Amino Acid-Induced Chemotaxis Plays a Key Role in the Adaptation of Vibrio harveyi from Seawater to the Muscle of the Host Fish
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Materials
2.2. Preparation of Muscle Extract and 2216E Muscle Medium
2.3. Transcriptome Sequencing and Data Analysis
2.3.1. Bacterial Sample Preparation
2.3.2. Transcriptome Sequencing
2.4. qRT-PCR
2.5. Induction of V. harveyi by Host Fish Muscle and Its Dominant Amino Acids
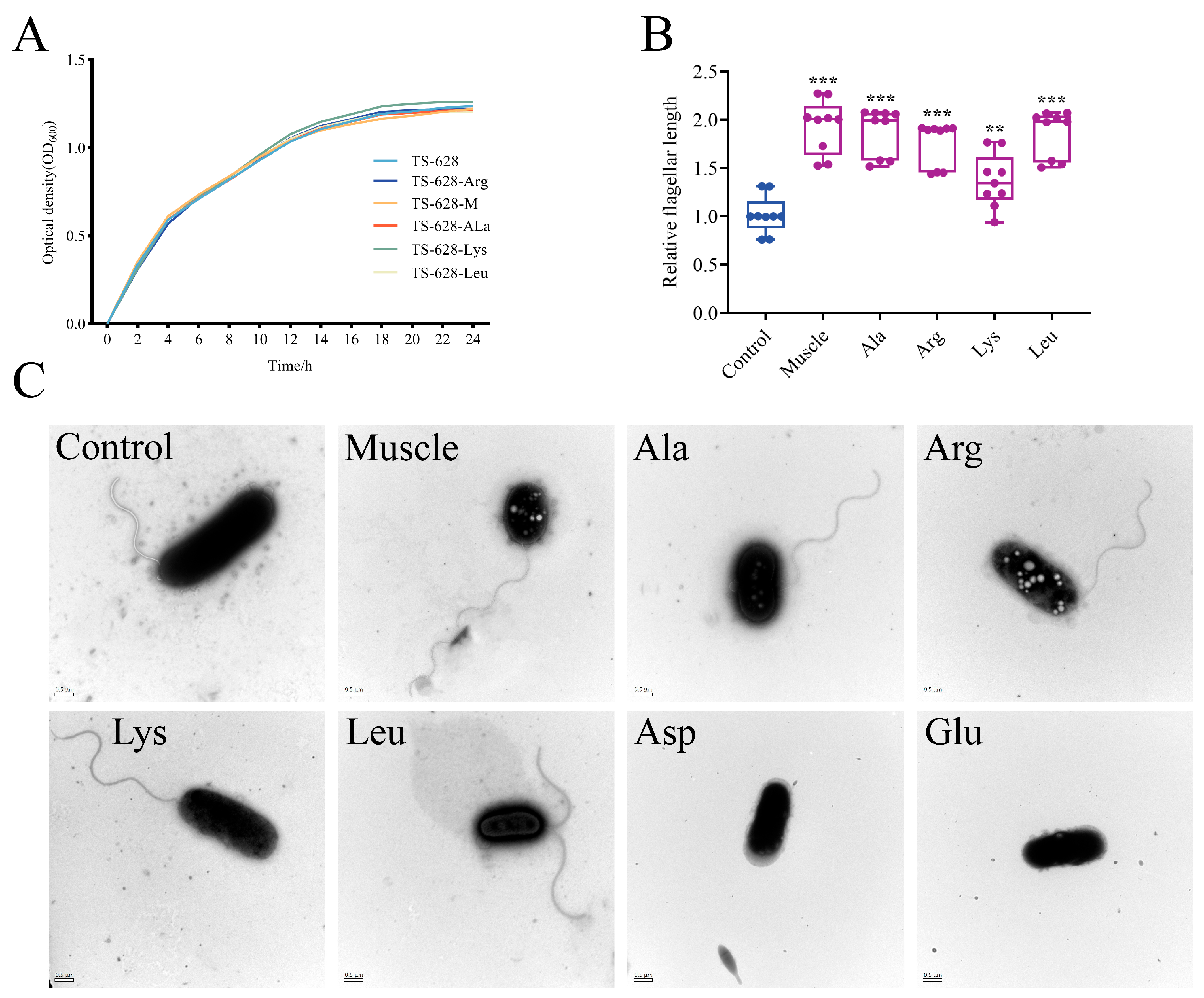
2.6. Bacterial Growth Curves
2.7. Bacterial Flagella Observation
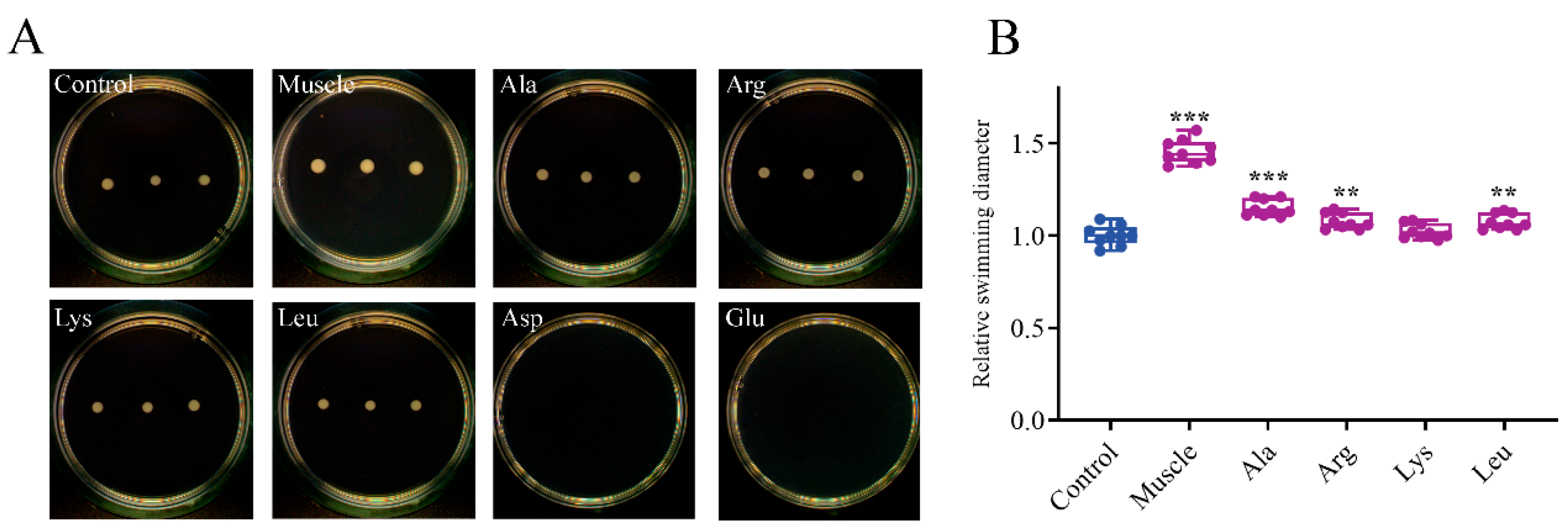
2.8. Bacterial Swimming, Swarming, and Twitching
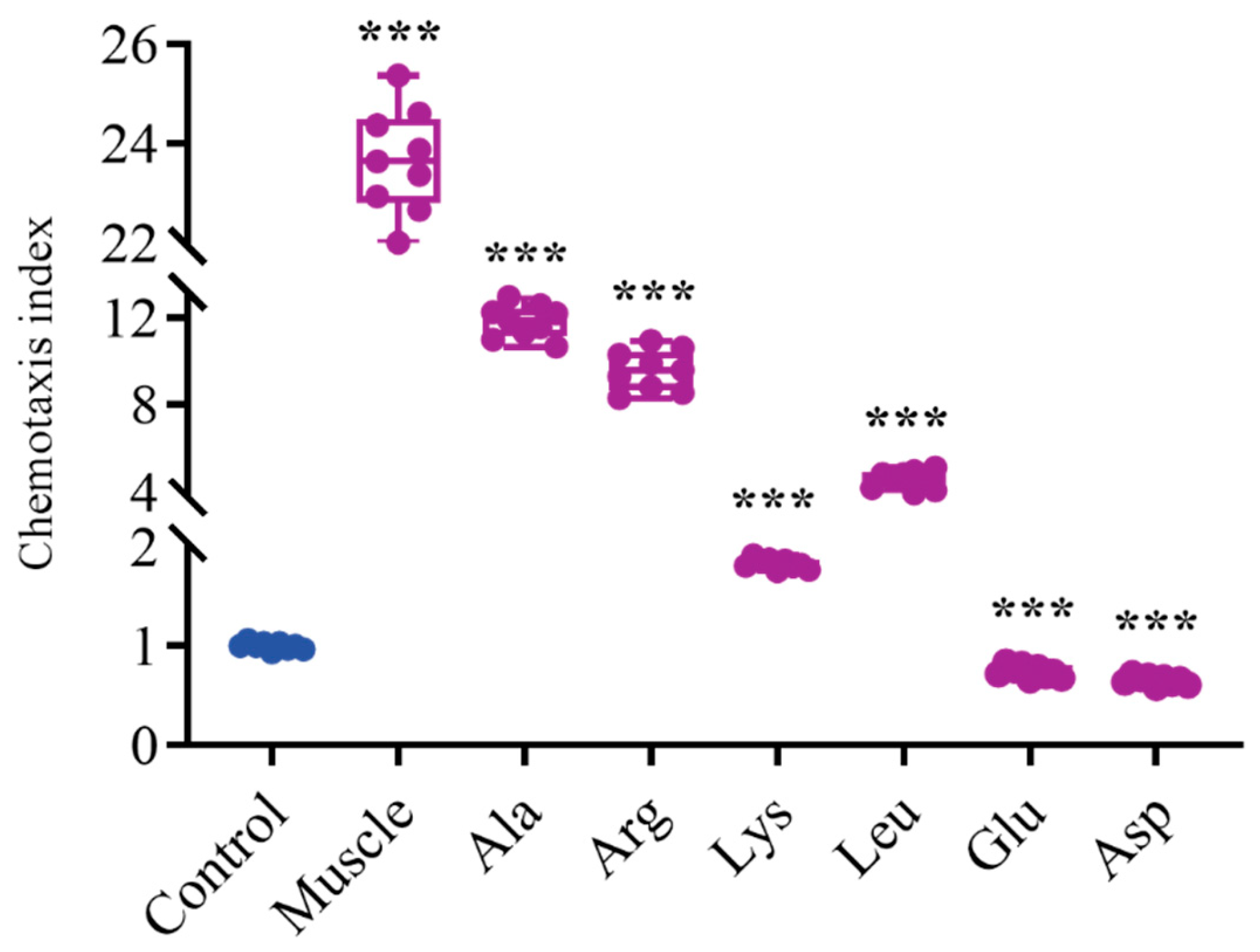
2.9. Bacterial Chemotaxis
2.10. Bacterial Adhesion
2.11. Bacterial Biofilm
2.12. Effects of Host Muscle and Its Dominant Amino Acids on Gene Expression Relative to the Chemotactic Process of V. harveyi
2.13. Data Analysis
3. Results
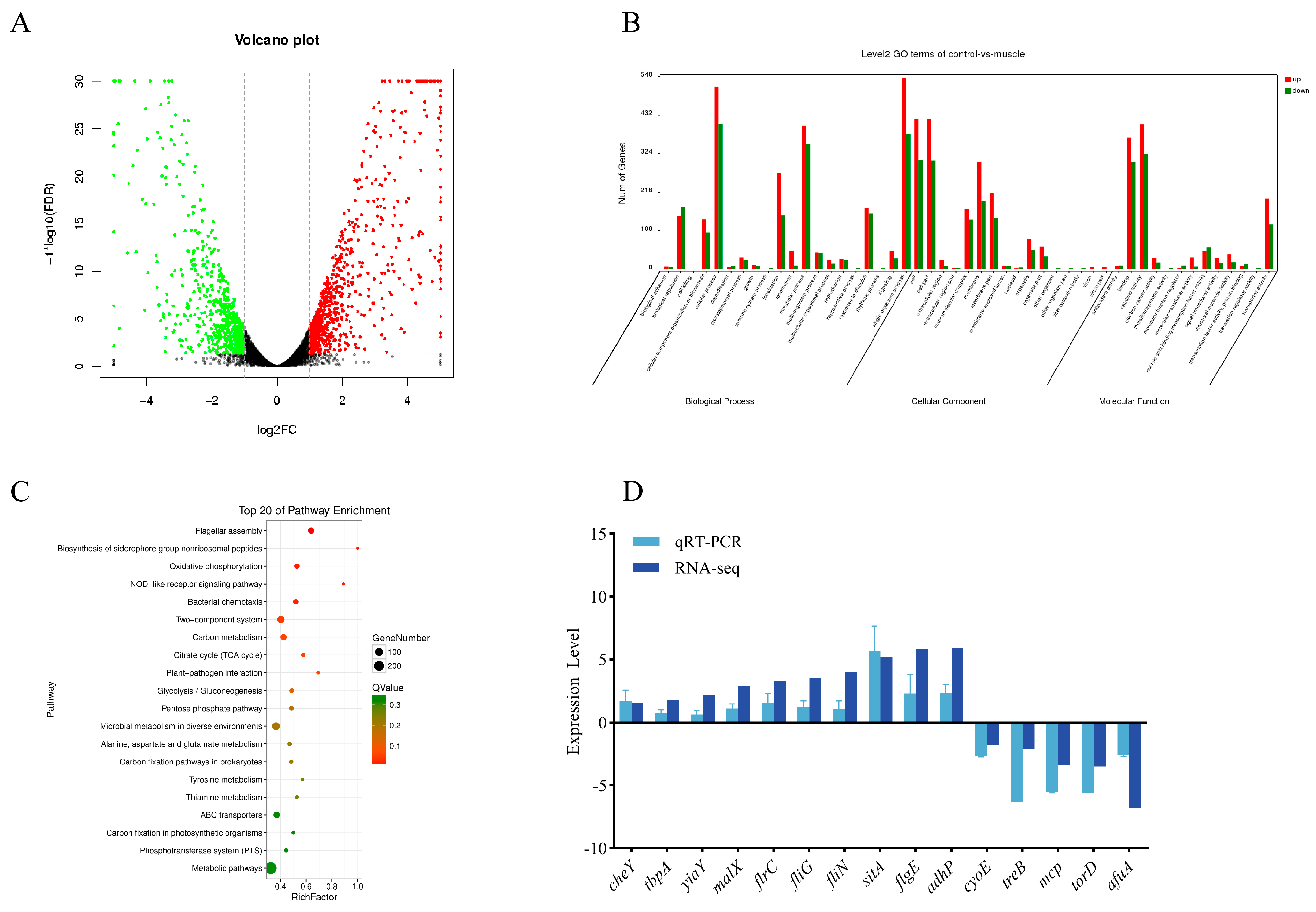
3.1. Transcriptome Analysis
3.2. Effects of Host Muscle and Its Dominant Amino Acids on the Flagellum of V. harveyi
3.3. Effects of Host Muscle and Its Dominant Amino Acids on Swimming, Swarming, and Twitching of V. harveyi
3.4. Effects of Host Muscle and Dominant Amino Acids on the Chemotaxis of V. harveyi
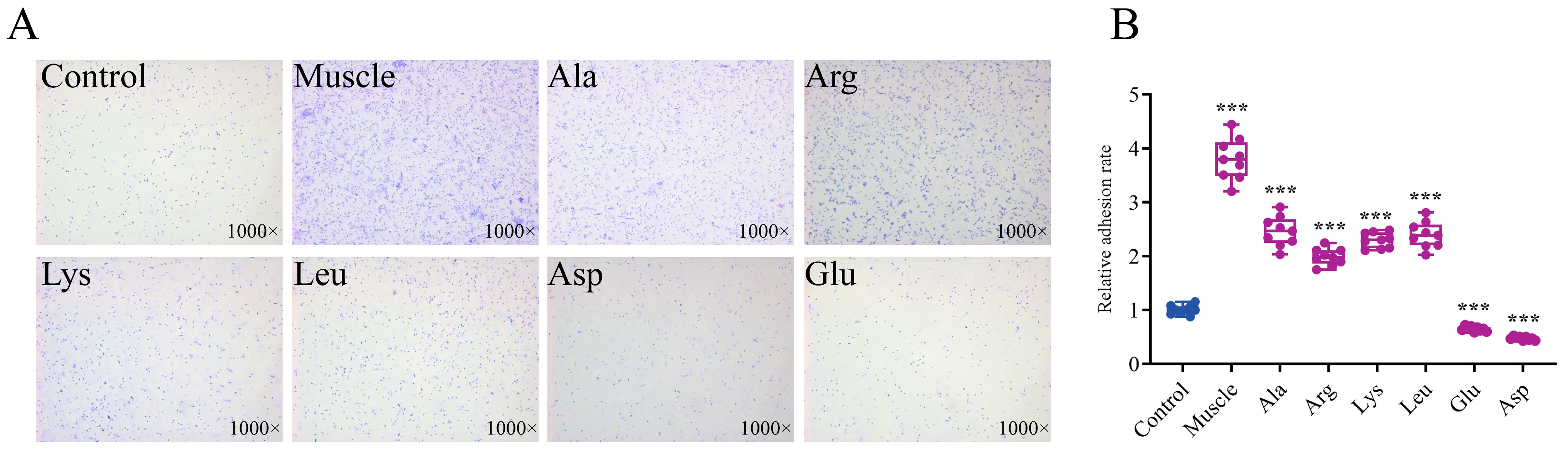
3.5. Effects of Host Muscle and Its Dominant Amino Acids on the Adhesion of V. harveyi
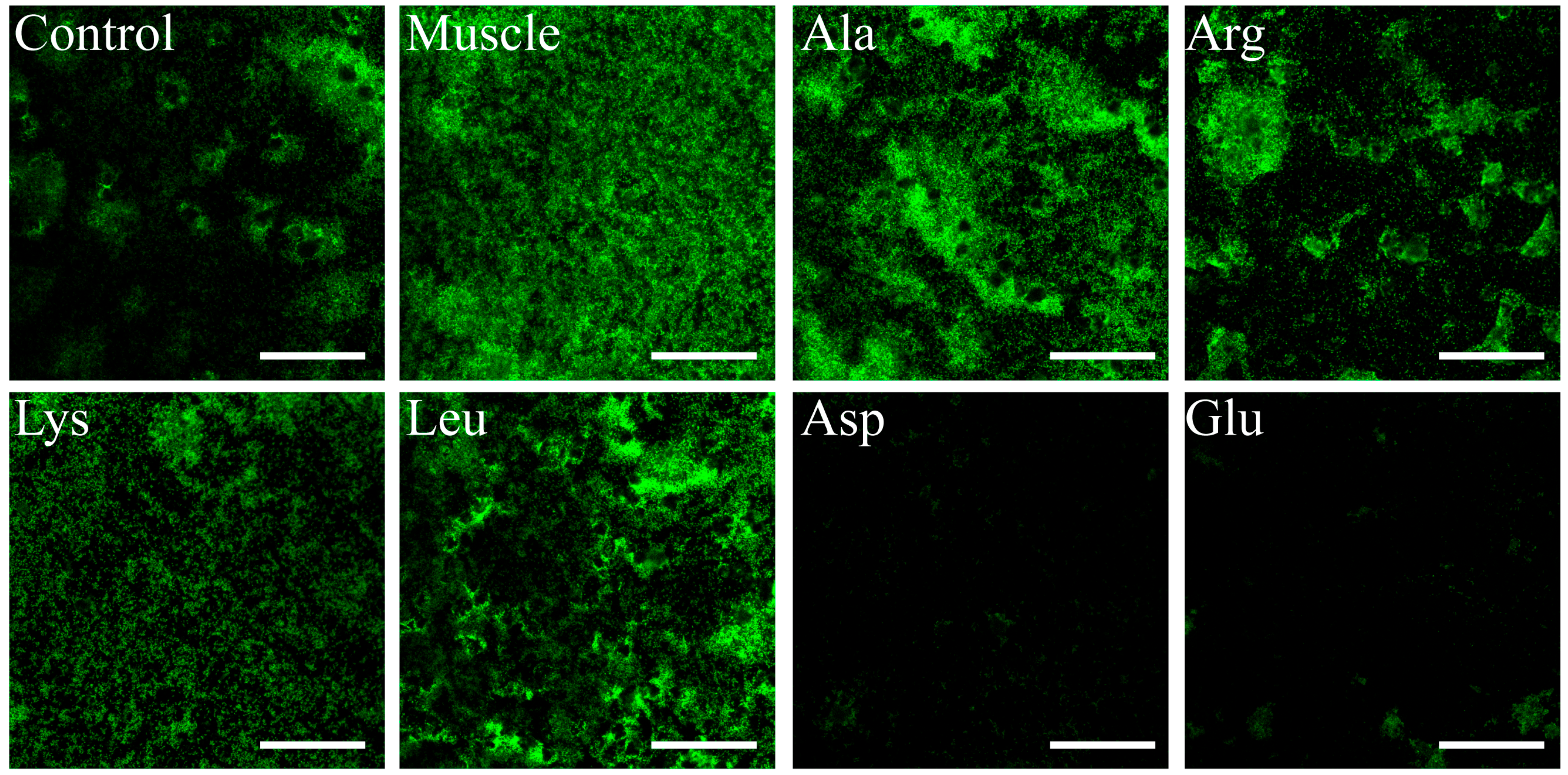
3.6. Effects of Host Muscle and Its Dominant Amino Acids on Biofilm Formation of V. harveyi
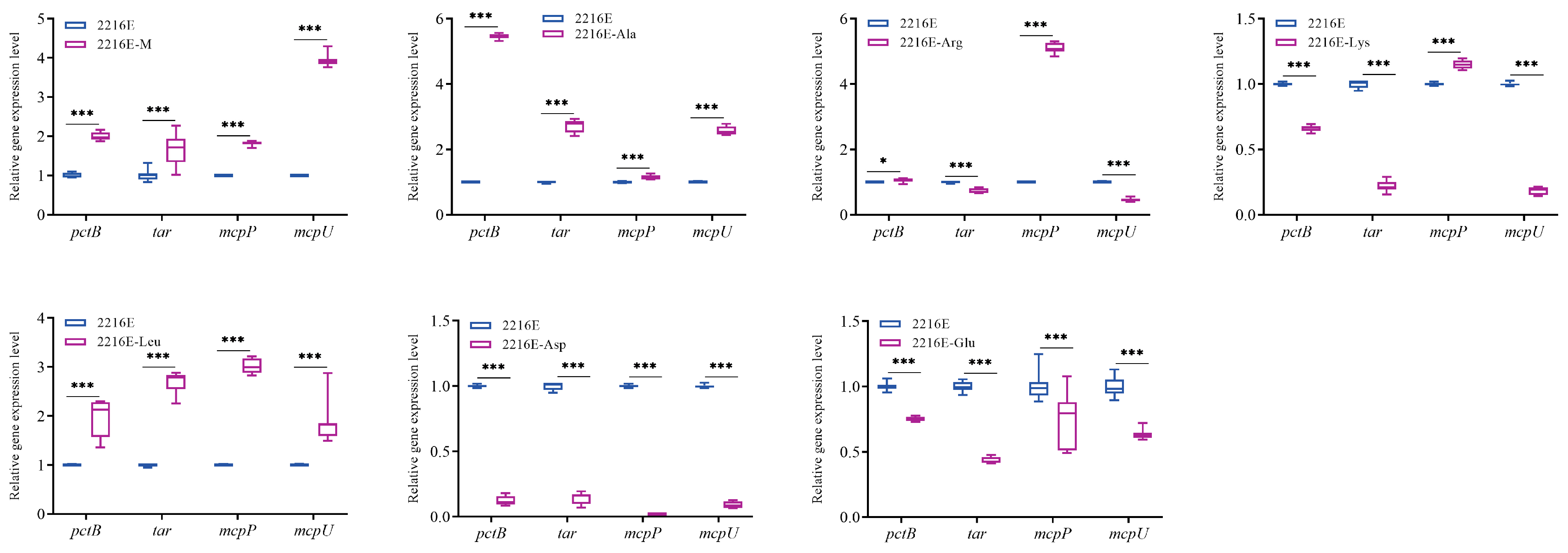
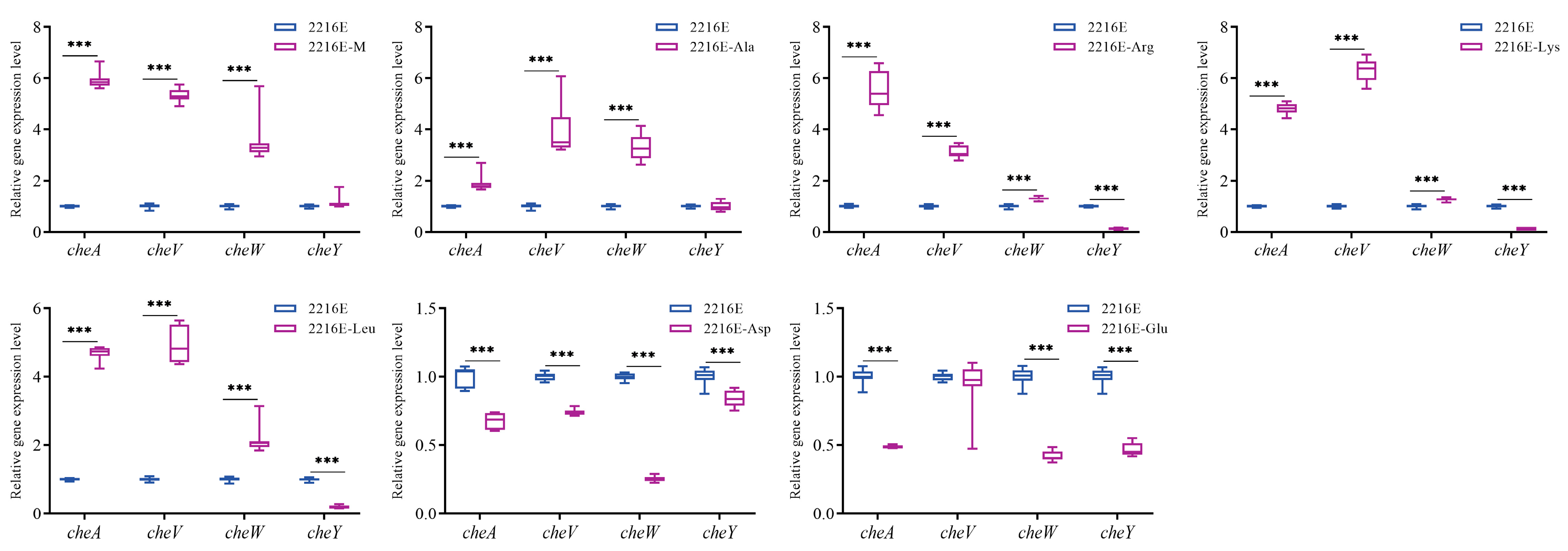
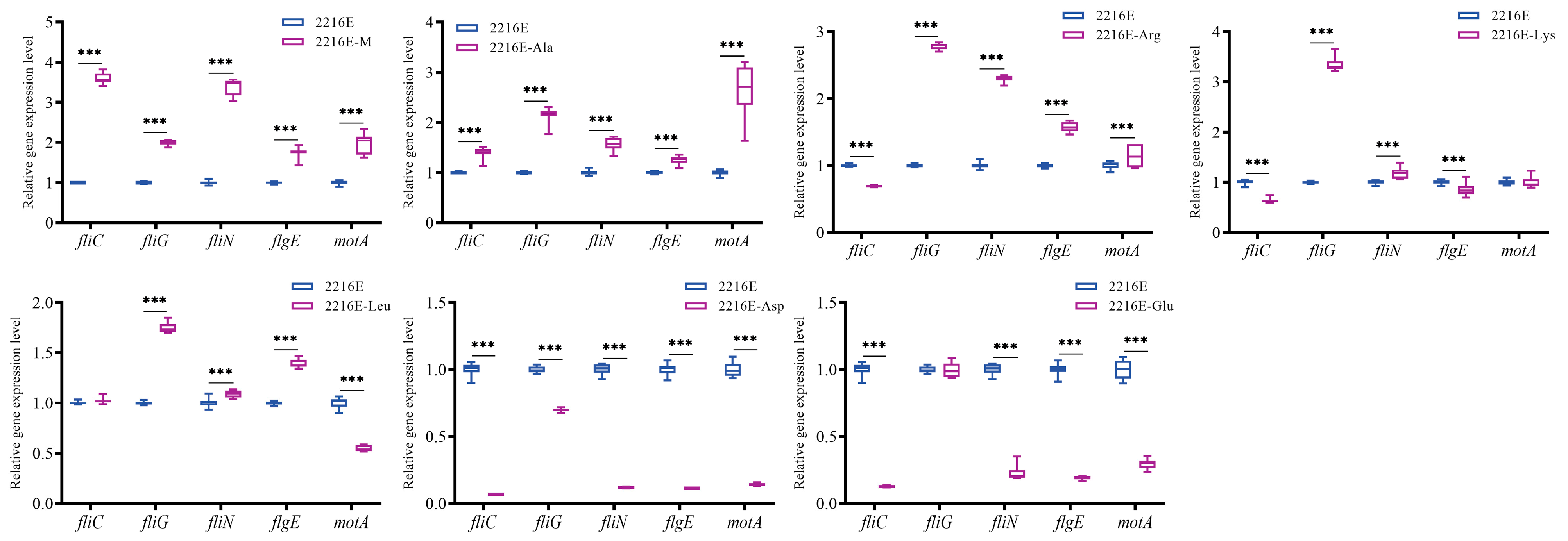
3.7. Effects of Host Muscle and Its Dominant Amino Acids on the Expression of Genes Related to the Bacterial Chemotaxis Process
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Triga, A.; Smyrli, M.; Katharios, P. Pathogenic and opportunistic Vibrio spp. Associated with Vibriosis incidences in the Greek aquaculture: The role of Vibrio harveyi as the principal cause of Vibriosis. Microorganisms 2023, 11, 1197. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Wu, H.; Guo, W.; Li, X.; Wang, J.; Duan, Y.; Zhang, P.; Huang, Z.; Li, Y.; Dong, G.; et al. Vibrio harveyi co-infected with Cryptocaryon irritans to orange-spotted groupers Epinephelus coioides. Fish Shellfish Immunol. 2023, 139, 108879. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Roh, H.; Kim, A.; Park, J.; Lee, J.Y.; Kim, Y.J.; Kang, H.G.; Kim, S.; Kim, H.S.; Cha, H.J.; et al. Molecular mechanisms underlying the vulnerability of Pacific abalone (Haliotis discus hannai) to Vibrio harveyi infection at higher water temperature. Fish Shellfish Immunol. 2023, 138, 108844. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; He, X.; Austin, B. Vibrio harveyi: A serious pathogen of fish and invertebrates in mariculture. Mar. Life Sci. Technol. 2020, 2, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.T.; Taengphu, S.; Sangsuriya, P.; Charoensapsri, W.; Phiwsaiya, K.; Sornwatana, T.; Khunrae, P.; Rattanarojpong, T.; Senapin, S. Recovery of Vibrio harveyi from scale drop and muscle necrosis disease in farmed barramundi, Lates calcarifer in Vietnam. Aquaculture 2017, 473, 89–96. [Google Scholar] [CrossRef]
- Shen, G.; Shi, C.; Fan, C.; Jia, D.; Wang, S.; Xie, G.; Li, G.; Mo, Z.; Huang, J. Isolation, identification and pathogenicity of Vibrio harveyi, the causal agent of skin ulcer disease in juvenile hybrid groupers Epinephelus fuscoguttatus × Epinephelus lanceolatus. J. Fish Dis. 2017, 40, 1351–1362. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Wang, L.; Wen, H.; Wang, K.; Qi, X.; Li, J.; He, F.; Li, Y. Genome-wide identification and characterization of toll-like receptor genes in spotted sea bass (Lateolabrax maculatus) and their involvement in the host immune response to Vibrio harveyi infection. Fish Shellfish Immunol. 2019, 92, 782–791. [Google Scholar] [CrossRef]
- Tian, Y.; Wen, H.; Qi, X.; Mao, X.; Shi, Z.; Li, J.; He, F.; Yang, W.; Zhang, X.; Li, Y. Analysis of apolipoprotein multigene family in spotted sea bass (Lateolabrax maculatus) and their expression profiles in response to Vibrio harveyi infection. Fish Shellfish Immunol. 2019, 92, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Alteri, C.J.; Mobley, H.L.T. Escherichia coli physiology and metabolism dictates adaptation to diverse host microenvironments. Curr. Opin. Microbiol. 2012, 15, 3–9. [Google Scholar] [CrossRef]
- Jones, S.A.; Gibson, T.; Maltby, R.C.; Chowdhury, F.Z.; Stewart, V.; Cohen, P.S.; Conway, T. Anaerobic respiration of Escherichia coli in the mouse intestine. Infect. Immun. 2011, 79, 4218–4226. [Google Scholar] [CrossRef]
- Jones, S.A.; Jorgensen, M.; Chowdhury, F.Z.; Rodgers, R.; Hartline, J.; Leatham, M.P.; Struve, C.; Krogfelt, K.A.; Cohen, P.S.; Conway, T. Glycogen and maltose utilization by Escherichia coli O157:H7 in the mouse intestine. Infect. Immun. 2008, 76, 2531–2540. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Kuskowski, M.A.; Gajewski, A.; Soto, S.; Horcajada, J.P.; de Anta, M.T.J.; Vila, J. Extended virulence genotypes and phylogenetic background of Escherichia coli isolates from patients with cystitis, pyelonephritis, or prostatitis. J. Infect. Dis. 2005, 191, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Russo, T.A. Molecular epidemiology of extraintestinal pathogenic Escherichia coli. EcoSal Plus 2018, 8, 26443356. [Google Scholar] [CrossRef] [PubMed]
- Russo, T.; Johnson, J.R. Medical and economic impact of extraintestinal infections due to Escherichia coli: Focus on an increasingly important endemic problem. Microbes Infect. 2003, 5, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Szymanski, C.M.; Baylink, A. Bacterial chemotaxis in human diseases. Trends Microbiol. 2023, 31, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Xi, D.; Wang, X.; Li, Y.; Li, Y.; Yan, J.; Cao, B. Vibrio cholerae VC1741 (PsrA) enhances the colonization of the pathogen in infant mice intestines in the presence of the long-chain fatty acid, oleic acid. Microb. Pathog. 2020, 147, 104443. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xing, X.; Wang, J.; Pang, B.; Liu, M.; Larios-Valencia, J.; Liu, T.; Liu, G.; Xie, S.; Hao, G.; et al. Hypermutation-induced in vivo oxidative stress resistance enhances Vibrio cholerae host adaptation. PLoS Pathog. 2018, 14, e1007413. [Google Scholar] [CrossRef]
- Patankar, Y.R.; Lovewell, R.R.; Poynter, M.E.; Jyot, J.; Kazmierczak, B.I.; Berwin, B. Flagellar motility is a key determinant of the magnitude of the inflammasome response to Pseudomonas aeruginosa. Infect. Immun. 2013, 81, 2043–2052. [Google Scholar] [CrossRef] [PubMed]
- Aso, H.; Miyoshi, S.I.; Nakao, H.; Okamoto, K.; Yamamoto, S. Induction of an outer membrane protein of 78 kda in Vibrio vulnificus cultured in the presence of desferrioxamine B under iron-limiting conditions. Fems Microbiol. Lett. 2002, 212, 65–70. [Google Scholar] [CrossRef]
- Lima, A.; Zunino, P.; D’Alessandro, B.; Piccini, C. An iron-regulated outer-membrane protein of Proteus mirabilis is a haem receptor that plays an important role in urinary tract infection and in vivo growth. J. Med. Microbiol. 2007, 56, 1600–1607. [Google Scholar] [CrossRef]
- Lan, Y.; Zhou, M.; Li, X.; Liu, X.; Li, J.; Liu, W. Preliminary investigation of iron acquisition in hypervirulent Klebsiella pneumoniae mediated by outer membrane vesicles. Infect. Drug Resist. 2022, 15, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Wang, J.; Wang, S.; Yan, Q. Study on the antigenicity of Vibrio harveyi TS-628 strain. Front. Biol. China 2007, 2, 263–267. [Google Scholar] [CrossRef]
- Meng, X.; Shen, Y.; Wang, S.; Xu, X.; Dang, Y.; Zhang, M.; Li, L.; Zhang, J.; Wang, R.; Li, J. Complement component 3 (C3): An important role in grass carp (Ctenopharyngodon idella) experimentally exposed to Aeromonas hydrophila. Fish Shellfish Immunol. 2019, 88, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Peng, K.; Wu, J.; Chen, J. Transgenic expression of salmon delta-5 and delta-6 desaturase in zebrafish muscle inhibits the growth of Vibrio alginolyticus and affects fish immunomodulatory activity. Fish Shellfish Immunol. 2014, 39, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lin, Y.; Lu, H.; Su, Y.; Liu, Z. Analysis and evaluation of nutritional components in the muscle of four grouper species. J. Fish. Res. 2020, 42, 463–472. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, D.; Ma, J.; Li, B.; Zhang, L. Nutritional components analysis and nutritive value evaluation of Epinephelus fuscoguttatus × Epinephelus lanceolatus muscles. Trans. Oceanol. Limnol. 2015, 4, 61–69. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, Q.; Yang, Z.; Li, Y.; Yan, Y.; Ping, S.; Zhang, L.; Lin, M.; Lu, W. Identification of the nitrogen-fixing Pseudomonas stutzeri major flagellar gene regulator FleQ and its role in biofilm formation and root colonization. Agriculture 2016, 15, 339–348. [Google Scholar] [CrossRef][Green Version]
- Zhang, M.; Yan, Q.; Mao, L.; Wang, S.; Huang, L.; Xu, X.; Qin, Y. KatG plays an important role in Aeromonas hydrophila survival in fish macrophages and escape for further infection. Gene 2018, 672, 156–164. [Google Scholar] [CrossRef]
- Jiao, J.; Zhao, L.; Huang, L.; Qin, Y.; Su, Y.; Zheng, W.; Zhang, J.; Yan, Q. The contributions of fliG gene to the pathogenicity of Pseudomonas plecoglossicida and pathogen-host interactions with Epinephelus coioides. Fish Shellfish Immunol. 2021, 119, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Badal, D.; Jayarani, A.V.; Kollaran, M.A.; Prakash, D.; Monisha, P.; Singh, V. Foraging signals promote swarming in starving Pseudomonas aeruginosa. mBio 2021, 12, e02033-21. [Google Scholar] [CrossRef]
- Huang, L.; Guo, L.; Xu, X.; Qin, Y.; Zhao, L.; Su, Y.; Yan, Q. The role of rpoS in the regulation of Vibrio alginolyticus virulence and the response to diverse stresses. J. Fish Dis. 2019, 42, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.H.; Arthur, K. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2000, 97, 4885–4890. [Google Scholar] [CrossRef]
- Zhang, Z.; Mao, L.; Qin, Y.; Zhao, L.; Huang, L.; Xu, X.; Qin, Y. Comparative transcriptome analysis revealed the role and mechanism of a FeoC-like LuxR-type regulator in intracellular survival of Aeromonas hydrophila. Aquaculture 2022, 556, 738287. [Google Scholar] [CrossRef]
- Wu, W.; Zhao, L.; Huang, L.; Qin, Y.; Zhang, J.; Zhao, J.; Yan, Q. Transcriptomic and metabolomic insights into the role of fliS in the pathogenicity of Pseudomonas plecoglossicida against Epinephelus coioides. Front. Mar. Sci. 2022, 9, 987825. [Google Scholar] [CrossRef]
- Cai, H.; Ma, Y.; Qin, Y.; Zhao, L.; Yan, Q.; Huang, L. Vvrr2: A new Vibrio ncRNA involved in dynamic synthesis of multiple biofilm matrix exopolusaccharides, biofilm structuring and virulence. Aquaculture 2023, 563, 738925. [Google Scholar] [CrossRef]
- Wang, J.; Wu, Z.; Wang, S.; Wang, X.; Zhang, D.; Wang, Q.; Lin, L.; Wang, G.; Guo, Z.; Chen, Y. Inhibitory effect of probiotic Bacillus spp. isolated from the digestive tract of Rhynchocypris Lagowskii on the adhesion of common pathogenic bacteria in the intestinal model. Microb. Pathog. 2022, 169, 105623. [Google Scholar] [CrossRef]
- Mao, L.; Qin, Y.; Kang, J.; Wu, B.; Huang, L.; Wang, S.; Zhang, M.; Zhang, J.; Zhang, R.; Yan, Q. Role of LuxR-type regulators in fish pathogenic Aeromonas hydrophila. J. Fish Dis. 2019, 43, 215–225. [Google Scholar] [CrossRef]
- Wooten, R.M.; Pakkulnan, R.; Anutrakunchai, C.; Kanthawong, S.; Taweechaisupapong, S.; Chareonsudjai, P.; Chareonsudjai, S. Extracellular DNA facilitates bacterial adhesion during Burkholderia pseudomallei biofilm formation. PLoS ONE 2019, 14, e0213288. [Google Scholar] [CrossRef]
- Qiu, X.; Cao, X.; Jian, H.; Wu, H.; Xu, G.; Tang, X. Transcriptomic analysis Reveals that changes in gene expression contribute to Microbacterium sediminis YLB-01 adaptation at low temperature under high hydrostatic pressure. Curr. Microbiol. 2022, 79, 95. [Google Scholar] [CrossRef]
- Cheng, Q.; McKeown, S.J.; Santos, L.; Santiago, F.S.; Khachigian, L.M.; Morand, E.F.; Hickey, M.J. Macrophage migration inhibitory factor increases leukocyte–endothelial interactions in human endothelial cells via promotion of expression of adhesion molecules. J. Immunol. 2010, 185, 1238–1247. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, G.F.; Malachowa, N.; Kobayashi, S.D.; Sturdevant, D.E.; Scott, D.P.; DeLeo, F.R. Insights into the Staphylococcus aureus-host interface: Global changes in host and pathogen gene expression in a rabbit skin infection model. PLoS ONE 2015, 10, e0117713. [Google Scholar] [CrossRef]
- Li, M.; Meng, H.; Li, H.; Gu, D. A novel transcription factor VPA0041 was identified to regulate the swarming motility in Vibrio parahaemolyticus. Pathogens 2022, 11, 453. [Google Scholar] [CrossRef]
- Dearns, D.B. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 2010, 8, 634–643. [Google Scholar] [CrossRef]
- Miller, L.D.; Russell, M.H.; Alexandre, G. Chapter 3 diversity in bacterial chemotactic responses and niche adaptation. Adv. Appl. Microbiol. 2009, 66, 53–75. [Google Scholar] [CrossRef]
- Lee, S.H.; Butler, S.M.; Camilli, A. Selection for in vivo regulators of bacterial virulence. Proc. Natl. Acad. Sci. USA 2001, 98, 6889–6894. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Y.; Niu, H.; Liu, Y.; Ma, Y.; Wang, X.; Li, Z.; Dong, Q. Different cellular fatty acid pattern and gene expression of planktonic and biofilm state Listeria monocytogenes under nutritional stress. Food Res. Int. 2023, 167, 112698. [Google Scholar] [CrossRef]
- Adachi, Y.; Sousa-Coelho, L.D.; Harata, I.; Aoun, C.; Weimer, S.; Shi, X.; Herrera, K.N.G.; Takahashi, H.; Doherty, C.; Noguchi, Y.; et al. l-Alanine activates hepatic AMP-activated protein kinase and modulates systemic glucose metabolism. Mol. Metab. 2018, 17, 61–70. [Google Scholar] [CrossRef]
- Kim, S.H.; Schneider, B.L.; Reitzer, L. Genetics and regulation of the major enzymes of Alanine synthesis in Escherichia coli. J. Bacteriol. 2010, 192, 5304–5311. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Su, Y.; Han, Y.; Guo, C.; Tian, Y.; Peng, X. Exogenous Alanine and/or Glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab. 2015, 21, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Xie, M.; Yang, Y.; Feng, J.; Tan, L.; Chen, C. The role of l-Alanine metabolism revealed by transcriptome analysis in Vibrio alginolyticus. Gene Rep. 2018, 10, 184–187. [Google Scholar] [CrossRef]
- Brijesh, K.; Cardona, S.T. Synthetic cystic fibrosis sputum medium regulates flagellar biosynthesis through the flhF gene in Burkholderia cenocepacia. Front. Cell. Infect. Microbiol. 2016, 6, 65. [Google Scholar] [CrossRef]
- Yang, Y.; Pollard, A.M.; Höfler, C.; Poschet, G.; Wirtz, M.; Hell, R.; Sourjik, V. Relation between chemotaxis and consumption of amino acids in bacteria. Mol. Microbiol. 2015, 96, 1272–1282. [Google Scholar] [CrossRef]



| Primers | Sequences |
|---|---|
| 16S rRNA-F/R | 5′-GGAGCAAACAGGATTAGATACCC-3′/5′-TTCTGATTCCCGAAGGCAC-3′ |
| cheY-F/R | 5′-CGGAAGGTGAGATGATTG-3′/5′-CTTTTCGCGTATGGATAA-3′ |
| tbpA-F/R | 5′-CTTAACCGCCTTCGCTTGG-3′/5′-GCTTTTTGGAGGATTCGCC-3′ |
| yiaY-F/R | 5′-GTCTTGCTTTGCTCCCTGTC-3′/5′-CTATCCTCACCTTGCGTTCC-3′ |
| malX-F/R | 5′-CTCGTCTTCGCTTGTCTGTT-3′/5′-GGCTTGCATTTCATTCTTCA-3′ |
| flrC-F/R | 5′-TTGTGGCATGTCATCAGAAG-3′/5′-CGAACGGACGAAGTAGGTAG-3′ |
| fliG-F/R | 5′-TCTGCTCGTGGTGTCATCC-3′/5′-TTCATCGCCATACAGGTTG-3′ |
| fliN-F/R | 5′-AAGAGCATTCCAGTGACCGTA-3′/5′-CGACTTCTGCTTGTCCCATTA-3′ |
| sitA-F/R | 5′-TGCTGAAGCATACAAAAAGC-3′/5′-TCACTGAATACCGCAGAGAT -3′ |
| flgE-F/R | 5′-TCAACTCTTCTTACACCACC-3′/5′-TCTACATTCACTTGCCATTC-3′ |
| adhP-F/R | 5′-GTGTTGGTGTTCCTTGGTTG-3′/5′-TGTTACTTTGAGCGCCTTAT-3′ |
| cyoE-F/R | 5′-ACCATGTGGTACAAACGAA-3′/5′-TAACGGGAAGAACAGGAAT-3′ |
| treB-F/R | 5′-ACAAAGCCGATACAAAACAG-3′/5′-TTCAGCAAGGTGGGAAATAC-3′ |
| mcp-F/R | 5′-TTGCTGCTGGTGACTCTGA-3′/5′-TTTCTTTTGCCGCTTCTTT-3′ |
| torD-F/R | 5′-GAGAAACGCGCAGAAATC-3′/5′-CCAAGACCCGCTAAAAAG-3′ |
| afuA-F/R | 5′-ACTCTTACCGTCAACCTTTC-3′/5′-GATTTTGCTGTCTACCTTTT-3′ |
| fliC-F/R | 5′-CAACGCAAACTCAGCACAA-3′/5′-TACGAACAGCCACATCCAA-3′ |
| motA-F/R | 5′-TGGGTTCGGTATTCTTGC-3′/5′-GTTTCGTTTCACTCGCTG-3′ |
| pctB-F/R | 5′-TGTGGTGTTCGTGGTGTTACTG-3′/5′-ACTCAATGTCATCACTTCGGTCAA-3′ |
| tar-F/R | 5′-CGGCAGCGATTGAGCAAGTAA-3′/5′-TTGAGTGCGATGAGCCAGTGTA-3′ |
| mcpP-F/R | 5′-CGTATTTGCGATGGGGAT-3′/5′-GGCGGACGATGATTTTTC-3′ |
| mcpU-F/R | 5′-GCTATCTCGCACCTTTCTTC-3′/5′-CAATCGTCTTTACGCTCACC-3′ |
| cheA-F/R | 5′-GCTTCCTATCATTGGCA-3′/5′-AGTGGTTCACGGTCTTG-3′ |
| cheV-F/R | 5′-TGGCACAAGAGGGTAGTAT-3′/5′-TCAATGAGGAGTGAAGGAT-3′ |
| cheW-F/R | 5′-CTACACAGAAATCGCTCC-3′/5′-CTTCACCTTCCATCAAAC-3′ |
| Gene Name | Description or Predicted Function | Gene Name | Description or Predicted Function |
|---|---|---|---|
| pctB | methyl-accepting chemotaxis protein | cheW | two-component system, chemotaxis family, purine-binding chemotaxis protein CheW |
| tar | methyl-accepting chemotaxis protein II, aspartate sensor receptor | cheY | two-component system, chemotaxis family, chemotaxis protein CheY |
| mcpP | methyl-accepting chemotaxis protein | fliC | flagellin |
| mcpU | methyl-accepting chemotaxis protein | fliG | flagellar motor switch protein FliG |
| cheA | two-component system, chemotaxis family, sensor kinase CheA | fliN | flagellar motor switch protein FliN |
| cheV | two-component system, chemotaxis family, chemotaxis protein CheV | flgE | flagellar hook protein FlgE |
| motA | chemotaxis protein MotA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Zhang, Z.; Yan, Q.; Du, Z.; Zhao, L.; Qin, Y. Amino Acid-Induced Chemotaxis Plays a Key Role in the Adaptation of Vibrio harveyi from Seawater to the Muscle of the Host Fish. Microorganisms 2024, 12, 1292. https://doi.org/10.3390/microorganisms12071292
Zhang X, Zhang Z, Yan Q, Du Z, Zhao L, Qin Y. Amino Acid-Induced Chemotaxis Plays a Key Role in the Adaptation of Vibrio harveyi from Seawater to the Muscle of the Host Fish. Microorganisms. 2024; 12(7):1292. https://doi.org/10.3390/microorganisms12071292
Chicago/Turabian StyleZhang, Xiaoxu, Zhe Zhang, Qingpi Yan, Ziyan Du, Lingmin Zhao, and Yingxue Qin. 2024. "Amino Acid-Induced Chemotaxis Plays a Key Role in the Adaptation of Vibrio harveyi from Seawater to the Muscle of the Host Fish" Microorganisms 12, no. 7: 1292. https://doi.org/10.3390/microorganisms12071292
APA StyleZhang, X., Zhang, Z., Yan, Q., Du, Z., Zhao, L., & Qin, Y. (2024). Amino Acid-Induced Chemotaxis Plays a Key Role in the Adaptation of Vibrio harveyi from Seawater to the Muscle of the Host Fish. Microorganisms, 12(7), 1292. https://doi.org/10.3390/microorganisms12071292






