Revealing the Viable Microbial Community of Biofilm in a Sewage Treatment System Using Propidium Monoazide Combined with Real-Time PCR and Metagenomics
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Cultivation Conditions, and Preparation of Viable and Dead Cells
2.1.1. Bacterial Strains and Cultivation Conditions
2.1.2. Preparation of Viable and Dead Cells
2.2. Biological Filter Operation and Biofilm Sampling
2.2.1. Biological Filter Operation
2.2.2. Biofilm Sampling
2.3. PMA Condition Optimization
2.4. PMA-qPCR Detection of Different Ratios of Viable/Dead Bacteria
2.5. PMA-qPCR Standard Curve
2.6. DNA Extraction
2.7. Primer Specificity Verification and Establishment of qPCR Detection System
2.7.1. Primer Specificity Verification
- Forward (F): 5’-GCGGCGTTGGAGAGTGATA-3’,
- Reverse (R): 5’-AGCAATGGAAAAAGCAGGATG-3’.
2.7.2. Establishment of qPCR Detection System
2.8. PMA Treatment for Viable Biofilm Quantification
2.9. Metagenomic Sequencing and Analysis
2.10. Statistical Analysis
3. Results
3.1. Optimization of PMA Conditions
3.1.1. Optimization of PMA Concentration
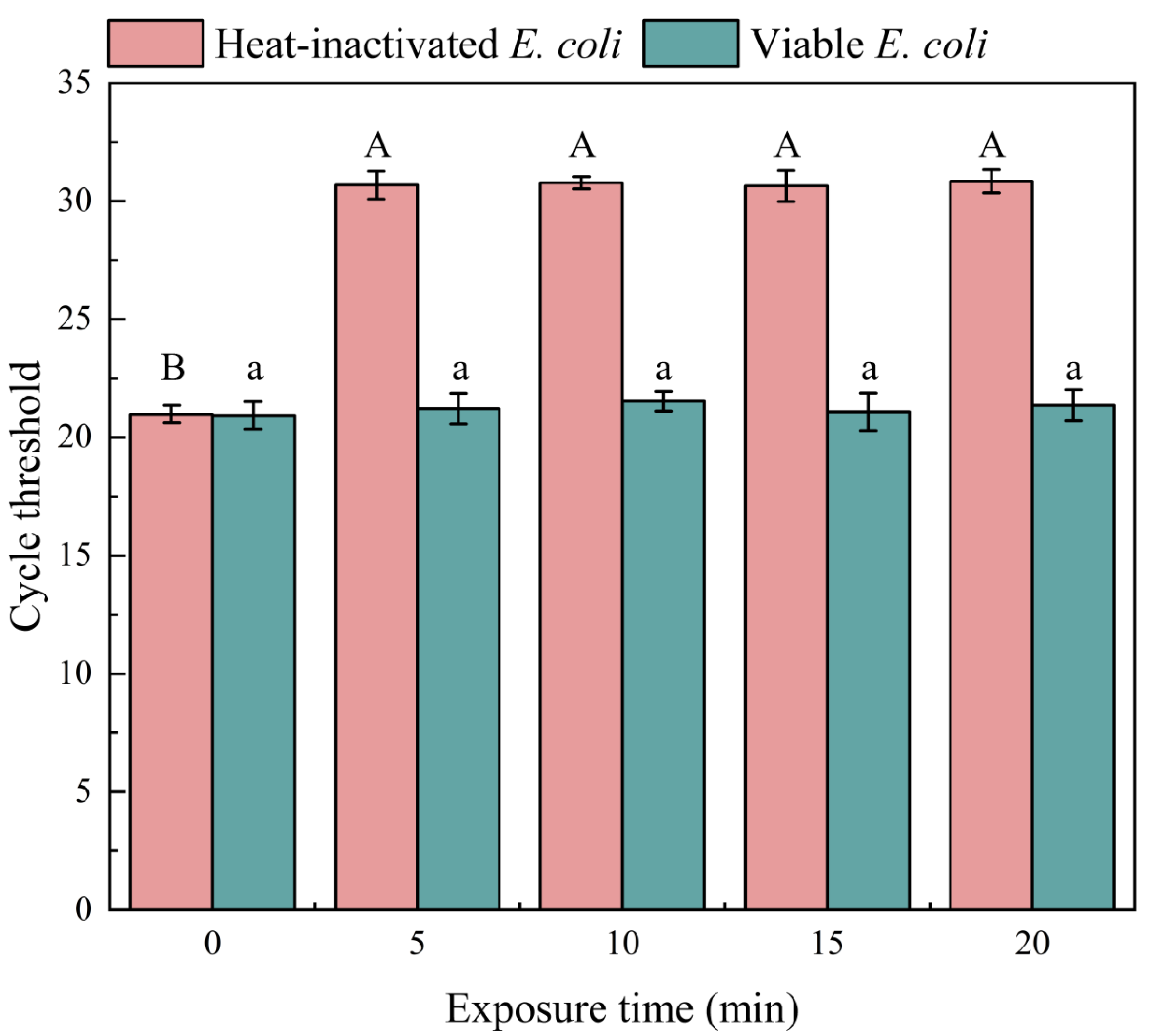
3.1.2. Optimization of Exposure Time
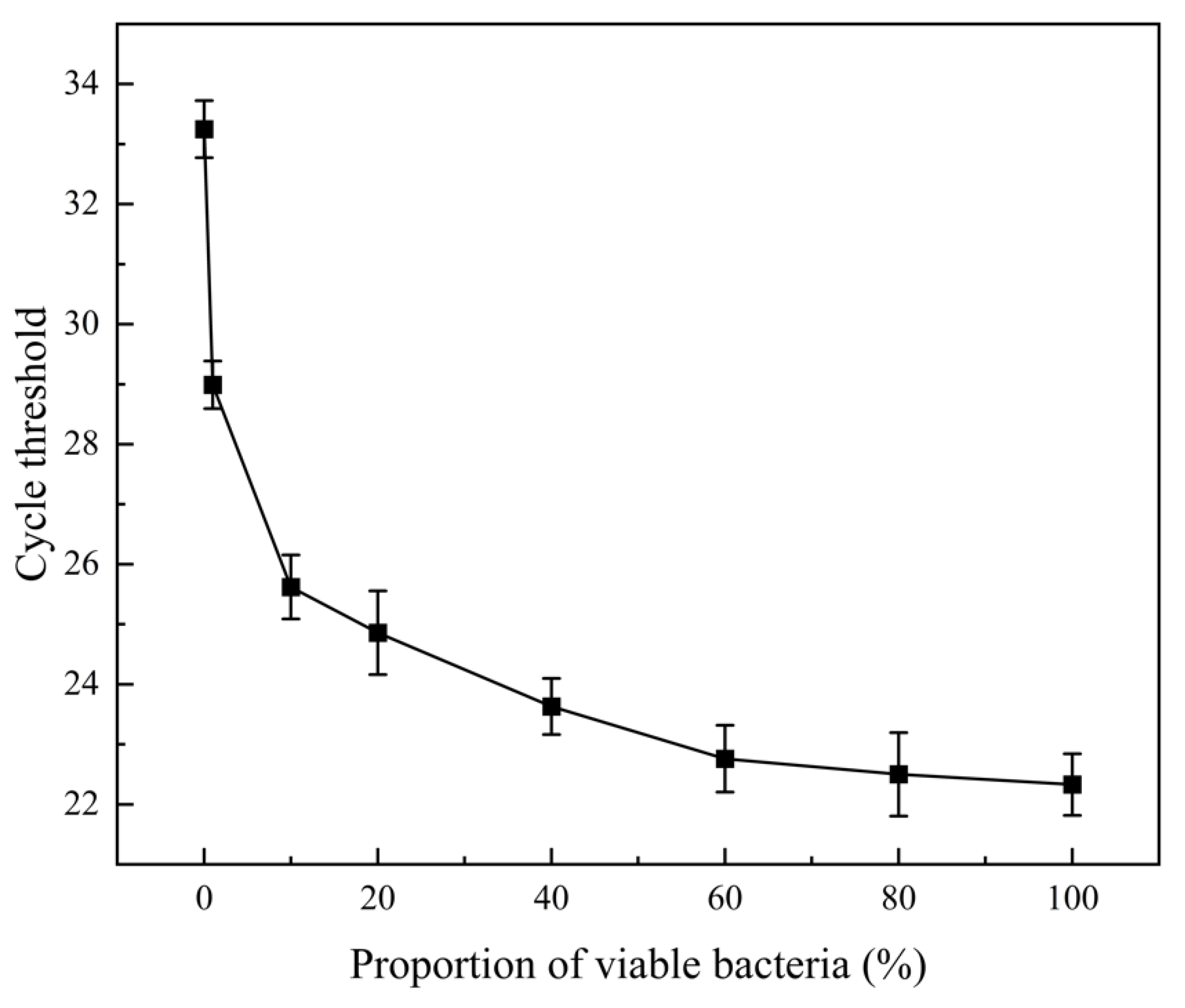
3.2. Selective Amplification of Living Cells by PMA-qPCR With Different Viable/Dead Bacterial Ratio Mixtures
3.3. Evaluation of PMA-qPCR Standard Curve and Detection Limit
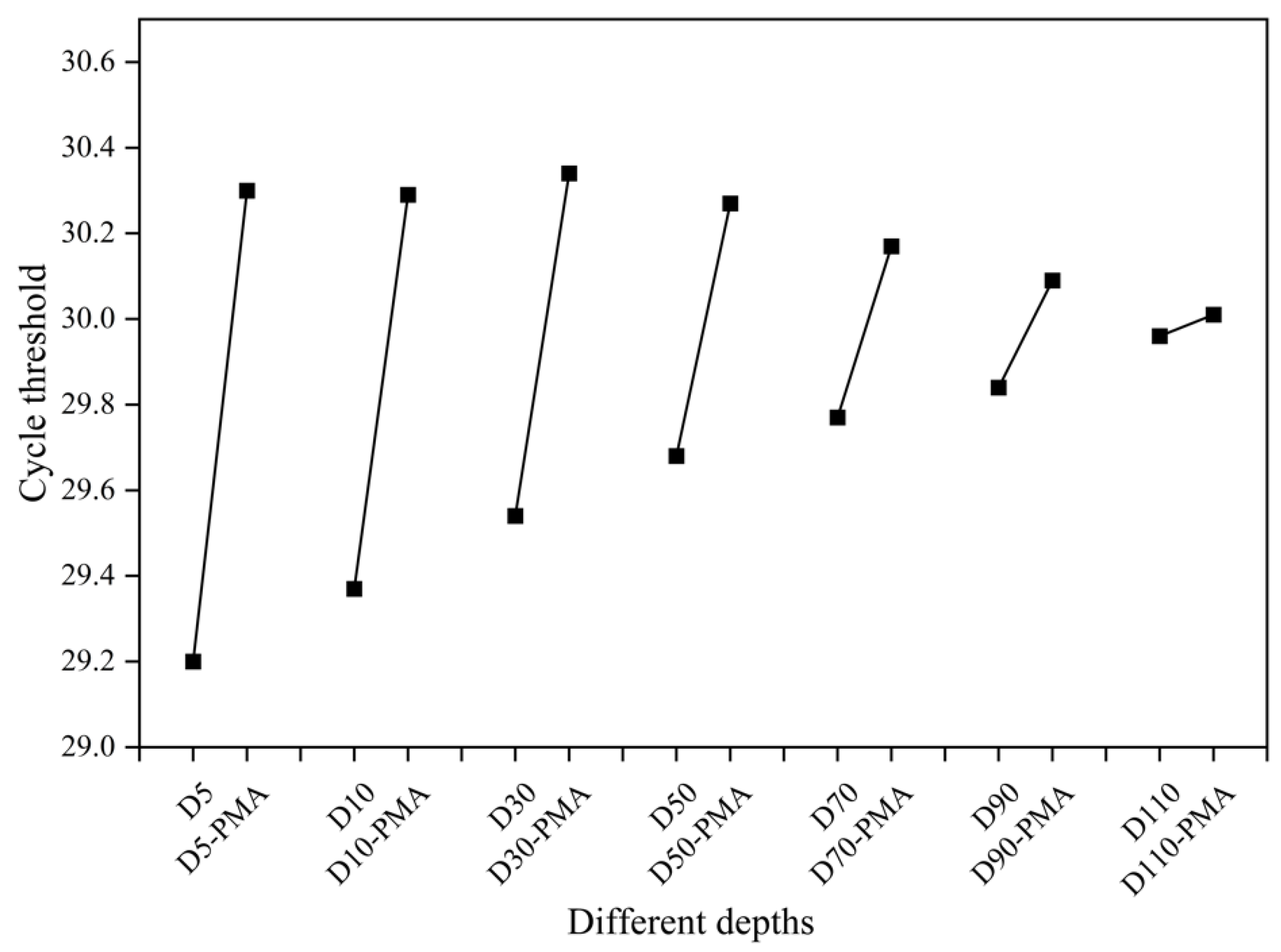
3.4. Detection of the Viability of Biofilm Samples
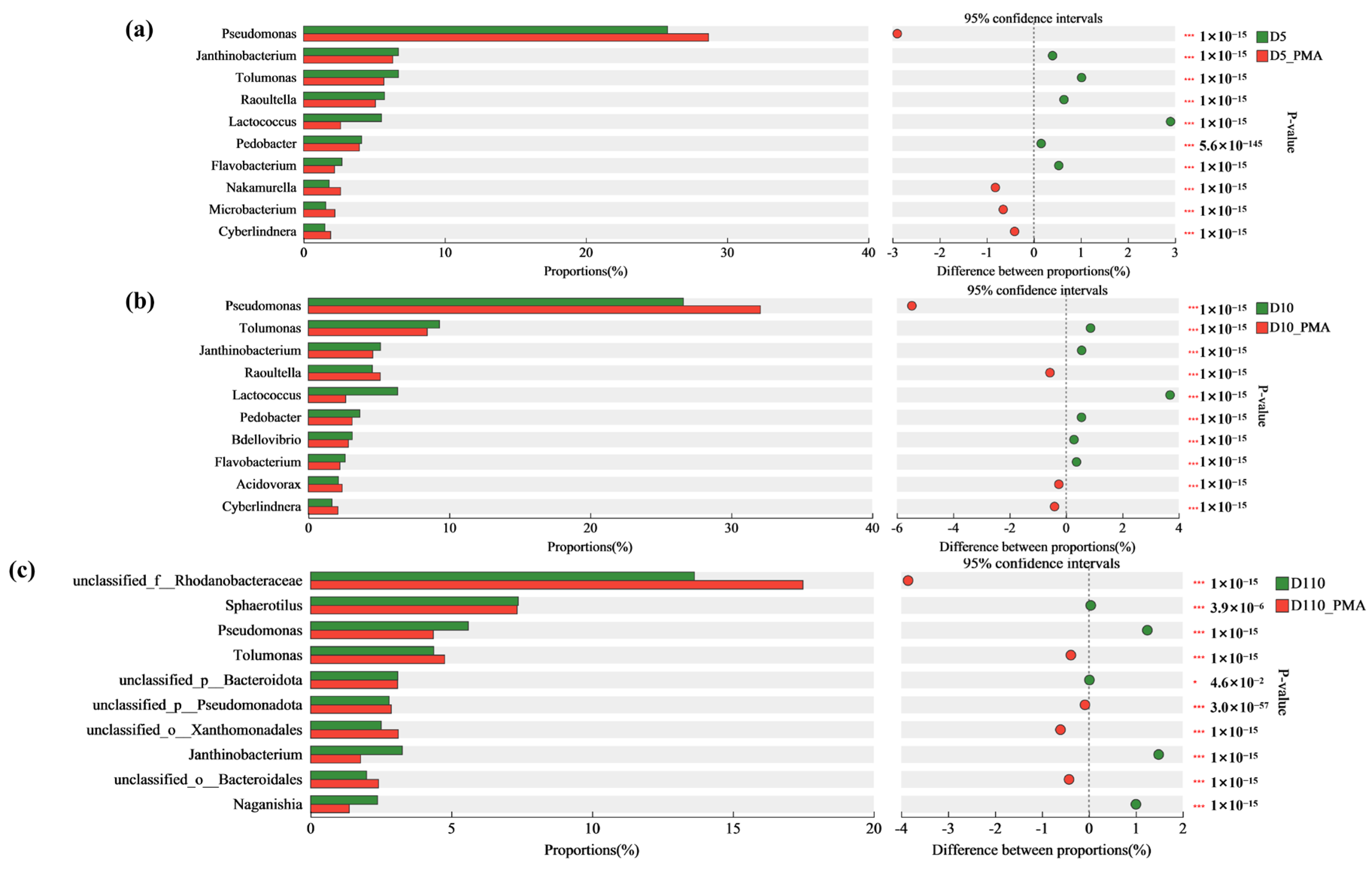
3.5. Microbial Community Structure and Function of Sewage Treatment Biofilm without and with PMA Treatment
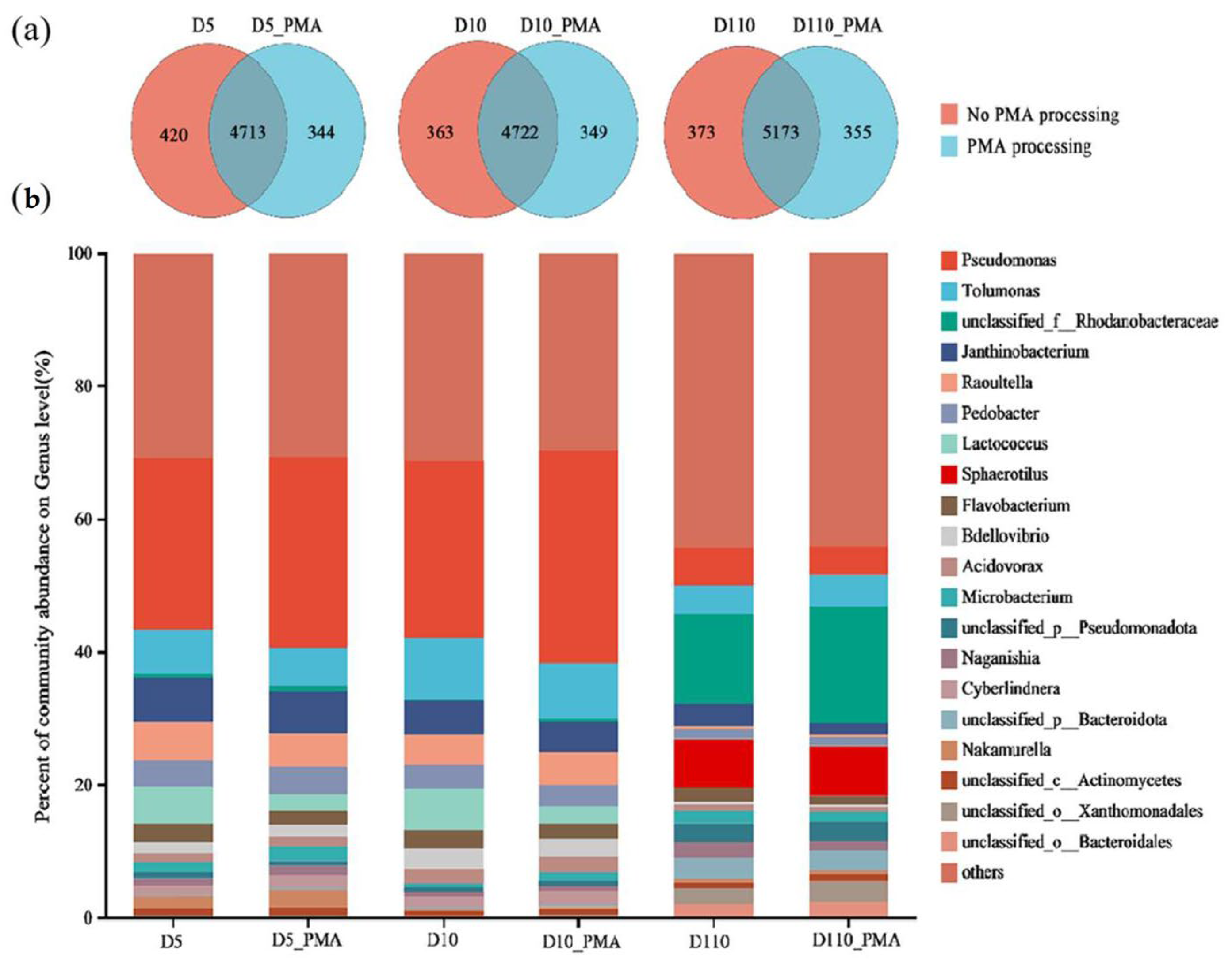
3.5.1. Microbial Community Structure of Sewage Treatment Biofilm without and with PMA Treatment
3.5.2. Microbial Community Function of Sewage Treatment Biofilm without and with PMA Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sanchez-Huerta, C.; Medina, J.S.; Wang, C.; Fortunato, L.; Hong, P.-Y. Understanding the role of sorption and biodegradation in the removal of organic micropollutants by membrane aerated biofilm reactor (MABR) with different biofilm thickness. Water Res. 2023, 236, 119935. [Google Scholar] [CrossRef]
- Liang, C.; de Jonge, N.; Carvalho, P.N.; Nielsen, J.L.; Bester, K. Biodegradation kinetics of organic micropollutants and microbial community dynamics in a moving bed biofilm reactor. Chem. Eng. J. 2021, 415, 128963. [Google Scholar] [CrossRef]
- Nelson, M.J.; Nakhla, G.; Zhu, J. Fluidized-Bed Bioreactor Applications for Biological Wastewater Treatment: A Review of Research and Developments. Engineering 2017, 3, 330–342. [Google Scholar] [CrossRef]
- Derlon, N.; Coufort-Saudejaud, C.; Queinnec, I.; Paul, E. Growth limiting conditions and denitrification govern extent and frequency of volume detachment of biofilms. Chem. Eng. J. 2013, 218, 368–375. [Google Scholar] [CrossRef]
- Huang, H.; Ren, H.; Ding, L.; Geng, J.; Xu, K.; Zhang, Y. Aging biofilm from a full-scale moving bed biofilm reactor: Characterization and enzymatic treatment study. Bioresour. Technol. 2014, 154, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Simões, M.; Simões, L.C.; Vieira, M.J. A review of current and emergent biofilm control strategies. LWT—Food Sci. Technol. 2010, 43, 573–583. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhu, S.; Fan, X.; Zhang, X.; Ren, H.; Huang, H. Precise portrayal of microscopic processes of wastewater biofilm formation: Taking SiO2 as the model carrier. Sci. Total Environ. 2022, 849, 157776. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, Q.; Wu, B.; Zhao, F.; Ma, S.; Hu, H.; Zhang, X.; Ren, H. Quorum sensing signaling distribution during the development of full-scale municipal wastewater treatment biofilms. Sci. Total Environ. 2019, 685, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Reboleiro-Rivas, P.; Martín-Pascual, J.; Juárez-Jiménez, B.; Poyatos, J.M.; Vílchez-Vargas, R.; Vlaeminck, S.E.; Rodelas, B.; González-López, J. Nitrogen removal in a moving bed membrane bioreactor for municipal sewage treatment: Community differentiation in attached biofilm and suspended biomass. Chem. Eng. J. 2015, 277, 209–218. [Google Scholar] [CrossRef]
- Qiu, Z.; Zhang, S.; Ding, Y.; Zhang, W.; Gong, L.; Yuan, Q.; Mu, X.; Fu, D. Comparison of Myriophyllum Spicatum and artificial plants on nutrients removal and microbial community in constructed wetlands receiving WWTPs effluents. Bioresour. Technol. 2021, 321, 124469. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Y.; Paez-Espino, D.; Polz, M.F.; Zhang, T. Prokaryotic viruses impact functional microorganisms in nutrient removal and carbon cycle in wastewater treatment plants. Nat. Commun. 2021, 12, 5398. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Wei, D.; Ngo, H.H.; Guo, W.; Xu, W.; Du, B.; Wei, Q. Performance, microbial community and fluorescent characteristic of microbial products in a solid-phase denitrification biofilm reactor for WWTP effluent treatment. J. Environ. Manag. 2018, 227, 375–385. [Google Scholar] [CrossRef]
- Roveto, P.M.; Gupta, A.; Schuler, A.J. Effects of surface skewness on local shear stresses, biofilm activity, and microbial communities for wastewater treatment. Bioresour. Technol. 2021, 320, 124251. [Google Scholar] [CrossRef]
- Campo, R.; Vassallo, A.; Rabbeni, G.; Arancio, W.; Gallo, G.; Di Bella, G. Reactivation of aerobic granular sludge for the treatment of industrial shipboard slop wastewater: Effects of long-term storage on granules structure, biofilm activity and microbial community. J. Water Process Eng. 2021, 42, 102101. [Google Scholar] [CrossRef]
- Wensel, C.R.; Pluznick, J.L.; Salzberg, S.L.; Sears, C.L. Next-generation sequencing: Insights to advance clinical investigations of the microbiome. J. Clin. Investig. 2022, 132, e154944. [Google Scholar] [CrossRef] [PubMed]
- Nagler, M.; Podmirseg, S.M.; Mayr, M.; Ascher-Jenull, J.; Insam, H. The masking effect of extracellular DNA and robustness of intracellular DNA in anaerobic digester NGS studies: A discriminatory study of the total DNA pool. Mol. Ecol. 2021, 30, 438–450. [Google Scholar] [CrossRef] [PubMed]
- De Vrieze, J.; Pinto, A.J.; Sloan, W.T.; Ijaz, U.Z. The active microbial community more accurately reflects the anaerobic digestion process: 16S rRNA (gene) sequencing as a predictive tool. Microbiome 2018, 6, 63. [Google Scholar] [CrossRef]
- De Vrieze, J.; Raport, L.; Roume, H.; Vilchez-Vargas, R.; Jáuregui, R.; Pieper, D.H.; Boon, N. The full-scale anaerobic digestion microbiome is represented by specific marker populations. Water Res. 2016, 104, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Dai, J.; Zhao, Y.; Wu, J.; Ji, C.; Zhang, Y. Advances in preparation, application in contaminant removal, and environmental risks of biochar-based catalysts: A review. Biochar 2022, 4, 51. [Google Scholar] [CrossRef]
- Stiefel, P.; Schmidt-Emrich, S.; Maniura-Weber, K.; Ren, Q. Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiol. 2015, 15, 36. [Google Scholar] [CrossRef]
- Liu, L.; Liu, C.; Zhang, H.; He, J.; Zhai, J.; Yu, D.; Dong, S. How to Identify the “LIVE/DEAD” States of Microbes Related to Biosensing. ACS Sens. 2020, 5, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.-W.; Bao, Y.-W.; Wang, H.-Y.; Chen, Z.; Wu, F.-G. Bacteria-derived fluorescent carbon dots for microbial live/dead differentiation. Nanoscale 2017, 9, 2150–2161. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Breidt, F., Jr. Enumeration of viable Listeria monocytogenes cells by real-time PCR with propidium monoazide and ethidium monoazide in the presence of dead cells. Appl. Environ. Microbiol. 2007, 73, 8028–8031. [Google Scholar] [CrossRef] [PubMed]
- Vondrakova, L.; Turonova, H.; Scholtz, V.; Pazlarova, J.; Demnerova, K. Impact of various killing methods on EMA/PMA-qPCR efficacy. Food Control 2018, 85, 23–28. [Google Scholar] [CrossRef]
- Reyneke, B.; Ndlovu, T.; Khan, S.; Khan, W. Comparison of EMA-, PMA- and DNase qPCR for the determination of microbial cell viability. Appl. Microbiol. Biotechnol. 2017, 101, 7371–7383. [Google Scholar] [CrossRef] [PubMed]
- Nocker, A.; Cheung, C.-Y.; Camper, A.K. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 2006, 67, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, F.; Yang, G.; Aguilar, Z.P.; Lai, W.; Xu, H. Asymmetric polymerase chain assay combined with propidium monoazide treatment and unmodified gold nanoparticles for colorimetric detection of viable emetic Bacillus cereus in milk. Sens. Actuators B Chem. 2018, 255, 1455–1461. [Google Scholar] [CrossRef]
- Al-Daoud, F.; Gossen, B.D.; Robson, J.; McDonald, M.R. Propidium Monoazide Improves Quantification of Resting Spores of Plasmodiophora brassicae with qPCR. Plant Dis. 2017, 101, 442–447. [Google Scholar] [CrossRef][Green Version]
- Hanifian, S. Behavior of Mycobacterium avium paratuberculosis in Lighvan cheese tracked by propidium monoazide qPCR and culture. LWT 2020, 133, 109886. [Google Scholar] [CrossRef]
- Tantikachornkiat, M.; Sakakibara, S.; Neuner, M.; Durall, D.M. The use of propidium monoazide in conjunction with qPCR and Illumina sequencing to identify and quantify live yeasts and bacteria. Int. J. Food Microbiol. 2016, 234, 53–59. [Google Scholar] [CrossRef]
- Elizaquivel, P.; Azizkhani, M.; Sanchez, G.; Aznar, R. Evaluation of Zataria multiflora Boiss. essential oil activity against Escherichia coli O157:H7, Salmonella enterica and Listeria monocytogenes by propidium monoazide quantitative PCR in vegetables. Food Control 2013, 34, 770–776. [Google Scholar] [CrossRef]
- Jones, K.L.; Cunha, F.; Casaro, S.; Galvao, K.N. Optimization and Testing of a Commercial Viability PCR Protocol to Detect Escherichia coli in Whole Blood. Microorganisms 2024, 12, 765. [Google Scholar] [CrossRef] [PubMed]
- Reichelt, B.; Szott, V.; Stingl, K.; Roesler, U.; Friese, A. Detection of Viable but Non-Culturable (VBNC)-Campylobacter in the Environment of Broiler Farms: Innovative Insights Delivered by Propidium Monoazide (PMA)-v-qPCR Analysis. Microorganisms 2023, 11, 2492. [Google Scholar] [CrossRef] [PubMed]
- Krohn, C.; Jansriphibul, K.; Dias, D.A.; Rees, C.A.; van den Akker, B.; Boer, J.C.; Plebanski, M.; Surapaneni, A.; O’Carroll, D.; Richard, S.; et al. Dead in the water—Role of relic DNA and primer choice for targeted sequencing surveys of anaerobic sewage sludge intended for biological monitoring. Water Res. 2024, 253, 121354. [Google Scholar] [CrossRef] [PubMed]
- Eramo, A.; Morales Medina, W.R.; Fahrenfeld, N.L. Viability-based quantification of antibiotic resistance genes and human fecal markers in wastewater effluent and receiving waters. Sci. Total Environ. 2019, 656, 495–502. [Google Scholar] [CrossRef]
- Li, D.; Tong, T.; Zeng, S.; Lin, Y.; Wu, S.; He, M. Quantification of viable bacteria in wastewater treatment plants by using propidium monoazide combined with quantitative PCR (PMA-qPCR). J. Environ. Sci. 2014, 26, 299–306. [Google Scholar] [CrossRef]
- Liu, W.; Xiang, P.; Ji, Y.; Chen, Z.; Lei, Z.; Huang, W.; Huang, W.; Liu, D. Response of viable bacteria to antibiotics in aerobic granular sludge: Resistance mechanisms and behaviors, bacterial communities, and driving factors. Water Res. 2023, 245, 120656. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, A.S.; Fahrenfeld, N.L. Influence of DNA from non-viable sources on the riverine water and biofilm microbiome, resistome, mobilome, and resistance gene host assignments. J. Hazard. Mater. 2023, 446, 130743. [Google Scholar] [CrossRef]
- Ni, J.; Hatori, S.; Wang, Y.; Li, Y.-Y.; Kubota, K. Uncovering Viable Microbiome in Anaerobic Sludge Digesters by Propidium Monoazide (PMA)-PCR. Microb. Ecol. 2019, 79, 925–932. [Google Scholar] [CrossRef]
- Zhang, X.; Lu, W.; Han, E.; Wang, S.; Shen, J. Hybrid Nanostructure-based Immunosensing for Electrochemical Assay of Escherichia coli as Indicator Bacteria Relevant to the Recycling of Urban Sludge. Electrochim. Acta 2014, 141, 384–390. [Google Scholar] [CrossRef]
- Cooksey, E.M.; Singh, G.; Scott, L.C.; Aw, T.G. Detection of coliphages and human adenoviruses in a subtropical estuarine lake. Science of The Total Environment 2019, 649, 1514–1521. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, V.; Elk, M.; Toledo-Hernandez, C.; Santo Domingo, J.W. Analysis of human mitochondrial DNA sequences from fecally polluted environmental waters as a tool to study population diversity. AIMS Environ. Sci. 2017, 4, 443. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, A.; Omino, N.; Nakamura, T.; Goto, S.; Matsumoto, R.; Yomogita, M.; Narisawa, N.; Kimijima, M.; Iritani, K. Evaluation of Cellular Responses of Heterotrophic Escherichia coli Cultured with Autotrophic Chlamydomonas reinhardtii as a Nutrient Source by Analyses Based on Microbiology and Transcriptome. Microorganisms 2024, 12, 452. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Schulze-Makuch, D.; de Vera, J.-P.; Cockell, C.; Leya, T.; Baque, M.; Walther-Antonio, M. The Development of an Effective Bacterial Single-Cell Lysis Method Suitable for Whole Genome Amplification in Microfluidic Platforms. Micromachines 2018, 9, 367. [Google Scholar] [CrossRef] [PubMed]
- Karni, M.; Zidon, D.; Polak, P.; Zalevsky, Z.; Shefi, O. Thermal Degradation of DNA. DNA Cell Biol. 2013, 32, 298–301. [Google Scholar] [CrossRef] [PubMed]
- Macori, G.; McCarthy, S.C.; Burgess, C.M.; Fanning, S.; Duffy, G. Investigation of the Causes of Shigatoxigenic Escherichia coli PCR Positive and Culture Negative Samples. Microorganisms. 2020, 8, 587. [Google Scholar] [CrossRef]
- Guo, L.; Ze, X.; Feng, H.; Liu, Y.; Ge, Y.; Zhao, X.; Song, C.; Jiao, Y.; Liu, J.; Mu, S.; et al. Identification and quantification of viable Lacticaseibacillus rhamnosus in probiotics using validated PMA-qPCR method. Front. Microbiol. 2024, 15, 1341884. [Google Scholar] [CrossRef]
- Vasquez, C.; Leyton-Carcaman, B.; Cid-Alda, F.P.; Segovia, I.; Pinto, F.; Abanto, M. Physical Pretreatments Applied in Three Commercial Kits for the Extraction of High-Quality DNA from Activated Sewage Sludge. Int. J. Mol. Sci. 2023, 24, 15243. [Google Scholar] [CrossRef] [PubMed]
- DB21/T 2734.1-2017; Method of PCR typing diagnosis for bacterial Part1: Mehtod of PCR detection for Escherichia coli. Standards Press of China: Liaoning, China, 2017.
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. Bmc Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, H.; Park, J.; Takagi, T. MetaGene: Prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006, 34, 5623–5630. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Fu, Y.; Ye, Z.; Jia, Y.; Fan, J.; Hashmi, M.Z.; Shen, C. An Optimized Method to Assess Viable Escherichia coli O157:H7 in Agricultural Soil Using Combined Propidium Monoazide Staining and Quantitative PCR. Front. Microbiol. 2020, 11, 1809. [Google Scholar] [CrossRef]
- Miwa, T.; Takimoto, Y.; Hatamoto, M.; Kuratate, D.; Watari, T.; Yamaguchi, T. Role of live cell colonization in the biofilm formation process in membrane bioreactors treating actual sewage under low organic loading rate conditions. Appl. Microbiol. Biotechnol. 2021, 105, 1721–1729. [Google Scholar] [CrossRef]
- Kang, D.; Zhang, L.; Yang, S.; Li, J.; Peng, Y. Linking morphological features to anammox communities in a partial nitritation and anammox (PN/A) biofilm reactor. J. Environ. Manag. 2023, 341, 118038. [Google Scholar] [CrossRef]
- Liu, M.; Hata, A.; Katayama, H.; Kasuga, I. Consecutive ultrafiltration and silica adsorption for recovery of extracellular antibiotic resistance genes from an urban river. Environ. Pollut. 2020, 260, 114062. [Google Scholar] [CrossRef]
- Caro, D.M.; Horstmann, L.; Ganzert, L.; Oses, R.; Friedl, T.; Wagner, D. An improved method for intracellular DNA (iDNA) recovery from terrestrial environments. Microbiologyopen 2023, 12, e1369. [Google Scholar] [CrossRef]
- Sivalingam, P.; Sabatino, R.; Sbaffi, T.; Fontaneto, D.; Corno, G.; Di Cesare, A. Extracellular DNA includes an important fraction of high-risk antibiotic resistance genes in treated wastewaters. Environ. Pollut. 2023, 323, 121325. [Google Scholar] [CrossRef] [PubMed]
- Bairoliya, S.; Xiang, J.K.Z.; Cao, B. Extracellular DNA in Environmental Samples: Occurrence, Extraction, Quantification, and Impact on Microbial Biodiversity Assessment. Appl. Environ. Microbiol. 2022, 88, e01845-21. [Google Scholar] [CrossRef] [PubMed]
- Grande, R.; Di Marcantonio, M.C.; Robuffo, I.; Pompilio, A.; Celia, C.; Di Marzio, L.; Paolino, D.; Codagnone, M.; Muraro, R.; Stoodley, P.; et al. Helicobacter pylori ATCC 43629/NCTC 11639 Outer Membrane Vesicles (OMVs) from Biofilm and Planktonic Phase Associated with Extracellular DNA (eDNA). Front. Microbiol. 2015, 6, 1369. [Google Scholar] [CrossRef]
- Tang, L.; Schramm, A.; Neu, T.R.; Revsbech, N.P.; Meyer, R.L. Extracellular DNA in adhesion and biofilm formation of four environmental isolates: A quantitative study. Fems Microbiol. Ecol. 2013, 86, 394–403. [Google Scholar] [CrossRef]
- Ni, J.; Ji, J.; Li, Y.-Y.; Kubota, K. Propidium monoazide—Polymerase chain reaction reveals viable microbial community shifts in anaerobic membrane bioreactors treating domestic sewage at low temperature. Bioresour. Technol. 2023, 387, 129564. [Google Scholar] [CrossRef] [PubMed]
- Codony, F.; Dinh-Thanh, M.; Agusti, G. Key Factors for Removing Bias in Viability PCR-Based Methods: A Review. Curr. Microbiol. 2020, 77, 682–687. [Google Scholar] [CrossRef]
- Van Holm, W.; Ghesquiere, J.; Boon, N.; Verspecht, T.; Bernaerts, K.; Zayed, N.; Chatzigiannidou, I.; Teughels, W. A Viability Quantitative PCR Dilemma: Are Longer Amplicons Better? Appl. Environ. Microbiol. 2021, 87, e02653-20. [Google Scholar] [CrossRef]
- Shi, Q.; Chen, Z.; Yan, H.; Xu, M.; Cao, K.-F.; Mao, Y.; Chen, X.; Hu, H.-Y. Identification of significant live bacterial community shifts in different reclaimed waters during ozone and chlorine disinfection. Sci. Total Environ. 2023, 896, 165199. [Google Scholar] [CrossRef]
- Sun, Y.-Q.; Ge, Y. Relic DNA effects on the estimates of bacterial community composition and taxa dynamics in soil. Appl. Microbiol. Biotechnol. 2023, 107, 4109–4117. [Google Scholar] [CrossRef]
- Gensberger, E.T.; Polt, M.; Konrad-Köszler, M.; Kinner, P.; Sessitsch, A.; Kostić, T. Evaluation of quantitative PCR combined with PMA treatment for molecular assessment of microbial water quality. Water Res. 2014, 67, 367–376. [Google Scholar] [CrossRef]
- Kapoor, V.; Gupta, I.; Pasha, A.B.M.T.; Duc, P. Real-Time Quantitative PCR Measurements of Fecal Indicator Bacteria and Human-Associated Source Tracking Markers in a Texas River following Hurricane Harvey. Environ. Sci. Technol. Lett. 2018, 5, 322–328. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, J.; Zheng, X.; Ning, T.; Wang, J.; Wei, X.; Tan, L.; Shen, F. Revealing the Viable Microbial Community of Biofilm in a Sewage Treatment System Using Propidium Monoazide Combined with Real-Time PCR and Metagenomics. Microorganisms 2024, 12, 1508. https://doi.org/10.3390/microorganisms12081508
Liang J, Zheng X, Ning T, Wang J, Wei X, Tan L, Shen F. Revealing the Viable Microbial Community of Biofilm in a Sewage Treatment System Using Propidium Monoazide Combined with Real-Time PCR and Metagenomics. Microorganisms. 2024; 12(8):1508. https://doi.org/10.3390/microorganisms12081508
Chicago/Turabian StyleLiang, Jiayin, Xiangqun Zheng, Tianyang Ning, Jiarui Wang, Xiaocheng Wei, Lu Tan, and Feng Shen. 2024. "Revealing the Viable Microbial Community of Biofilm in a Sewage Treatment System Using Propidium Monoazide Combined with Real-Time PCR and Metagenomics" Microorganisms 12, no. 8: 1508. https://doi.org/10.3390/microorganisms12081508
APA StyleLiang, J., Zheng, X., Ning, T., Wang, J., Wei, X., Tan, L., & Shen, F. (2024). Revealing the Viable Microbial Community of Biofilm in a Sewage Treatment System Using Propidium Monoazide Combined with Real-Time PCR and Metagenomics. Microorganisms, 12(8), 1508. https://doi.org/10.3390/microorganisms12081508






