Protein Kinase A Negatively Regulates the Acetic Acid Stress Response in S. cerevisiae
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Growth Media, and Growth Conditions
| Strain | Genotype | Source | Application |
|---|---|---|---|
| BY4741 | MATa his3 Δ1 leu2Δ0 met17Δ0 ura3Δ0 | Lab. stock | Figure 1A–D, Figure 2, Figure 3, Figure 4, Figure 5 and Figure 6 |
| ZLY4370 | BY4741 ras1Δ::kanMX4 | This study | Figure 1A–D |
| ZLY4376 | BY4741 ras2Δ::kanMX4-2D | This study | Figure 1A–D, Figure 2 and Figure 3 |
| ZLY4373 | BY4741 pde1Δ::kanMX4 | This study | Figure 1A–D |
| ZLY4368 | BY4741 pde2Δ::kanMX4 | This study | Figure 1A–D and Figure 3 |
| ZLY4162 | BY4741 YRO2–lacZ::kanMX4 | This study | Figure 1E,F |
| ZLY4153 | BY4741 YGP1–lacZ::kanMX4 | This study | Figure 1E,F |
| ZLY4238 | BY4741 TPO2–lacZ::LEU2 | This study | Figure 1E,F |
| ZLY4322 | BY4741 YRO2–lacZ::kanMX4 RAS2::RAS2Ala18Val19::HIS3 | This study | Figure 1E,F |
| ZLY4319 | BY4741 YGP1–lacZ::kanMX4 RAS2::RAS2Ala18Val19::HIS3 | This study | Figure 1E,F and Figure 3 |
| ZLY4323 | BY4741 TPO2–lacZ::LEU2 RAS2::RAS2Ala18Val19::HIS3 | This study | Figure 1E,F |
| ZLY4043 | BY4741 haa1Δ::kanMX4 | SGDP * | Figure 2, Figure 3, Figure 5 and Figure 6 |
| ZLY4470 | BY4741 haa1::HIS3 ras2::kanMX4 | This study | Figure 2 |
| ZLY4419 ** | MATa his3 Δ1 leu2Δ0 met17Δ0 ura3Δ0 | This study | Figure 3C |
| ZLY4424 ** | MATa his3 Δ1 leu2Δ0 met17Δ0 ura3Δ0 | This study | Figure 3C |
| ZLY4411 | BY4741 ras2Δ::kanMX4-1D | This study | Figure 3C |
| ZLY4445 | BY4741 tpk1::kanMX4 | This study | Figure 4 |
| ZLY4438 | BY4741 tpk2::kanMX4 | This study | Figure 4 |
| ZLY4441 | BY4741 tpk3::kanMX4 | This study | Figure 4 |
| ZLY4516 | MATa ura3 Δ leu2Δ his3Δ tpk1::kanMX4 tpk2::kanMX4 | This study | Figure 4 and Figure 5 |
| ZLY4518 | MATα ura3 Δ leu2Δ his3Δ met17Δ lys2Δ tpk1::kanMX4 tpk2::kanMX4 | This study | Figure 4 |
| ZLY4520 | MATa ura3 Δ leu2Δ his3Δ lys2Δ tpk1::kanMX4 tpk3::kanMX4 | This study | Figure 4, Figure 5 and Figure 6 |
| ZLY4521 | MATα ura3 Δ leu2Δ his3Δ met17Δ tpk1::kanMX4 tpk3::kanMX4 | This study | Figure 4 |
| ZLY4522 | MATα ura3 Δ leu2Δ his3Δ met17Δ tpk2::kanMX4 tpk3::kanMX4 | This study | Figure 4 and Figure 5 |
| ZLY4524 | BY4741 tpk2::kanMX4 tpk3::kanMX4 | This study | Figure 4 |
| ZDY166 | BY4741 haa1::HIS3 | This study | Figure 5F |
| ZLY5264 | MATa ura3 Δ leu2Δ his3Δ lys2Δ tpk1::kanMX4 tpk3::kanMX4 haa1::LEU2 | This study | Figure 5 and Figure 6 |
2.2. Plasmid Constructs
| Plasmid | Description | Reference | Application |
|---|---|---|---|
| pZL3164 | pRS416-YRO2–lacZ, expressing YRO2–lacZ reporter gene with 1929-bp YRO2 promoter sequence fused to lacZ coding sequence. | [19] | Figure 1A–D, Figure 2, Figure 4 and Figure 5 |
| pZL3170 | pRS416-YGP1–lacZ, expressing YGP1–lacZ reporter gene with 1980 bp YGP1 promoter sequence fused to lacZ coding sequence. | This study | Figure 1A–D, Figure 2, Figure 4 and Figure 5 |
| YCp50-RAS2A18V19 | A hyperactive RAS2 mutant allele, RAS2A18V19, cloned in centromeric plasmid with a URA3 selection marker. | [35] | |
| pZL3326 | pRS303-RAS2A18V19, encoding the dominant active RAS2A18V19 allele on an integrative plasmid carrying a HIS3 selection marker. | This study | Figure 1E,F and Figure 3 |
| YIp356-kanMX4 | An integrative plasmid carrying a lacZ reporter gene and the kanMX4 selection marker. | This study | |
| pZL3203 | YIp356-kanMX4-YRO2, encoding an YRO2–lacZ reporter gene for integration into the genome. | This study | Figure 1E,F |
| pZL3201 | YIp356-kanMX4-YGP1, encoding an YGP1–lacZ reporter gene for integration into the genome. | This study | Figure 1E,F |
| pZL3307 | YIp366-TPO2, encoding an TPO2–lacZ reporter gene for integration into the genome. | This study | Figure 1E,F |
| pZL3158 | pRS416-TPO2–lacZ, expressing TPO2–lacZ reporter gene with 1926 bp TPO2 promoter sequence fused to the lacZ coding sequence. | [19] | Figure 2 and Figure 5 |
| pZL3161 | pRS416-TPO3–lacZ, expressing TPO3–lacZ reporter gene with 1976 bp TPO3 promoter sequence fused to the lacZ coding sequence. | This study | Figure 5 |
| pZL3155 | pRS416-SPI1–lacZ, expressing SPI1–lacZ reporter gene with 980 bp SPI1 promoter sequence fused to the lacZ coding sequence. | This study | Figure 5 |
2.3. Yeast Transformation and β-Galactosidase Activity Assays
2.4. Generation of Growth Curves
2.5. Serial Dilution of Cells for Growth Analysis
2.6. Differential Interference Contrast Microscopy of Yeast Cultures
3. Results
3.1. ras2Δ and pde2Δ Increase and Decrease the Expression of YRO2 and YGP1–lacZ Reporter Gene, Respectively

3.2. A Hyperactive Allele of RAS2, RAS2A18V19, Reduces the Expression of YRO2, YGP1, and TPO2–lacZ Reporter Genes
3.3. Increased Expression of YRO2–, YGP1–, and TPO2–lacZ Reporter Genes Due to ras2Δ Requires Haa1
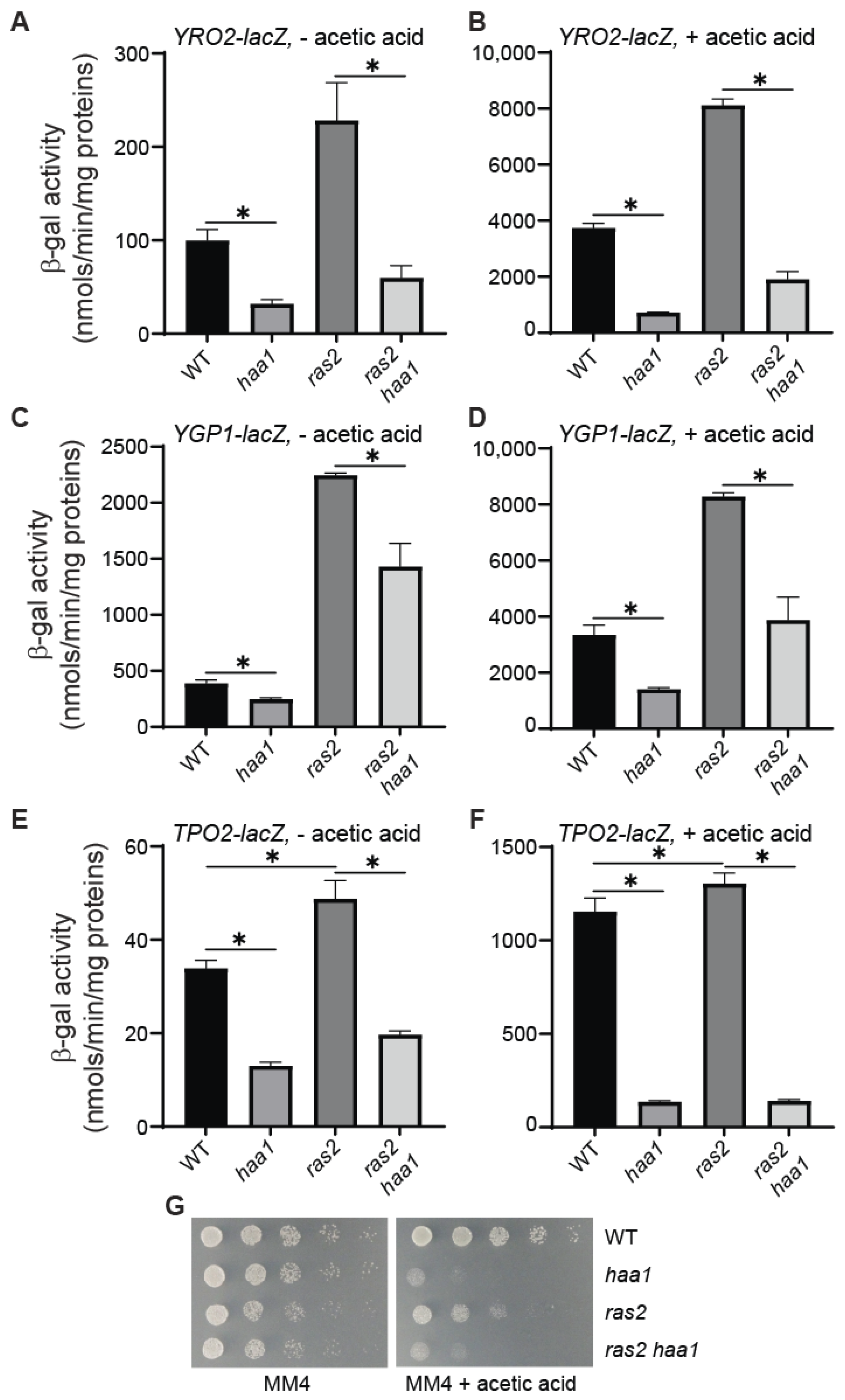
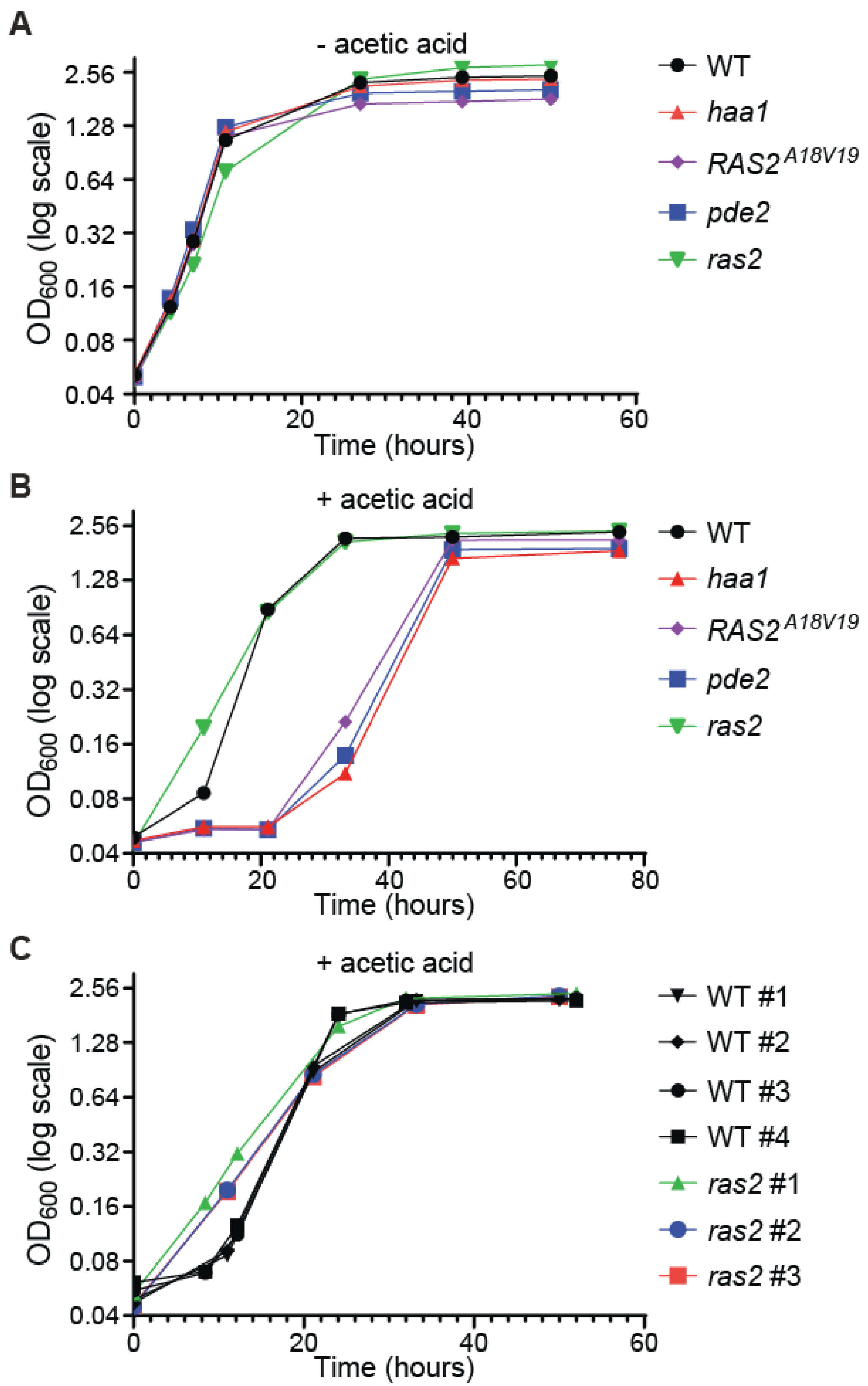
3.4. pde2Δ and RAS2A18V19 Increase Acetic Acid Sensitivity While ras2Δ Improves Cellular Fitness in Response to Acetic Acid Stress
3.5. Mutations in TPK1 (TPK3) and TPK2 Have Opposite Effects on YRO2 and YGP1 Expression

3.6. haa1Δ Largely Abolishes the Increased Expression of YRO2, TPO2, TPO3, and SPI1 in tpk1Δ tpk3Δ Double Mutant Cells
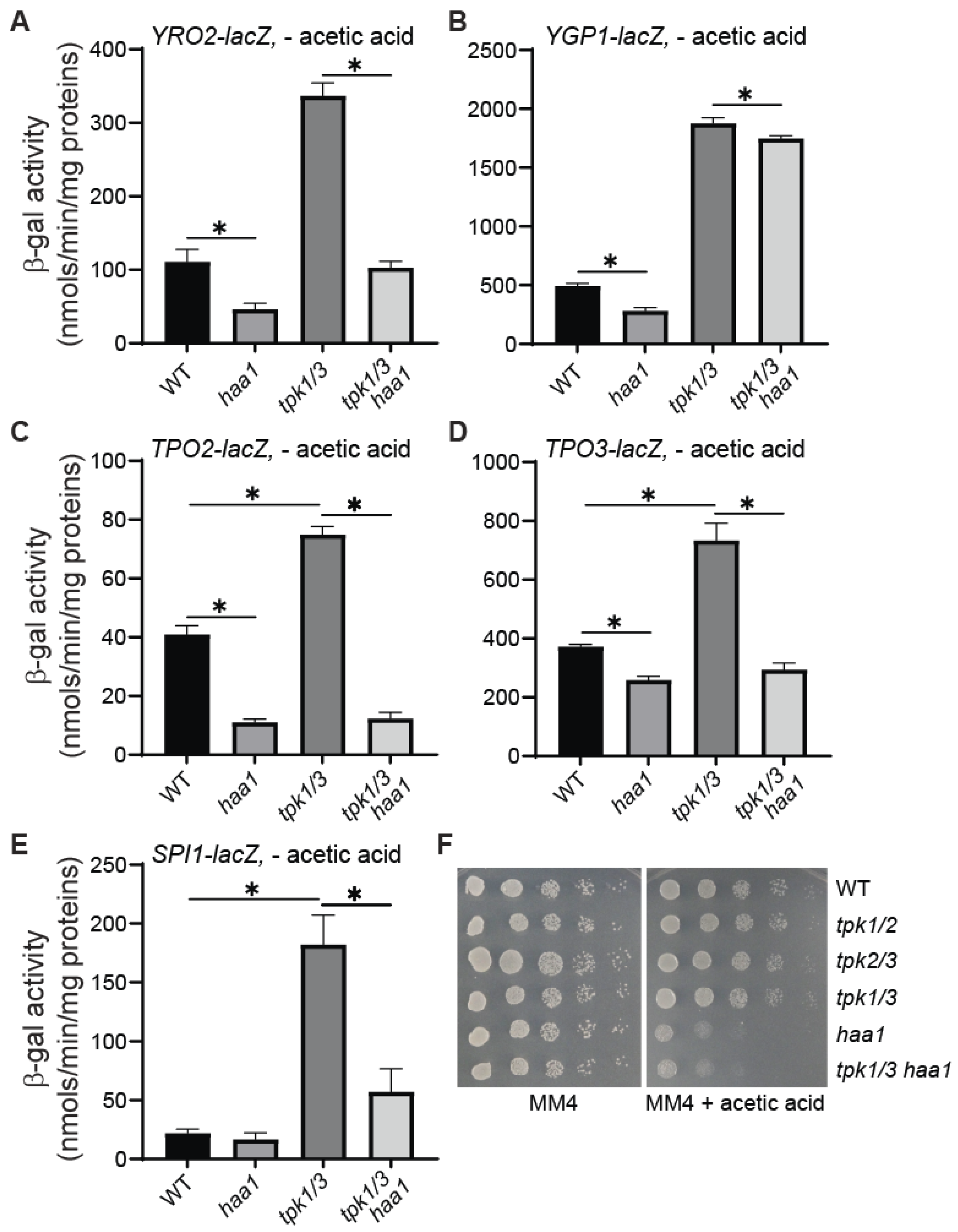
3.7. tpk1Δ tpk3Δ Double Mutants Yield Flocculent Cultures in Response to Acetic Acid Stress

4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mira, N.P.; Teixeira, M.C.; Sa-Correia, I. Adaptive Response and Tolerance to Weak Acids in Saccharomyces cerevisiae: A Genome-Wide View. Omics J. Integr. Biol. 2010, 14, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Kren, A.; Mamnun, Y.M.; Bauer, B.E.; Schuller, C.; Wolfger, H.; Hatzixanthis, K.; Mollapour, M.; Gregori, C.; Piper, P.; Kuchler, K. War1p, a Novel Transcription Factor Controlling Weak Acid Stress Response in Yeast. Mol. Cell Biol. 2003, 23, 1775–1785. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, A.R.; Mira, N.P.; Vargas, R.C.; Canelhas, I.; Sa-Correia, I. Saccharomyces cerevisiae Adaptation to Weak Acids Involves the Transcription Factor Haa1p and Haa1p-Regulated Genes. Biochem. Biophys. Res. Commun. 2005, 337, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Mira, N.P.; Becker, J.D.; Sa-Correia, I. Genomic Expression Program Involving the Haa1p-Regulon in Saccharomyces cerevisiae Response to Acetic Acid. Omics J. Integr. Biol. 2010, 14, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Mira, N.P.; Henriques, S.F.; Keller, G.; Teixeira, M.C.; Matos, R.G.; Arraiano, C.M.; Winge, D.R.; Sa-Correia, I. Identification of a DNA-Binding Site for the Transcription Factor Haa1, Required for Saccharomyces cerevisiae Response to Acetic Acid Stress. Nucleic Acids Res. 2011, 39, 6896–6907. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Ishii, Y.; Ogawa, J.; Shima, J. Enhancement of Acetic Acid Tolerance in Saccharomyces cerevisiae by Overexpression of the HAA1 Gene, Encoding a Transcriptional Activator. Appl. Environ. Microbiol. 2012, 78, 8161–8163. [Google Scholar] [CrossRef] [PubMed]
- Palma, M.; Guerreiro, J.F.; Sa-Correia, I. Adaptive Response and Tolerance to Acetic Acid in Saccharomyces cerevisiae and Zygosaccharomyces bailii: A Physiological Genomics Perspective. Front. Microbiol. 2018, 9, 274. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Cho, K.H.; Park, K.H.; Jang, J.; Hahn, J.S. Activation of Haa1 and War1 Transcription Factors by Differential Binding of Weak Acid Anions in Saccharomyces cerevisiae. Nucleic Acids Res. 2019, 47, 1211–1224. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Nijland, J.G.; Driessen, A.J.M. Combined Roles of Exporters in Acetic Acid Tolerance in Saccharomyces cerevisiae. Biotechnol. Biofuels Bioprod. 2022, 15, 67. [Google Scholar] [CrossRef] [PubMed]
- Chaves, S.R.; Rego, A.; Martins, V.M.; Santos-Pereira, C.; Sousa, M.J.; Corte-Real, M. Regulation of Cell Death Induced by Acetic Acid in Yeasts. Front. Cell Dev. Biol. 2021, 9, 642375. [Google Scholar] [CrossRef]
- Guaragnella, N.; Antonacci, L.; Passarella, S.; Marra, E.; Giannattasio, S. Achievements and Perspectives in Yeast Acetic Acid-induced Programmed Cell Death Pathways. Biochem. Soc. Trans. 2011, 39, 1538–1543. [Google Scholar] [CrossRef] [PubMed]
- Ludovico, P.; Sousa, M.J.; Silva, M.T.; Leao, C.L.; Corte-Real, M. Saccharomyces cerevisiae Commits to a Programmed Cell Death Process in Response to Acetic Acid. Microbiology 2001, 147, 2409–2415. [Google Scholar] [CrossRef] [PubMed]
- Swinnen, S.; Henriques, S.F.; Shrestha, R.; Ho, P.W.; Sa-Correia, I.; Nevoigt, E. Improvement of Yeast Tolerance to Acetic Acid through Haa1 Transcription Factor Engineering: Towards the Underlying Mechanisms. Microb. Cell Fact. 2017, 16, 7. [Google Scholar] [CrossRef] [PubMed]
- Stratford, M.; Nebe-von-Caron, G.; Steels, H.; Novodvorska, M.; Ueckert, J.; Archer, D.B. Weak-Acid Preservatives: pH and proton Movements in the Yeast Saccharomyces cerevisiae. Int. J. Food Microbiol. 2013, 161, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Antunes, M.; Kale, D.; Sychrova, H.; Sa-Correia, I. The Hrk1 Kinase is a Determinant of Acetic Acid Tolerance in Yeast by Modulating H(+) and K(+) Homeostasis. Microb. Cell 2023, 10, 261–276. [Google Scholar] [CrossRef] [PubMed]
- Simoes, T.; Mira, N.P.; Fernandes, A.R.; Sa-Correia, I. The SPI1 Gene, Encoding a Glycosylphosphatidylinositol-Anchored Cell Wall Protein, Plays a Prominent Role in the Development of Yeast Resistance to Lipophilic Weak-Acid Food Preservatives. Appl. Environ. Microbiol. 2006, 72, 7168–7175. [Google Scholar] [CrossRef]
- Takabatake, A.; Kawazoe, N.; Izawa, S. Plasma Membrane Proteins Yro2 and Mrh1 are Required for Acetic Acid Tolerance in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2015, 99, 2805–2814. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, M.; Akase, S.P.; Nakanishi, R.; Horie, H.; Kaneko, Y.; Harashima, S. Nuclear Localization of Haa1, which Is Linked to Its Phosphorylation Status, Mediates Lactic Acid Tolerance in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2014, 80, 3488–3495. [Google Scholar] [CrossRef]
- Collins, M.E.; Black, J.J.; Liu, Z. Casein Kinase I Isoform Hrr25 Is a Negative Regulator of Haa1 in the Weak Acid Stress Response Pathway in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2017, 83, e00672-17. [Google Scholar] [CrossRef]
- Cook, A.; Bono, F.; Jinek, M.; Conti, E. Structural Biology of Nucleocytoplasmic Transport. Annu. Rev. Biochem. 2007, 76, 647–671. [Google Scholar] [CrossRef]
- Thevelein, J.M.; de Winde, J.H. Novel Sensing Mechanisms and Targets for the cAMP-Protein Kinase A Pathway in the Yeast Saccharomyces cerevisiae. Mol. Microbiol. 1999, 33, 904–918. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.; Struhl, K. Protein Kinase A Mediates Growth-Regulated Expression of Yeast Ribosomal Protein Genes by Modulating RAP1 Transcriptional Activity. Mol. Cell Biol. 1994, 14, 1920–1928. [Google Scholar] [PubMed]
- Jorgensen, P.; Rupes, I.; Sharom, J.R.; Schneper, L.; Broach, J.R.; Tyers, M. A Dynamic Transcriptional Network Communicates Growth Potential to Ribosome Synthesis and Critical Cell Size. Genes Dev. 2004, 18, 2491–2505. [Google Scholar] [CrossRef] [PubMed]
- Zurita-Martinez, S.A.; Cardenas, M.E. Tor and cyclic AMP-Protein Kinase A: Two Parallel Pathways Regulating Expression of Genes Required for Cell Growth. Eukaryot. Cell 2005, 4, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.C.; Powers, T. Coordinate Regulation of Multiple and Distinct Biosynthetic Pathways by TOR and PKA Kinases in S. cerevisiae. Curr. Genet. 2006, 49, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Bonomelli, B.; Busti, S.; Martegani, E.; Colombo, S. Active Ras2 in Mitochondria Promotes Regulated Cell Death in a cAMP/PKA Pathway-Dependent Manner in Budding Yeast. FEBS Lett. 2023, 597, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Burtner, C.R.; Murakami, C.J.; Kennedy, B.K.; Kaeberlein, M. A Molecular Mechanism of Chronological Aging in Yeast. Cell Cycle 2009, 8, 1256–1270. [Google Scholar] [CrossRef] [PubMed]
- Malcher, M.; Schladebeck, S.; Mosch, H.U. The Yak1 Protein Kinase Lies at the Center of a Regulatory Cascade Affecting Adhesive Growth and Stress Resistance in Saccharomyces cerevisiae. Genetics 2011, 187, 717–730. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Salas-Navarrete, P.C.; de Oca Miranda, A.I.M.; Martinez, A.; Caspeta, L. Evolutionary and Reverse Engineering to Increase Saccharomyces cerevisiae Tolerance to Acetic Acid, Acidic pH, and High Temperature. Appl. Microbiol. Biotechnol. 2022, 106, 383–399. [Google Scholar] [CrossRef]
- Garrett, S.; Menold, M.M.; Broach, J.R. The Saccharomyces cerevisiae YAK1 Gene Encodes a Protein Kinase that Is Induced by Arrest Early in the Cell Cycle. Mol. Cell Biol. 1991, 11, 4045–4052. [Google Scholar] [CrossRef]
- Smith, A.; Ward, M.P.; Garrett, S. Yeast PKA Represses Msn2p/Msn4p-Dependent Gene Expression to Regulate Growth, Stress Response and Glycogen Accumulation. EMBO J. 1998, 17, 3556–3564. [Google Scholar] [CrossRef] [PubMed]
- Amberg, D.C.; Burke, D.J.; Strathern, J.N. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2005. [Google Scholar]
- Giaever, G.; Chu, A.M.; Ni, L.; Connelly, C.; Riles, L.; Veronneau, S.; Dow, S.; Lucau-Danila, A.; Anderson, K.; Andre, B.; et al. Functional Profiling of the Saccharomyces cerevisiae Genome. Nature 2002, 418, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Chelstowska, A.; Liu, Z.; Jia, Y.; Amberg, D.; Butow, R.A. Signalling between Mitochondria and the Nucleus Regulates the Expression of a New D-Lactate Dehydrogenase Activity in Yeast. Yeast 1999, 15, 1377–1391. [Google Scholar] [CrossRef]
- Xue, Y.; Batlle, M.; Hirsch, J.P. GPR1 Encodes a Putative G Protein-Coupled Receptor that Associates with the Gpa2p Galpha Subunit and Functions in a Ras-Independent Pathway. EMBO J. 1998, 17, 1996–2007. [Google Scholar] [CrossRef]
- Myers, A.M.; Tzagoloff, A.; Kinney, D.M.; Lusty, C.J. Yeast Shuttle and Integrative Vectors with Multiple Cloning Sites Suitable for Construction of lacZ Fusions. Gene 1986, 45, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Gietz, D.; St Jean, A.; Woods, R.A.; Schiestl, R.H. Improved Method for High Efficiency Transformation of Intact Yeast Cells. Nucleic Acids Res. 1992, 20, 1425. [Google Scholar] [CrossRef] [PubMed]
- Keller, G.; Ray, E.; Brown, P.O.; Winge, D.R. Haa1, a Protein Homologous to the Copper-Regulated Transcription Factor Ace1, Is a Novel Transcriptional Activator. J. Biol. Chem. 2001, 276, 38697–38702. [Google Scholar] [CrossRef] [PubMed]
- Broek, D.; Samiy, N.; Fasano, O.; Fujiyama, A.; Tamanoi, F.; Northup, J.; Wigler, M. Differential Activation of Yeast Adenylate Cyclase by Wild-Type and Mutant RAS Proteins. Cell 1985, 41, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Nikawa, J.; Sass, P.; Wigler, M. Cloning and Characterization of the Low-Affinity Cyclic AMP Phosphodiesterase Gene of Saccharomyces cerevisiae. Mol. Cell Biol. 1987, 7, 3629–3636. [Google Scholar] [CrossRef]
- Francois, J.; Neves, M.J.; Hers, H.G. The Control of Trehalose Biosynthesis in Saccharomyces cerevisiae: Evidence for a Catabolite Inactivation and Repression of Trehalose-6-Phosphate Synthase and Trehalose-6-Phosphate Phosphatase. Yeast 1991, 7, 575–587. [Google Scholar] [CrossRef]
- Kataoka, T.; Powers, S.; McGill, C.; Fasano, O.; Strathern, J.; Broach, J.; Wigler, M. Genetic Analysis of Yeast RAS1 and RAS2 Genes. Cell 1984, 37, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Toda, T.; Cameron, S.; Sass, P.; Zoller, M.; Wigler, M. Three Different Genes in S. cerevisiae Encode the Catalytic Subunits of the cAMP-Dependent Protein Kinase. Cell 1987, 50, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Creamer, D.R.; Hubbard, S.J.; Ashe, M.P.; Grant, C.M. Yeast Protein Kinase A Isoforms: A Means of Encoding Specificity in the Response to Diverse Stress Conditions? Biomolecules 2022, 12, 958. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Heitman, J. Cyclic AMP-Dependent Protein Kinase Regulates Pseudohyphal Differentiation in Saccharomyces cerevisiae. Mol. Cell Biol. 1999, 19, 4874–4887. [Google Scholar] [CrossRef] [PubMed]
- Robertson, L.S.; Fink, G.R. The Three Yeast A Kinases Have Specific Signaling Functions in Pseudohyphal Growth. Proc. Natl. Acad. Sci. USA 1998, 95, 13783–13787. [Google Scholar] [CrossRef] [PubMed]
- Cullen, P.J.; Sprague, G.F., Jr. The Regulation of Filamentous Growth in Yeast. Genetics 2012, 190, 23–49. [Google Scholar] [CrossRef] [PubMed]
- Soares, E.V. Flocculation in Saccharomyces cerevisiae: A review. J. Appl. Microbiol. 2011, 110, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Garrett, S.; Broach, J. Loss of Ras activity in Saccharomyces cerevisiae Is Suppressed by Disruptions of a New Kinase Gene, YAKI, Whose Product May Act Downstream of the cAMP-Dependent Protein Kinase. Genes Dev. 1989, 3, 1336–1348. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.E.; Soulard, A.; Hall, M.N. TOR Regulates Ribosomal Protein Gene Expression via PKA and the Forkhead Transcription Factor FHL1. Cell 2004, 119, 969–979. [Google Scholar] [CrossRef]
- Dechant, R.; Binda, M.; Lee, S.S.; Pelet, S.; Winderickx, J.; Peter, M. Cytosolic pH Is a Second Messenger for Glucose and Regulates the PKA Pathway through V-ATPase. EMBO J. 2010, 29, 2515–2526. [Google Scholar] [CrossRef]
- Colombo, S.; Ma, P.; Cauwenberg, L.; Winderickx, J.; Crauwels, M.; Teunissen, A.; Nauwelaers, D.; de Winde, J.H.; Gorwa, M.F.; Colavizza, D.; et al. Involvement of Distinct G-Proteins, Gpa2 and Ras, in Glucose- and Intracellular Acidification-Induced cAMP Signalling in the Yeast Saccharomyces cerevisiae. EMBO J. 1998, 17, 3326–3341. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bourgeois, N.M.; Black, J.J.; Bhondeley, M.; Liu, Z. Protein Kinase A Negatively Regulates the Acetic Acid Stress Response in S. cerevisiae. Microorganisms 2024, 12, 1452. https://doi.org/10.3390/microorganisms12071452
Bourgeois NM, Black JJ, Bhondeley M, Liu Z. Protein Kinase A Negatively Regulates the Acetic Acid Stress Response in S. cerevisiae. Microorganisms. 2024; 12(7):1452. https://doi.org/10.3390/microorganisms12071452
Chicago/Turabian StyleBourgeois, Natasha M., Joshua J. Black, Manika Bhondeley, and Zhengchang Liu. 2024. "Protein Kinase A Negatively Regulates the Acetic Acid Stress Response in S. cerevisiae" Microorganisms 12, no. 7: 1452. https://doi.org/10.3390/microorganisms12071452
APA StyleBourgeois, N. M., Black, J. J., Bhondeley, M., & Liu, Z. (2024). Protein Kinase A Negatively Regulates the Acetic Acid Stress Response in S. cerevisiae. Microorganisms, 12(7), 1452. https://doi.org/10.3390/microorganisms12071452






