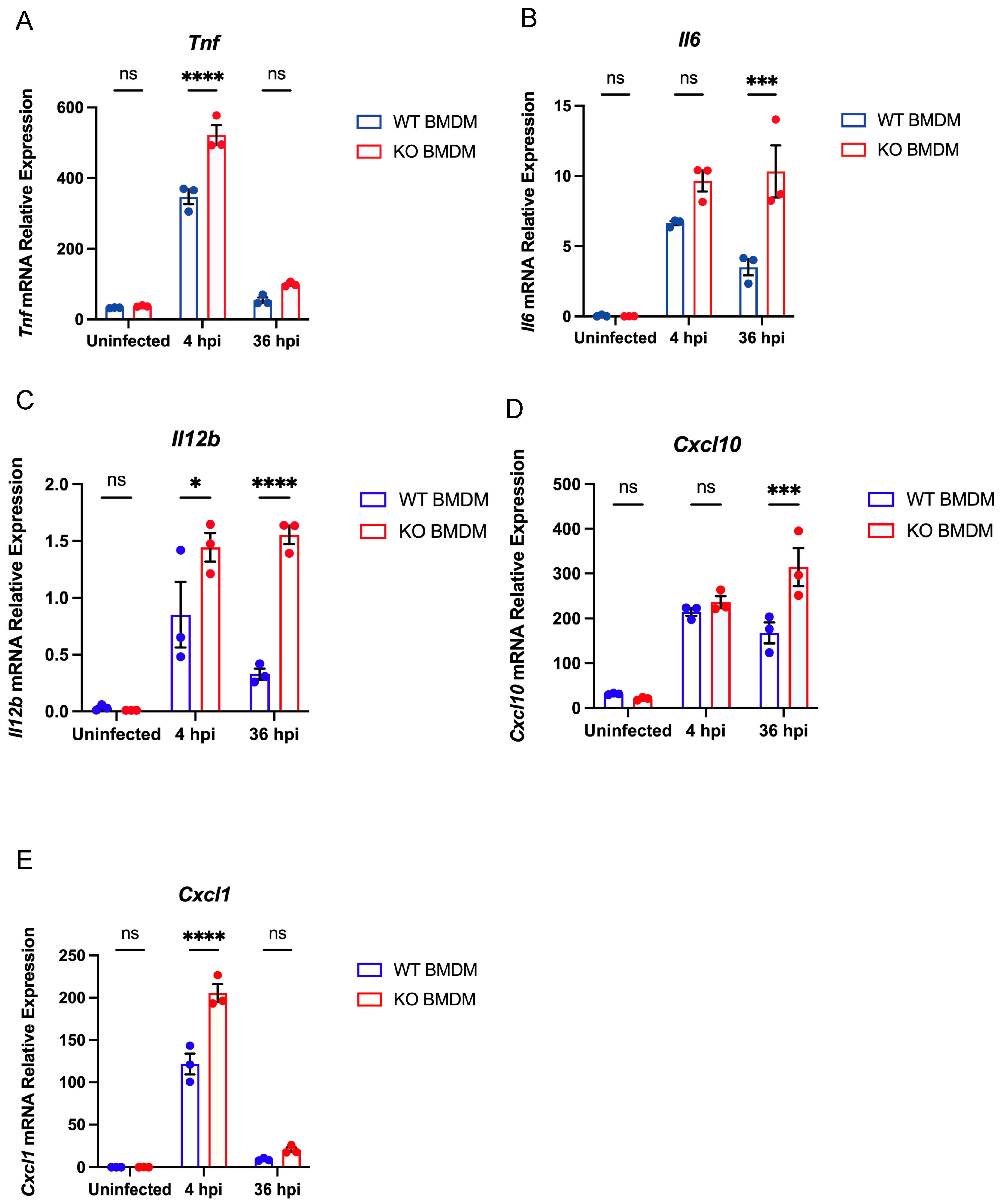
3.4. Gpr84−/− BMDMs Promotes the Expression of Pro-Inflammatory Cytokine
4. Discussion
5. Limitations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Global Tuberculosis Report 2024; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Kumar, A.; Rani, M.; Ehtesham, N.Z.; Hasnain, S.E. Commentary: Modification of Host Responses by Mycobacteria. Front. Immunol. 2017, 8, 466. [Google Scholar] [CrossRef] [PubMed]
- Young, C.; Walzl, G.; Du Plessis, N. Therapeutic host-directed strategies to improve outcome in tuberculosis. Mucosal Immunol. 2020, 13, 190–204. [Google Scholar] [CrossRef] [PubMed]
- Baskakova, K.O.; Kuzmichev, P.K.; Karbyshev, M.S. Advanced applications of Nanodiscs-based platforms for antibodies discovery. Biophys. Chem. 2024, 313, 107290. [Google Scholar] [CrossRef] [PubMed]
- Gulezian, E.; Crivello, C.; Bednenko, J.; Zafra, C.; Zhang, Y.; Colussi, P.; Hussain, S. Membrane protein production and formulation for drug discovery. Trends Pharmacol. Sci. 2021, 42, 657–674. [Google Scholar] [CrossRef]
- Das, S.; Banerjee, S.; Majumder, S.; Chowdhury, B.P.; Goswami, A.; Halder, K.; Chakraborty, U.; Pal, N.K.; Majumdar, S. Immune subversion by Mycobacterium tuberculosis through CCR5 mediated signaling: Involvement of IL-10. PLoS ONE 2014, 9, e92477. [Google Scholar] [CrossRef]
- Singh, V.; Jamwal, S.; Jain, R.; Verma, P.; Gokhale, R.; Rao, K.V. Mycobacterium tuberculosis-driven targeted recalibration of macrophage lipid homeostasis promotes the foamy phenotype. Cell Host Microbe 2012, 12, 669–681. [Google Scholar] [CrossRef]
- Yang, H.; Liu, H.; Chen, H.; Mo, H.; Chen, J.; Huang, X.; Zheng, R.; Liu, Z.; Feng, Y.; Liu, F.; et al. G protein-coupled receptor160 regulates mycobacteria entry into macrophages by activating ERK. Cell Signal 2016, 28, 1145–1151. [Google Scholar] [CrossRef]
- Bartlett, S.; Gemiarto, A.T.; Ngo, M.D.; Sajiir, H.; Hailu, S.; Sinha, R.; Foo, C.X.; Kleynhans, L.; Tshivhula, H.; Webber, T.; et al. GPR183 Regulates Interferons, Autophagy, and Bacterial Growth During Mycobacterium tuberculosis Infection and Is Associated With TB Disease Severity. Front. Immunol. 2020, 11, 601534. [Google Scholar] [CrossRef]
- Stanley, S.A.; Barczak, A.K.; Silvis, M.R.; Luo, S.S.; Sogi, K.; Vokes, M.; Bray, M.A.; Carpenter, A.E.; Moore, C.B.; Siddiqi, N.; et al. Identification of host-targeted small molecules that restrict intracellular Mycobacterium tuberculosis growth. PLoS Pathog. 2014, 10, e1003946. [Google Scholar] [CrossRef]
- Yousefi, S.; Cooper, P.R.; Potter, S.L.; Mueck, B.; Jarai, G. Cloning and expression analysis of a novel G-protein-coupled receptor selectively expressed on granulocytes. J. Leukoc. Biol. 2001, 69, 1045–1052. [Google Scholar] [CrossRef]
- Wang, J.; Wu, X.; Simonavicius, N.; Tian, H.; Ling, L. Medium-chain fatty acids as ligands for orphan G protein-coupled receptor GPR84. J. Biol. Chem. 2006, 281, 34457–34464. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chen, L.H.; Yang, H.; Fang, Y.C.; Wang, S.W.; Wang, M.; Yuan, Q.T.; Wu, W.; Zhang, Y.M.; Liu, Z.J.; et al. GPR84 signaling promotes intestinal mucosal inflammation via enhancing NLRP3 inflammasome activation in macrophages. Acta Pharmacol. Sin. 2022, 43, 2042–2054. [Google Scholar] [CrossRef]
- Yin, C.; Cheng, L.; Pan, J.; Chen, L.; Xue, Q.; Qin, J.; Wang, S.; Du, B.; Liu, M.; Zhang, Y.; et al. Regulatory role of Gpr84 in the switch of alveolar macrophages from CD11b(lo) to CD11b(hi) status during lung injury process. Mucosal Immunol. 2020, 13, 892–907. [Google Scholar] [CrossRef] [PubMed]
- Recio, C.; Lucy, D.; Purvis, G.S.D.; Iveson, P.; Zeboudj, L.; Iqbal, A.J.; Lin, D.; O’Callaghan, C.; Davison, L.; Griesbach, E.; et al. Activation of the Immune-Metabolic Receptor GPR84 Enhances Inflammation and Phagocytosis in Macrophages. Front. Immunol. 2018, 9, 1419. [Google Scholar] [CrossRef] [PubMed]
- Reyes, A.W.B.; Kim, H.; Huy, T.X.N.; Vu, S.H.; Nguyen, T.T.; Kang, C.K.; Min, W.; Lee, H.J.; Lee, J.H.; Kim, S. Immune-metabolic receptor GPR84 surrogate and endogenous agonists, 6-OAU and lauric acid, alter Brucella abortus 544 infection in both in vitro and in vivo systems. Microb. Pathog. 2021, 158, 105079. [Google Scholar] [CrossRef]
- Torraca, V.; White, R.J.; Sealy, I.M.; Mazon-Moya, M.; Duggan, G.; Willis, A.R.; Busch-Nentwich, E.M.; Mostowy, S. Transcriptional profiling of zebrafish identifies host factors controlling susceptibility to Shigella flexneri. Dis. Model. Mech. 2024, 17, dmm050431. [Google Scholar] [CrossRef]
- Didangelos, A. COVID-19 Hyperinflammation: What about Neutrophils? mSphere 2020, 5, e00367-20. [Google Scholar] [CrossRef]
- Ding, Y.; Bei, C.; Xue, Q.; Niu, L.; Tong, J.; Chen, Y.; Takiff, H.E.; Gao, Q.; Yan, B. Transcriptomic Analysis of Mycobacterial Infected Macrophages Reveals a High MOI Specific Type I IFN Signaling. Infect. Immun. 2023, 91, e0015523. [Google Scholar] [CrossRef]
- Sadee, W.; Cheeseman, I.H.; Papp, A.; Pietrzak, M.; Seweryn, M.; Zhou, X.; Lin, S.; Williams, A.M.; Wewers, M.D.; Curry, H.M.; et al. Human alveolar macrophage response to Mycobacterium tuberculosis: Immune characteristics underlying large inter-individual variability. bioRxiv 2022. [Google Scholar] [CrossRef]
- Takaki, K.; Davis, J.M.; Winglee, K.; Ramakrishnan, L. Evaluation of the pathogenesis and treatment of Mycobacterium marinum infection in zebrafish. Nat. Protoc. 2013, 8, 1114–1124. [Google Scholar] [CrossRef]
- Kam, J.Y.; Cheng, T.; Garland, D.C.; Britton, W.J.; Tobin, D.M.; Oehlers, S.H. Inhibition of infection-induced vascular permeability modulates host leukocyte recruitment to Mycobacterium marinum granulomas in zebrafish. Pathog. Dis. 2022, 80, ftac009. [Google Scholar] [CrossRef] [PubMed]
- Mittal, E.; Roth, A.T.; Seth, A.; Singamaneni, S.; Beatty, W.; Philips, J.A. Single cell preparations of Mycobacterium tuberculosis damage the mycobacterial envelope and disrupt macrophage interactions. eLife 2023, 12, e85416. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Tong, J.; Luo, G.; Sun, R.; Bei, C.; Feng, Z.; Meng, L.; Wang, F.; Zhou, J.; Chen, Z.; et al. Mycobacterial CpsA activates type I IFN signaling in macrophages via cGAS-mediated pathway. iScience 2024, 27, 109807. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Meng, L.; Bei, C.; Liu, Q.; Wang, M.; Yang, T.; Takiff, H.E.; Zhang, S.; Gao, Q.; Wang, C.; et al. Modern Beijing sublineage of Mycobacterium tuberculosis shift macrophage into a hyperinflammatory status. Emerg. Microbes Infect. 2022, 11, 715–724. [Google Scholar] [CrossRef]
- Wang, Q.; Zhu, L.; Jones, V.; Wang, C.; Hua, Y.; Shi, X.; Feng, X.; Jackson, M.; Niu, C.; Gao, Q. CpsA, a LytR-CpsA-Psr Family Protein in Mycobacterium marinum, Is Required for Cell Wall Integrity and Virulence. Infect. Immun. 2015, 83, 2844–2854. [Google Scholar] [CrossRef]
- Wang, S.; Chen, L.L.; Yin, C.C.; Du, B.; Qian, M.; Ren, H. Establishment of Gpr84 knockout mice and its effect on secretion of TNF-α of macrophage. Highlights Sci. Online 2017, 10, 242–247. [Google Scholar]
- Kamber, R.A.; Nishiga, Y.; Morton, B.; Banuelos, A.M.; Barkal, A.A.; Vences-Catalan, F.; Gu, M.; Fernandez, D.; Seoane, J.A.; Yao, D.; et al. Inter-cellular CRISPR screens reveal regulators of cancer cell phagocytosis. Nature 2021, 597, 549–554. [Google Scholar] [CrossRef]
- Greenspan, P.; Mayer, E.P.; Fowler, S.D. Nile red: A selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 1985, 100, 965–973. [Google Scholar] [CrossRef]
- Sun, W.; Zhang, X.; Qiao, Y.; Griffin, N.; Zhang, H.; Wang, L.; Liu, H. Exposure to PFOA and its novel analogs disrupts lipid metabolism in zebrafish. Ecotoxicol. Environ. Saf. 2023, 259, 115020. [Google Scholar] [CrossRef]
- Pedrosa, L.F.; Kouzounis, D.; Schols, H.; de Vos, P.; Fabi, J.P. Assessing high-temperature and pressure extraction of bioactive water-soluble polysaccharides from passion fruit mesocarp. Carbohydr. Polym. 2024, 335, 122010. [Google Scholar] [CrossRef]
- Listenberger, L.L.; Studer, A.M.; Brown, D.A.; Wolins, N.E. Fluorescent Detection of Lipid Droplets and Associated Proteins. Curr. Protoc. Cell Biol. 2016, 71, 4–31. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- The Gene Ontology, C. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Huang, Q.; Feng, D.; Liu, K.; Wang, P.; Xiao, H.; Wang, Y.; Zhang, S.; Liu, Z. A medium-chain fatty acid receptor Gpr84 in zebrafish: Expression pattern and roles in immune regulation. Dev. Comp. Immunol. 2014, 45, 252–258. [Google Scholar] [CrossRef]
- Krampert, M.; Kuenzle, S.; Thai, S.N.; Lee, N.; Iruela-Arispe, M.L.; Werner, S. ADAMTS1 proteinase is up-regulated in wounded skin and regulates migration of fibroblasts and endothelial cells. J. Biol. Chem. 2005, 280, 23844–23852. [Google Scholar] [CrossRef]
- Martens, C.R.; Dorn, L.E.; Kenney, A.D.; Bansal, S.S.; Yount, J.S.; Accornero, F. BEX1 is a critical determinant of viral myocarditis. PLoS Pathog. 2022, 18, e1010342. [Google Scholar] [CrossRef]
- Ledoult, E.; Jendoubi, M.; Collet, A.; Guerrier, T.; Largy, A.; Speca, S.; Vivier, S.; Bray, F.; Figeac, M.; Hachulla, E.; et al. Simple gene signature to assess murine fibroblast polarization. Sci. Rep. 2022, 12, 11748. [Google Scholar] [CrossRef]
- Collison, A.; Hatchwell, L.; Verrills, N.; Wark, P.A.; de Siqueira, A.P.; Tooze, M.; Carpenter, H.; Don, A.S.; Morris, J.C.; Zimmermann, N.; et al. The E3 ubiquitin ligase midline 1 promotes allergen and rhinovirus-induced asthma by inhibiting protein phosphatase 2A activity. Nat. Med. 2013, 19, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Woo, K.V.; Qu, X.; Babaev, V.R.; Linton, M.F.; Guzman, R.J.; Fazio, S.; Baldwin, H.S. Tie1 attenuation reduces murine atherosclerosis in a dose-dependent and shear stress-specific manner. J. Clin. Investig. 2011, 121, 1624–1635. [Google Scholar] [CrossRef] [PubMed]
- Fader Kaiser, C.M.; Romano, P.S.; Vanrell, M.C.; Pocognoni, C.A.; Jacob, J.; Caruso, B.; Delgui, L.R. Biogenesis and Breakdown of Lipid Droplets in Pathological Conditions. Front. Cell Dev. Biol. 2021, 9, 826248. [Google Scholar] [CrossRef]
- Jin, Y.; McFie, P.J.; Banman, S.L.; Brandt, C.; Stone, S.J. Diacylglycerol acyltransferase-2 (DGAT2) and monoacylglycerol acyltransferase-2 (MGAT2) interact to promote triacylglycerol synthesis. J. Biol. Chem. 2014, 289, 28237–28248. [Google Scholar] [CrossRef]
- Cheng, Y.C.; Acedera, J.D.; Li, Y.J.; Shieh, S.Y. A keratinocyte-adipocyte signaling loop is reprogrammed by loss of BTG3 to augment skin carcinogenesis. Cell Death Differ. 2024, 31, 970–982. [Google Scholar] [CrossRef]
- Li, Y.; Chen, L.; Sottas, C.; Raul, M.C.; Patel, N.D.; Bijja, J.R.; Ahmed, S.K.; Kapelanski-Lamoureux, A.; Lazaris, A.; Metrakos, P.; et al. The mitochondrial TSPO ligand Atriol mitigates metabolic-associated steatohepatitis by downregulating CXCL1. Metabolism 2024, 159, 155942. [Google Scholar] [CrossRef]
- Okuda, J.; Arikawa, Y.; Takeuchi, Y.; Mahmoud, M.M.; Suzaki, E.; Kataoka, K.; Suzuki, T.; Okinaka, Y.; Nakai, T. Intracellular replication of Edwardsiella tarda in murine macrophage is dependent on the type III secretion system and induces an up-regulation of anti-apoptotic NF-kappaB target genes protecting the macrophage from staurosporine-induced apoptosis. Microb. Pathog. 2006, 41, 226–240. [Google Scholar] [CrossRef]
- Mikkelsen, R.B.; Arora, T.; Trost, K.; Dmytriyeva, O.; Jensen, S.K.; Meijnikman, A.S.; Olofsson, L.E.; Lappa, D.; Aydin, O.; Nielsen, J.; et al. Type 2 diabetes is associated with increased circulating levels of 3-hydroxydecanoate activating GPR84 and neutrophil migration. iScience 2022, 25, 105683. [Google Scholar] [CrossRef]
- Marsango, S.; Milligan, G. Regulation of the pro-inflammatory G protein-coupled receptor GPR84. Br. J. Pharmacol. 2024, 181, 1500–1508. [Google Scholar] [CrossRef]
- Wang, S.W.; Zhang, Q.; Lu, D.; Fang, Y.C.; Yan, X.C.; Chen, J.; Xia, Z.K.; Yuan, Q.T.; Chen, L.H.; Zhang, Y.M.; et al. GPR84 regulates pulmonary inflammation by modulating neutrophil functions. Acta Pharmacol. Sin. 2023, 44, 1665–1675. [Google Scholar] [CrossRef]
- Ohue-Kitano, R.; Nonaka, H.; Nishida, A.; Masujima, Y.; Takahashi, D.; Ikeda, T.; Uwamizu, A.; Tanaka, M.; Kohjima, M.; Igarashi, M.; et al. Medium-chain fatty acids suppress lipotoxicity-induced hepatic fibrosis via the immunomodulating receptor GPR84. JCI Insight 2023, 8, e165469. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Ma, L.; Ding, Y.; He, L.; Chang, M.; Shan, Y.; Siwko, S.; Chen, G.; Liu, Y.; Jin, Y.; et al. Fatty acid sensing GPCR (GPR84) signaling safeguards cartilage homeostasis and protects against osteoarthritis. Pharmacol. Res. 2021, 164, 105406. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Tokizane, K.; Konishi, H.; Yu, H.R.; Kiyama, H. Agonists for G-protein-coupled receptor 84 (GPR84) alter cellular morphology and motility but do not induce pro-inflammatory responses in microglia. J. Neuroinflamm. 2017, 14, 198. [Google Scholar] [CrossRef] [PubMed]
- Russell, D.G.; Huang, L.; VanderVen, B.C. Immunometabolism at the interface between macrophages and pathogens. Nat. Rev. Immunol. 2019, 19, 291–304. [Google Scholar] [CrossRef]
- Guerrini, V.; Gennaro, M.L. Foam Cells: One Size Doesn’t Fit All. Trends Immunol. 2019, 40, 1163–1179. [Google Scholar] [CrossRef]
- Guerrini, V.; Prideaux, B.; Blanc, L.; Bruiners, N.; Arrigucci, R.; Singh, S.; Ho-Liang, H.P.; Salamon, H.; Chen, P.Y.; Lakehal, K.; et al. Storage lipid studies in tuberculosis reveal that foam cell biogenesis is disease-specific. PLoS Pathog. 2018, 14, e1007223. [Google Scholar] [CrossRef]
- Ouimet, M.; Koster, S.; Sakowski, E.; Ramkhelawon, B.; van Solingen, C.; Oldebeken, S.; Karunakaran, D.; Portal-Celhay, C.; Sheedy, F.J.; Ray, T.D.; et al. Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism. Nat. Immunol. 2016, 17, 677–686. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, X.; Zhang, S.; Liu, Z. Zebrafish fatty acids receptor Gpr84 enhances macrophage phagocytosis. Fish. Shellfish. Immunol. 2019, 84, 1098–1099. [Google Scholar] [CrossRef]
- Li, C.; Wang, J.; Xu, J.F.; Pi, J.; Zheng, B. Roles of HIF-1alpha signaling in Mycobacterium tuberculosis infection: New targets for anti-TB therapeutics? Biochem. Biophys. Res. Commun. 2024, 711, 149920. [Google Scholar] [CrossRef]


| Gene Name | GeneBank (No.) | Forward and Reverse Primer Sequence (5′-3′) |
|---|---|---|
| Human GPR84 | MT536737.1 | F: TGAAGCCTAACTGTCCACCAG R: CCACATGATAGAGGCTGAGT |
| Human β-actin | PQ040393.1 | F: TCACCATGGATGATGATATCGC R: ATAGGAATCCTTCTGACCCATGC |
| Mouse Gpr84 | AF272948.1 | F: CCACCGCTTTTGCAAGGATGT R: AACGGTAGCCCTCAACAGAG |
| Mouse β-actin | BC138611.1 | F: GCCGGGACCTGACAGACTAC R: TGGCCA TCTCCTGCTCGAAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wumaier, R.; Zhang, K.; Zhou, J.; Wen, Z.; Chen, Z.; Luo, G.; Wang, H.; Qin, J.; Du, B.; Ren, H.; et al. Mycobacteria Exploit Host GPR84 to Dampen Pro-Inflammatory Responses and Promote Infection in Macrophages. Microorganisms 2025, 13, 110. https://doi.org/10.3390/microorganisms13010110
Wumaier R, Zhang K, Zhou J, Wen Z, Chen Z, Luo G, Wang H, Qin J, Du B, Ren H, et al. Mycobacteria Exploit Host GPR84 to Dampen Pro-Inflammatory Responses and Promote Infection in Macrophages. Microorganisms. 2025; 13(1):110. https://doi.org/10.3390/microorganisms13010110
Chicago/Turabian StyleWumaier, Reziya, Ke Zhang, Jing Zhou, Zilu Wen, Zihan Chen, Geyang Luo, Hao Wang, Juliang Qin, Bing Du, Hua Ren, and et al. 2025. "Mycobacteria Exploit Host GPR84 to Dampen Pro-Inflammatory Responses and Promote Infection in Macrophages" Microorganisms 13, no. 1: 110. https://doi.org/10.3390/microorganisms13010110
APA StyleWumaier, R., Zhang, K., Zhou, J., Wen, Z., Chen, Z., Luo, G., Wang, H., Qin, J., Du, B., Ren, H., Song, Y., Gao, Q., & Yan, B. (2025). Mycobacteria Exploit Host GPR84 to Dampen Pro-Inflammatory Responses and Promote Infection in Macrophages. Microorganisms, 13(1), 110. https://doi.org/10.3390/microorganisms13010110







