Rapid and Highly Sensitive Detection of Mycobacterium tuberculosis Utilizing the Recombinase Aided Amplification-Based CRISPR-Cas13a System
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Samples Collection
2.2. DNA Rapid Extraction
2.3. Preparation of the MTB Gene and H37Rv DNA
2.4. Preparation of Cas13a Protein
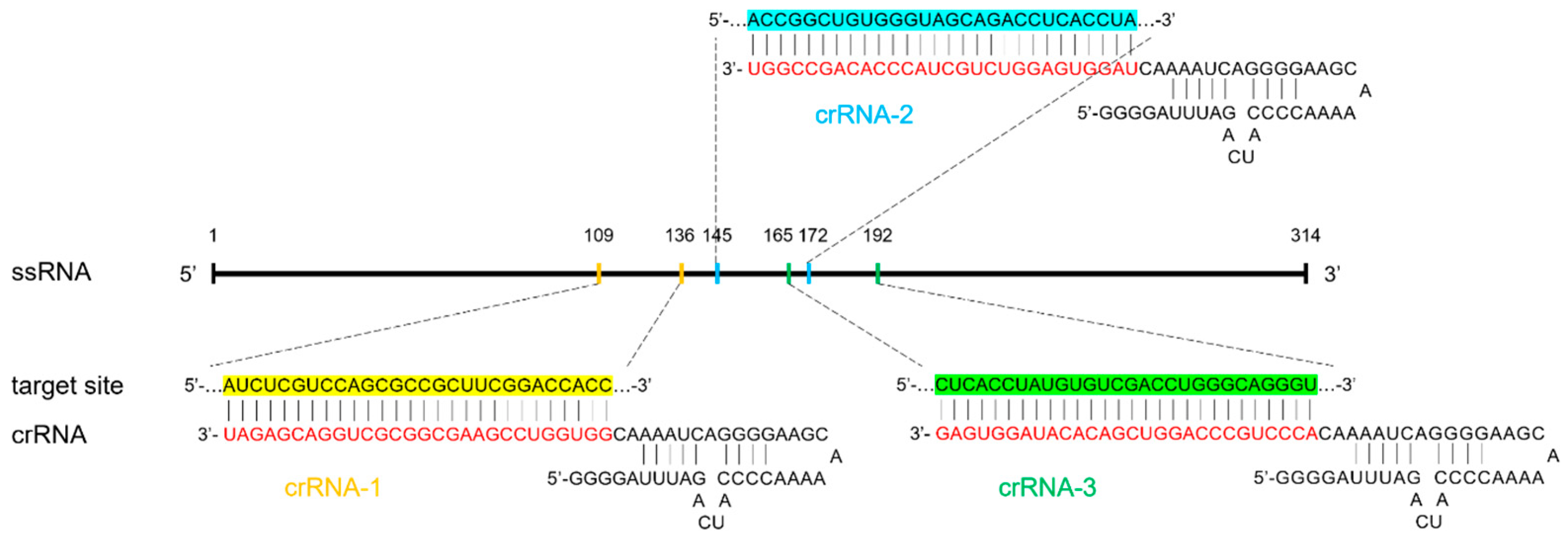
2.5. Preparation of CrRNA
2.6. RAA Coupled with CRISPR-Cas13a Assay
2.7. Statistical Analysis
3. Results
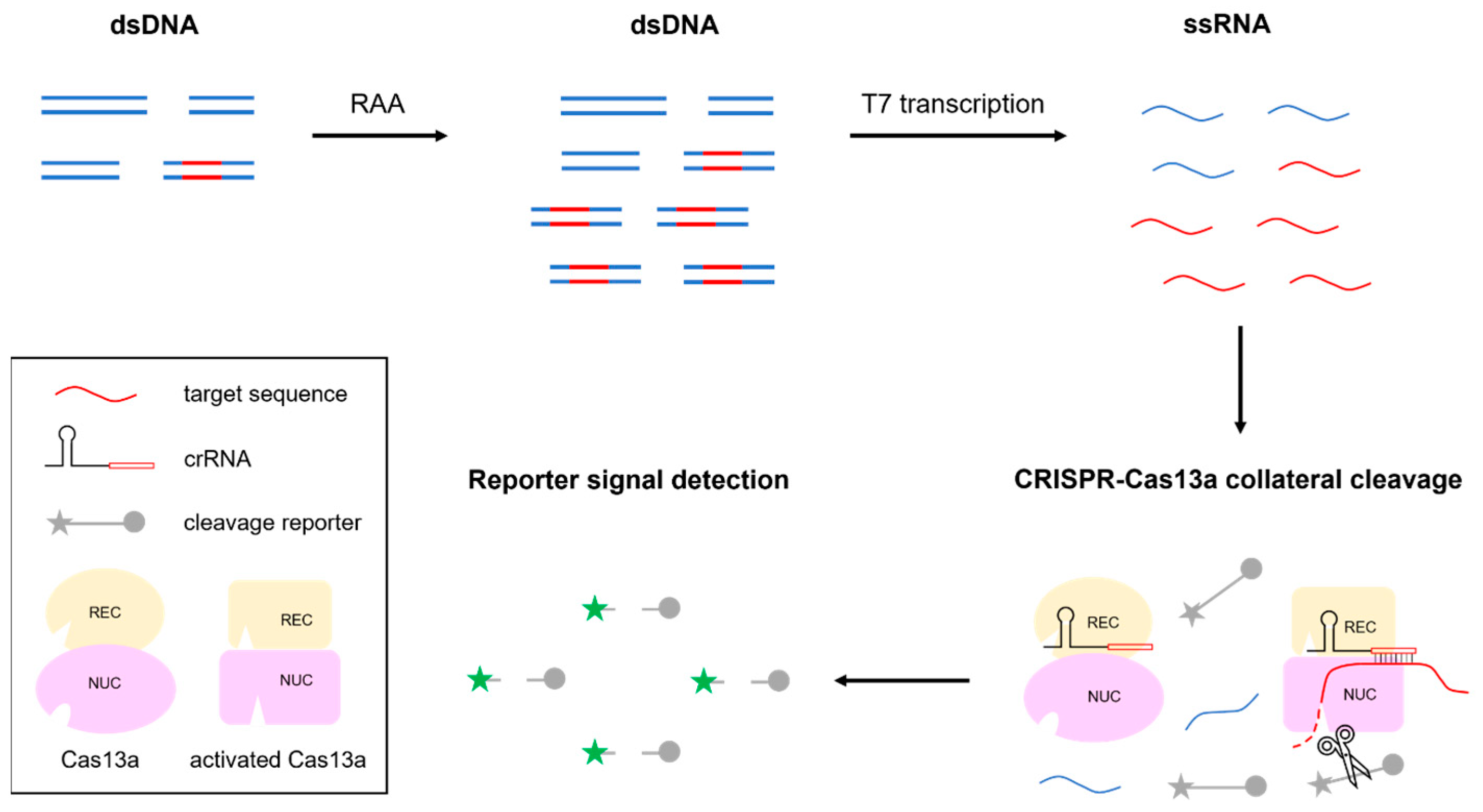
3.1. Schematic of RAA-CRISPR-MTB
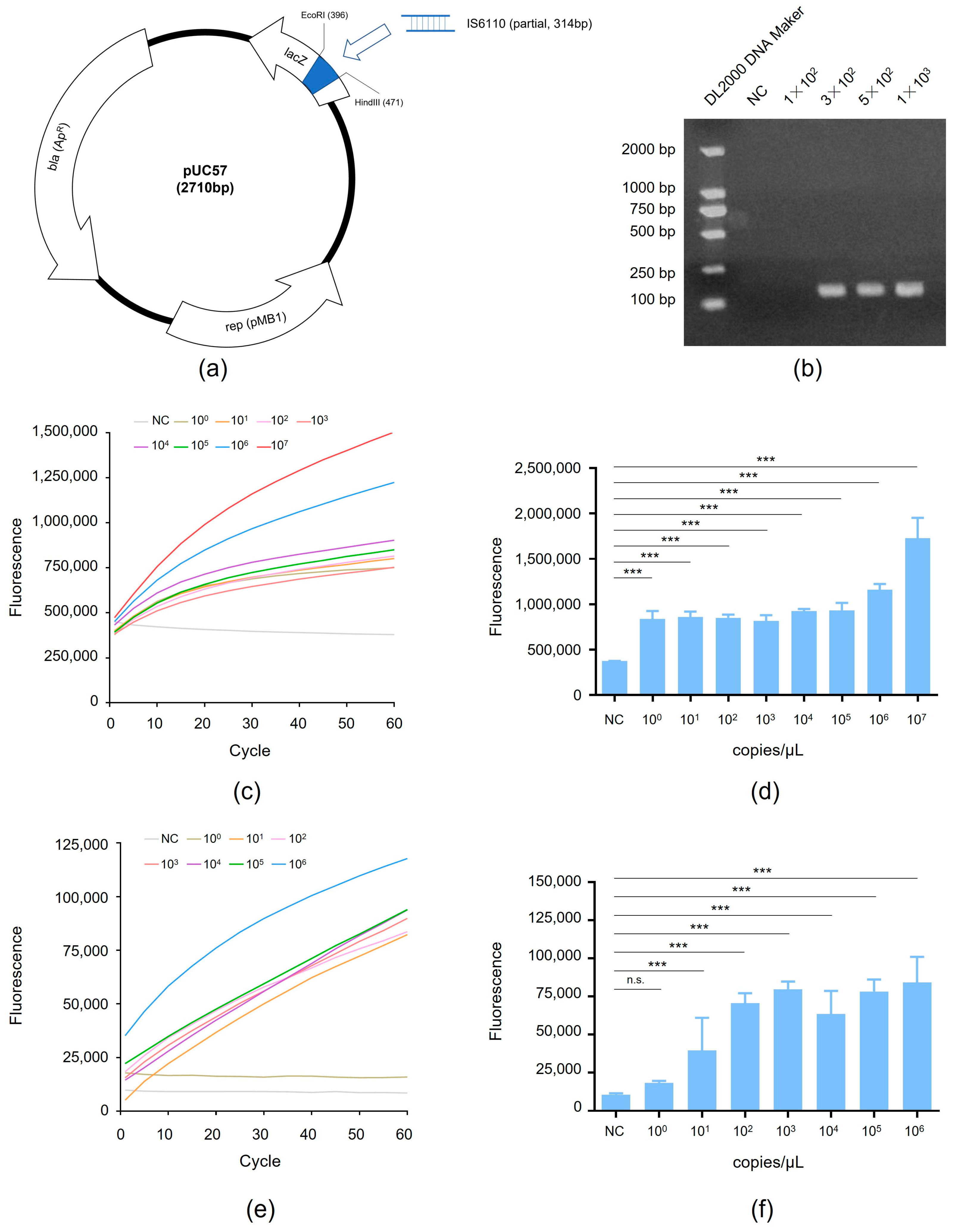
3.2. Detection of RAA-CRISPR Method Using the MTB Gene
3.3. Detection of RAA-CRISPR Method Using the Standard Strain H37Rv
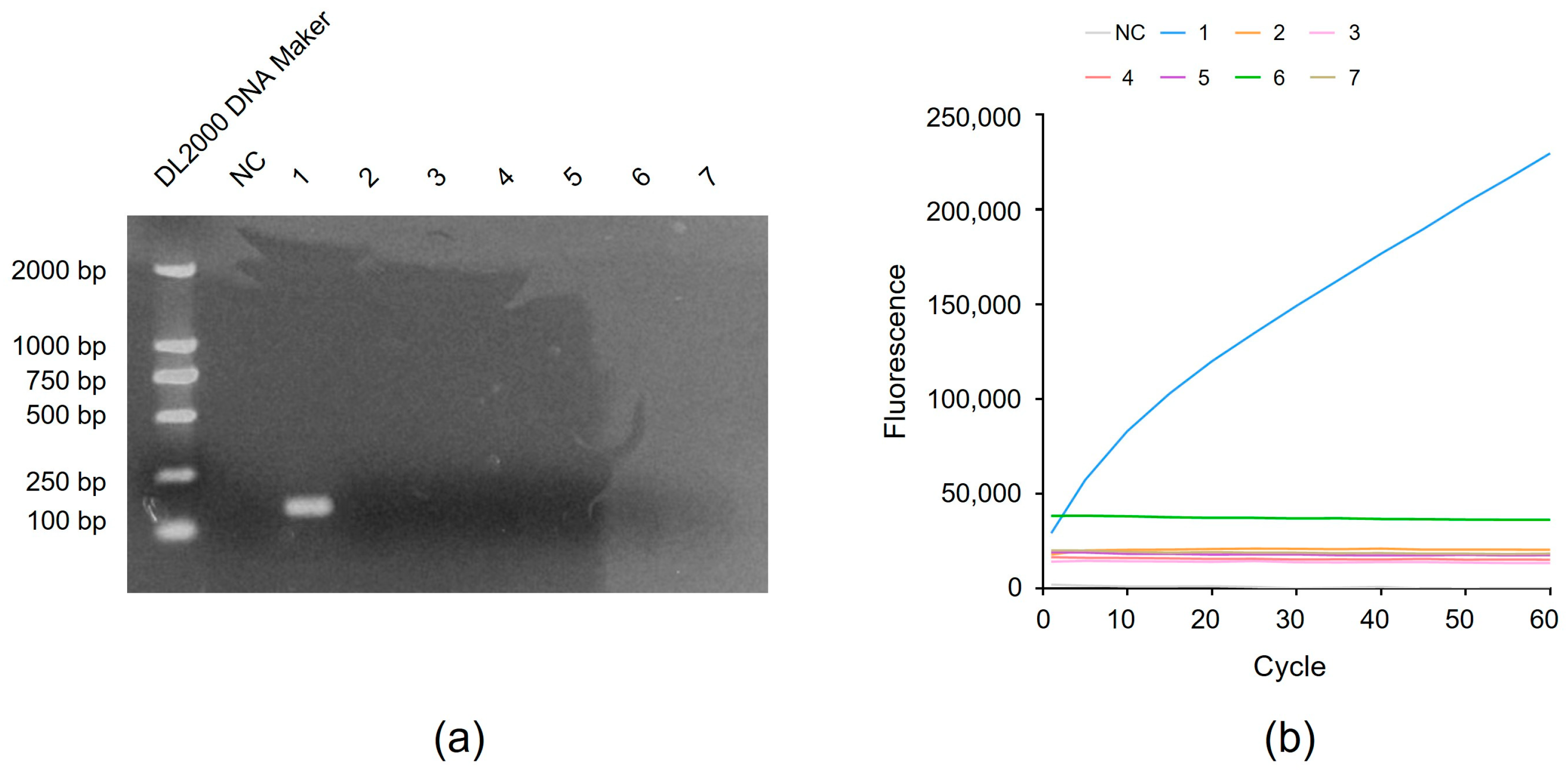
3.4. Detection of the Specificity of RAA-CRISPR Method
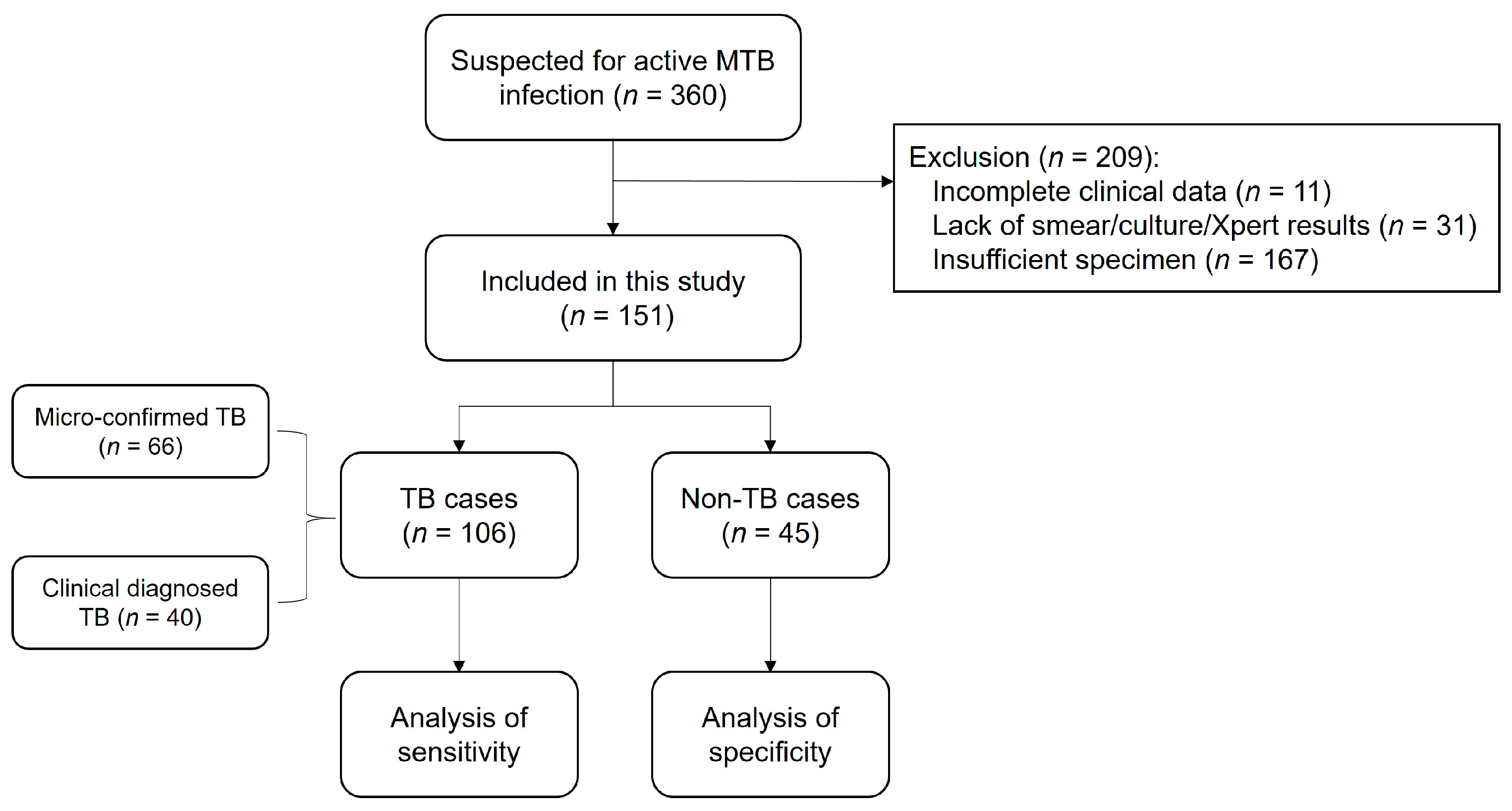
3.5. Flowchart of the Study Population
3.6. Application of RAA-CRISPR in Clinical Tuberculosis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report 2023. Available online: https://www.who.int/publications/i/item/9789240083851 (accessed on 13 June 2024).
- Furin, J.; Cox, H.; Pai, M. Tuberculosis. Lancet 2019, 393, 1642–1656. [Google Scholar] [CrossRef] [PubMed]
- Suarez, I.; Funger, S.M.; Kroger, S.; Rademacher, J.; Fatkenheuer, G.; Rybniker, J. The Diagnosis and Treatment of Tuberculosis. Dtsch. Arztebl. Int. 2019, 116, 729–735. [Google Scholar] [CrossRef]
- Kohli, M.; Schiller, I.; Dendukuri, N.; Yao, M.; Dheda, K.; Denkinger, C.M.; Schumacher, S.G.; Steingart, K.R. Xpert MTB/RIF Ultra and Xpert MTB/RIF assays for extrapulmonary tuberculosis and rifampicin resistance in adults. Cochrane Database Syst. Rev. 2021, 1, CD012768. [Google Scholar]
- Yan, L.; Xiao, H.; Zhang, Q. Systematic review: Comparison of Xpert MTB/RIF, LAMP and SAT methods for the diagnosis of pulmonary tuberculosis. Tuberculosis 2016, 96, 75–86. [Google Scholar] [CrossRef]
- Walzl, G.; McNerney, R.; du Plessis, N.; Bates, M.; McHugh, T.D.; Chegou, N.N.; Zumla, A. Tuberculosis: Advances and challenges in development of new diagnostics and biomarkers. Lancet Infect. Dis. 2018, 18, e199–e210. [Google Scholar] [CrossRef] [PubMed]
- Kostyusheva, A.; Brezgin, S.; Babin, Y.; Vasilyeva, I.; Glebe, D.; Kostyushev, D.; Chulanov, V. CRISPR-Cas systems for diagnosing infectious diseases. Methods 2022, 203, 431–446. [Google Scholar] [CrossRef]
- Wang, H.; Wang, T.; Gao, F.; Ren, W. Application of CRISPR/Cas Technology in Spermatogenesis Research and Male Infertility Treatment. Genes 2022, 13, 1000. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Man, S.; Ye, S.; Liu, G.; Ma, L. CRISPR-Cas based virus detection: Recent advances and perspectives. Biosens. Bioelectron. 2021, 193, 113541. [Google Scholar] [CrossRef]
- Kim, S.; Ji, S.; Koh, H.R. CRISPR as a Diagnostic Tool. Biomolecules 2021, 11, 1162. [Google Scholar] [CrossRef]
- Puig-Serra, P.; Casado-Rosas, M.C.; Martinez-Lage, M.; Olalla-Sastre, B.; Alonso-Yanez, A.; Torres-Ruiz, R.; Rodriguez-Perales, S. CRISPR Approaches for the Diagnosis of Human Diseases. Int. J. Mol. Sci. 2022, 23, 1757. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Zhou, Y.; Li, H.; Shang, Y.; Zhang, X.; Yuan, J.; Li, S.; Li, C.; Pang, Y. Development and clinical evaluation of a CRISPR/Cas13a-based diagnostic test to detect Mycobacterium tuberculosis in clinical specimens. Front. Microbiol. 2023, 14, 1117085. [Google Scholar] [CrossRef] [PubMed]
- Boyle, D.S.; McNerney, R.; Teng Low, H.; Leader, B.T.; Perez-Osorio, A.C.; Meyer, J.C.; O’Sullivan, D.M.; Brooks, D.G.; Piepenburg, O.; Forrest, M.S. Rapid detection of Mycobacterium tuberculosis by recombinase polymerase amplification. PLoS ONE 2014, 9, e103091. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Qi, Y.; He, P.; Wan, W.; OuYang, X.; Yu, Y.; Wen, B.; Xiong, X. Development of a Lateral Flow Strip-Based Recombinase-Aided Amplification for Active Chlamydia psittaci Infection. Front. Microbiol. 2022, 13, 928025. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhang, X.; Zhang, Y.; Ren, W.; Yao, C.; Li, C.; Liu, Y.; Tang, S. Preliminary Study on Detection Method of MTB DNA by PCR Amplification Combined with CRISPR-Cas13a System. Clin. J. Antituberc. 2020, 42, 1280–1288. [Google Scholar]
- Gao, D.; Zhu, X.; Lu, B. Development and application of sensitive, specific, and rapid CRISPR-Cas13-based diagnosis. J. Med. Virol. 2021, 93, 4198–4204. [Google Scholar]
- East-Seletsky, A.; O’Connell, M.R.; Knight, S.C.; Burstein, D.; Cate, J.H.; Tjian, R.; Doudna, J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016, 538, 270–273. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Qiu, M.; Li, X.; Yang, J.; Li, J. CRISPR-Cas13a system: A novel tool for molecular diagnostics. Front. Microbiol. 2022, 13, 1060947. [Google Scholar] [CrossRef]
- Li, H.; Dong, X.; Wang, Y.; Yang, L.; Cai, K.; Zhang, X.; Kou, Z.; He, L.; Sun, S.; Li, T.; et al. Sensitive and Easy-Read CRISPR Strip for COVID-19 Rapid Point-of-Care Testing. CRISPR J. 2021, 4, 392–399. [Google Scholar] [CrossRef]
- Arizti-Sanz, J.; Freije, C.A.; Stanton, A.C.; Petros, B.A.; Boehm, C.K.; Siddiqui, S.; Shaw, B.M.; Adams, G.; Kosoko-Thoroddsen, T.F.; Kemball, M.E.; et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun. 2020, 11, 5921. [Google Scholar] [CrossRef]
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149. [Google Scholar] [CrossRef] [PubMed]
- Qin, P.; Park, M.; Alfson, K.J.; Tamhankar, M.; Carrion, R.; Patterson, J.L.; Griffiths, A.; He, Q.; Yildiz, A.; Mathies, R.; et al. Rapid and Fully Microfluidic Ebola Virus Detection with CRISPR-Cas13a. ACS Sens. 2019, 4, 1048–1054. [Google Scholar] [CrossRef]
- Zhang, X.; Tian, Y.; Xu, L.; Fan, Z.; Cao, Y.; Ma, Y.; Li, H.; Ren, F. CRISPR/Cas13-assisted hepatitis B virus covalently closed circular DNA detection. Hepatol. Int. 2022, 16, 306–315. [Google Scholar] [CrossRef]
- Kaminski, M.M.; Alcantar, M.A.; Lape, I.T.; Greensmith, R.; Huske, A.C.; Valeri, J.A.; Marty, F.M.; Klambt, V.; Azzi, J.; Akalin, E.; et al. A CRISPR-based assay for the detection of opportunistic infections post-transplantation and for the monitoring of transplant rejection. Nat. Biomed. Eng. 2020, 4, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhou, X.; Shan, Y.; Yue, H.; Huang, R.; Hu, J.; Xing, D. Sensitive detection of a bacterial pathogen using allosteric probe-initiated catalysis and CRISPR-Cas13a amplification reaction. Nat. Commun. 2020, 11, 267. [Google Scholar] [CrossRef]
- Huang, Z.; LaCourse, S.M.; Kay, A.W.; Stern, J.; Escudero, J.N.; Youngquist, B.M.; Zheng, W.; Vambe, D.; Dlamini, M.; Mtetwa, G.; et al. CRISPR detection of circulating cell-free Mycobacterium tuberculosis DNA in adults and children, including children with HIV: A molecular diagnostics study. Lancet Microbe 2022, 3, e482–e492. [Google Scholar] [CrossRef]
- Thabet, S.; Souissi, N. Transposition mechanism, molecular characterization and evolution of IS6110, the specific evolutionary marker of Mycobacterium tuberculosis complex. Mol. Biol. Rep. 2017, 44, 25–34. [Google Scholar] [CrossRef] [PubMed]
- McEvoy, C.R.; Falmer, A.A.; Gey van Pittius, N.C.; Victor, T.C.; van Helden, P.D.; Warren, R.M. The role of IS6110 in the evolution of Mycobacterium tuberculosis. Tuberculosis 2007, 87, 393–404. [Google Scholar] [CrossRef]
- Ai, J.W.; Zhou, X.; Xu, T.; Yang, M.; Chen, Y.; He, G.Q.; Pan, N.; Cai, Y.; Li, Y.; Wang, X.; et al. CRISPR-based rapid and ultra-sensitive diagnostic test for Mycobacterium tuberculosis. Emerg. Microbes Infect. 2019, 8, 1361–1369. [Google Scholar] [CrossRef]
- Jiang, C.; Tao, D.; Geng, Y.; Yang, H.; Xu, B.; Chen, Y.; Hu, C.; Chen, H.; Xie, S.; Guo, A. Sensitive and Specific Detection of Lumpy Skin Disease Virus in Cattle by CRISPR-Cas12a Fluorescent Assay Coupled with Recombinase Polymerase Amplification. Genes 2022, 13, 734. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Tang, H.; Li, R.; Xia, Z.; Yang, W.; Zhu, Y.; Liu, Z.; Lu, G.; Ni, S.; Shen, J. A New Method Based on LAMP-CRISPR-Cas12a-Lateral Flow Immunochromatographic Strip for Detection. Infect. Drug Resist. 2022, 15, 685–696. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Wang, N.; Pang, M.; Miao, H.; Dai, X.; Li, B.; Yang, X.; Li, C.; Liu, Y. Rapid and Highly Sensitive Detection of Mycobacterium tuberculosis Utilizing the Recombinase Aided Amplification-Based CRISPR-Cas13a System. Microorganisms 2024, 12, 1507. https://doi.org/10.3390/microorganisms12081507
Li Q, Wang N, Pang M, Miao H, Dai X, Li B, Yang X, Li C, Liu Y. Rapid and Highly Sensitive Detection of Mycobacterium tuberculosis Utilizing the Recombinase Aided Amplification-Based CRISPR-Cas13a System. Microorganisms. 2024; 12(8):1507. https://doi.org/10.3390/microorganisms12081507
Chicago/Turabian StyleLi, Qiao, Nenhan Wang, Mengdi Pang, Honghao Miao, Xiaowei Dai, Bo Li, Xinyu Yang, Chuanyou Li, and Yi Liu. 2024. "Rapid and Highly Sensitive Detection of Mycobacterium tuberculosis Utilizing the Recombinase Aided Amplification-Based CRISPR-Cas13a System" Microorganisms 12, no. 8: 1507. https://doi.org/10.3390/microorganisms12081507
APA StyleLi, Q., Wang, N., Pang, M., Miao, H., Dai, X., Li, B., Yang, X., Li, C., & Liu, Y. (2024). Rapid and Highly Sensitive Detection of Mycobacterium tuberculosis Utilizing the Recombinase Aided Amplification-Based CRISPR-Cas13a System. Microorganisms, 12(8), 1507. https://doi.org/10.3390/microorganisms12081507




