Oleanolic Acid Promotes the Formation of Probiotic Escherichia coli Nissle 1917 (EcN) Biofilm by Inhibiting Bacterial Motility
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Determination of EcN Biofilm Formation Curve
2.3. Determination of Minimal Inhibitory Concentration
2.4. Growth Curves
2.5. Biofilm Formation
2.6. MTT Assay
2.7. Extracellular Polysaccharide Determination
2.8. Viable Bacteria in Biofilms
2.9. Determination of Bacterial Surface Hydrophobicity
2.10. Confocal Laser Scanning Microscopy (CLSM)
2.11. Transcriptomic Analysis
2.11.1. RNA Extraction, Library Construction, and RNA Sequencing
2.11.2. Raw Data Processing, Differential Gene Expression, and Functional Enrichment Analysis
2.12. qRT-PCR Analysis
2.13. Motility Assay
2.13.1. Swimming Motility
2.13.2. Twitching Motility
2.14. Congo Red Agar (CRA) Plate Assay
2.15. Evaluation of the Effect of FeCl3 on EcN Biofilm
2.16. Statistical Analysis
3. Results
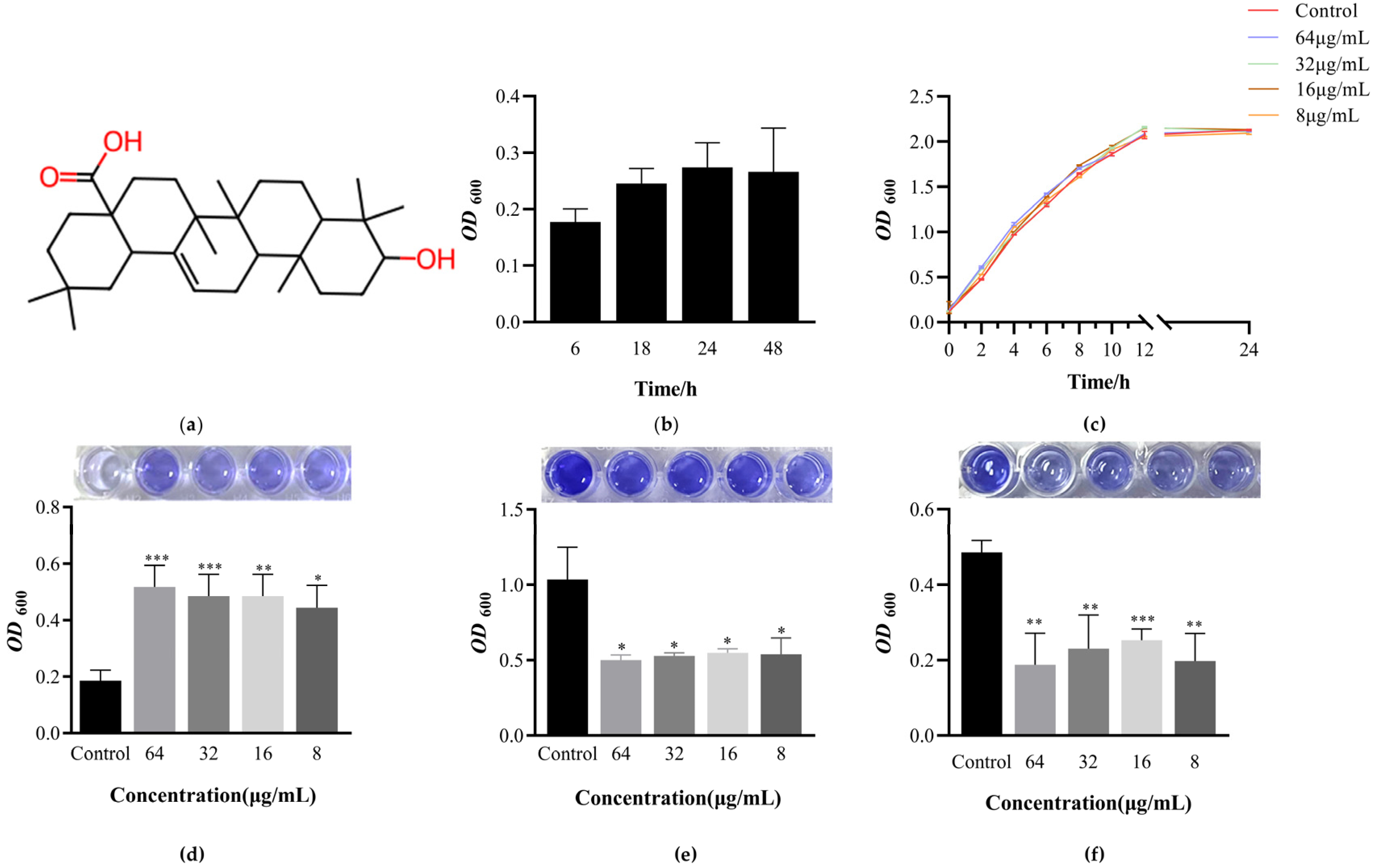
3.1. Determination of EcN Biofilm Formation Curve
3.2. Determination of Minimum Inhibitory Concentration of OA and Its Effect on EcN Growth
3.3. OA Can Promote the Formation of EcN Biofilm and Have the Ability to Clear Biofilm against Pathogens
3.4. The Effect of OA on the Metabolic Activity of EcN Biofilm Bacteria
3.5. The Effect of OA on the Growth of Live Bacteria and Extracellular Polysaccharides during Biofilm Formation Process
3.6. The Effect of OA on the Surface Hydrophobicity of EcN
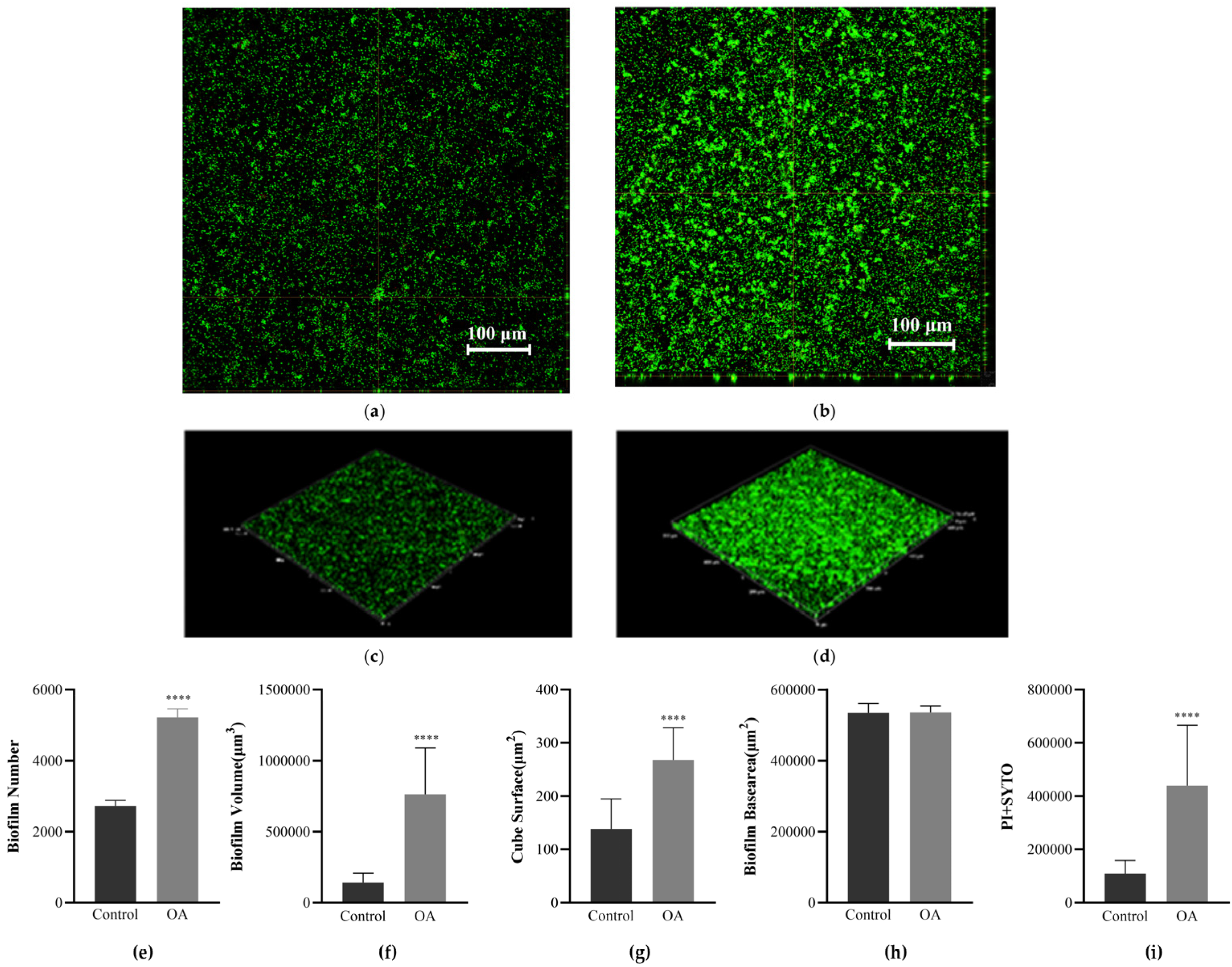
3.7. Observation of Biofilm Morphology Using Confocal Laser Scanning Microscopy (CLSM)
3.8. Transcriptomic Analysis
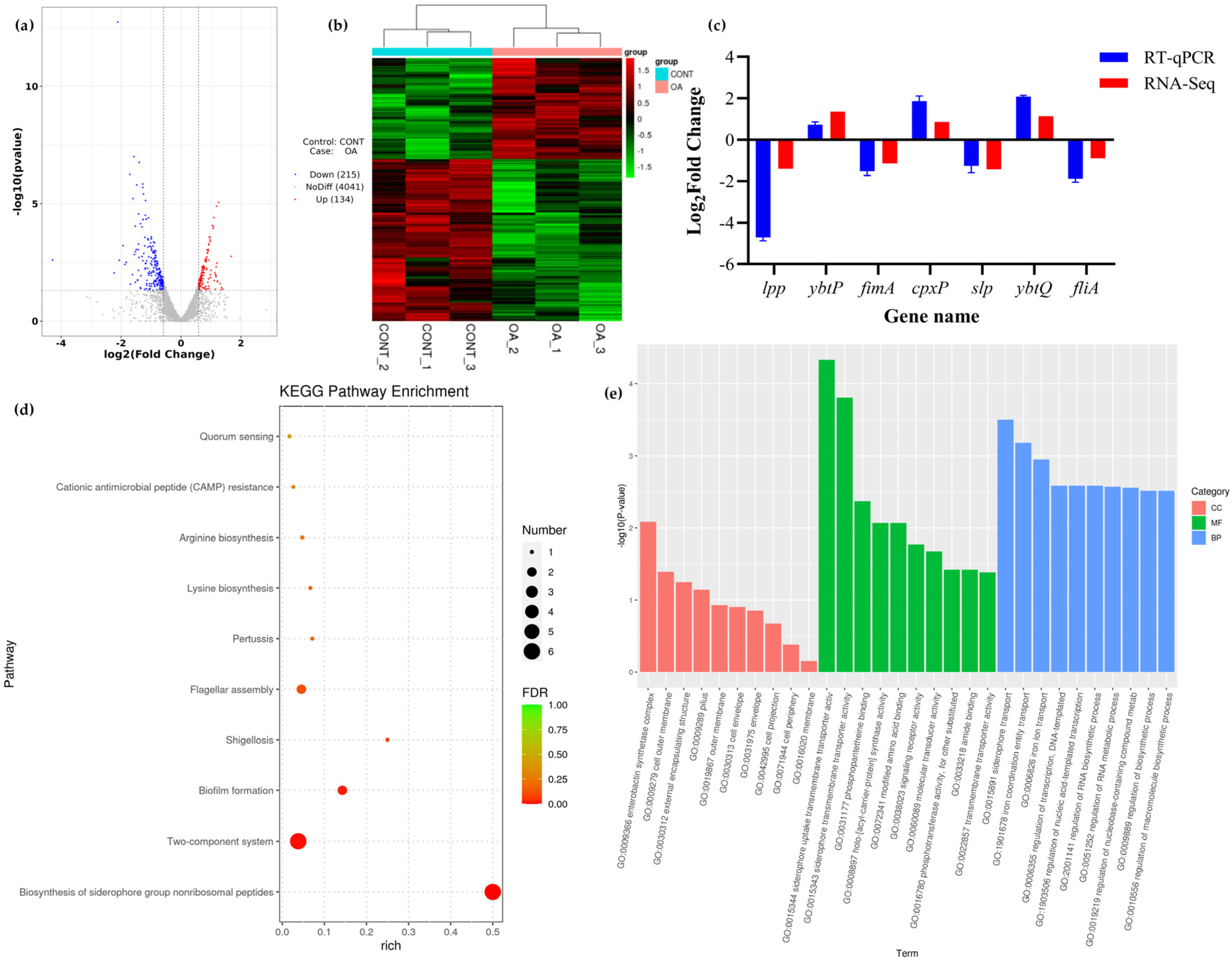
3.8.1. Differential Expression Analysis
3.8.2. KEGG Pathway Enrichment Analysis
3.8.3. GO Enrichment Analysis
3.9. QRT PCR Validation
3.10. Perform Relevant Phenotypic Tests on Transcriptomic Results
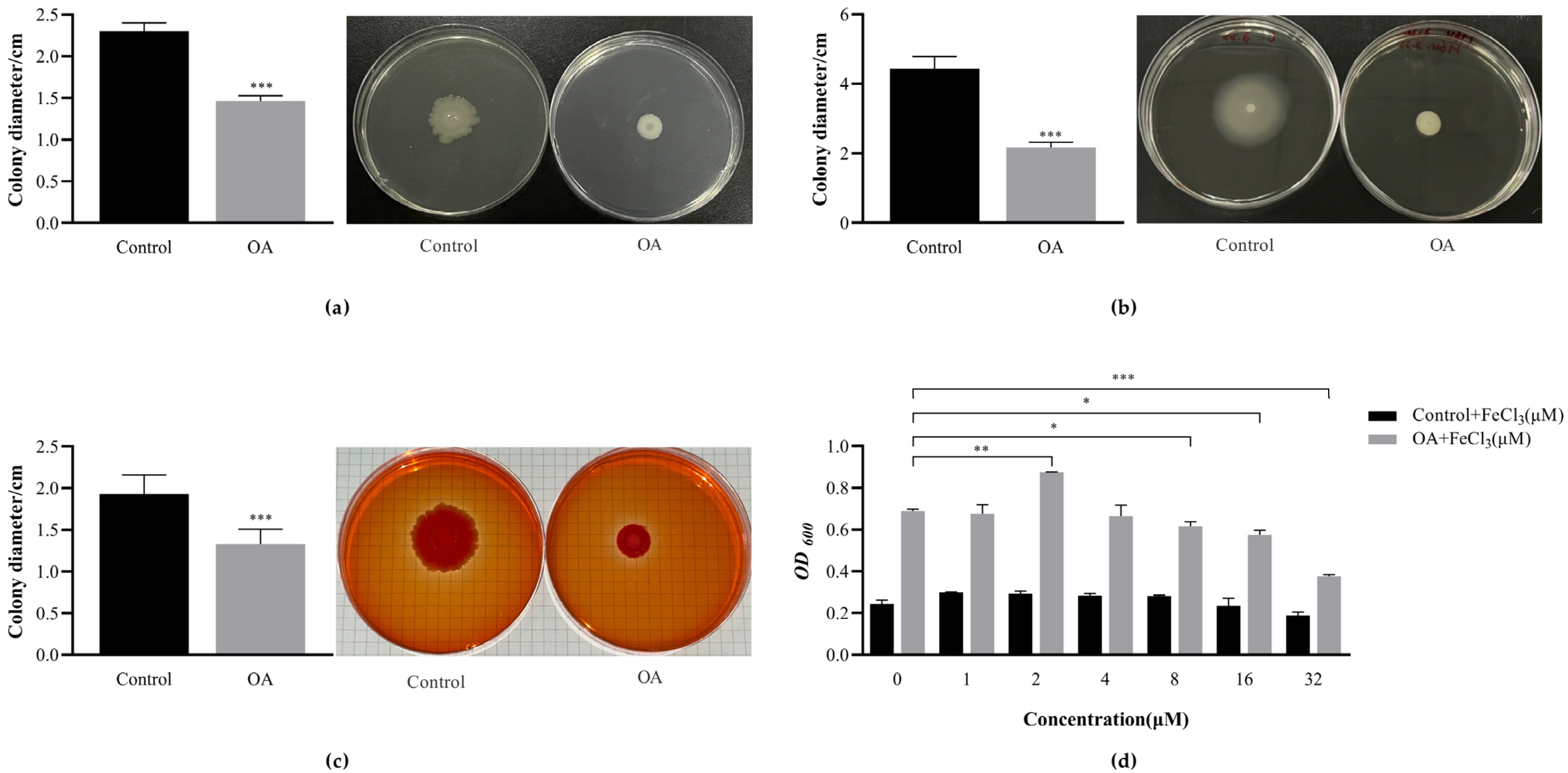
3.10.1. OA Affects the Exercise Phenotype of EcN
3.10.2. OA Affects the Production of EcN Curli
3.10.3. FeCl3 Promotes the Formation of EcN Biofilm
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [PubMed]
- Kailasapathy, K.; Chin, J. Survival and therapeutic potential of probiotic organisms with reference to Lactobacillus acidophilus and Bifidobacterium spp. Immunol. Cell Biol. 2000, 78, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Sanders, M.E.; Guarner, F.; Guerrant, R.; Holt, P.R.; Quigley, E.M.; Sartor, R.B.; Sherman, P.M.; Mayer, E.A. An update on the use and investigation of probiotics in health and disease. Gut 2013, 62, 787–796. [Google Scholar] [CrossRef]
- Luo, X.; Kong, Q.; Wang, Y.; Duan, X.; Wang, P.; Li, C.; Huan, Y. Colonization of Clostridium butyricum in Rats and Its Effect on Intestinal Microbial Composition. Microorganisms 2021, 9, 1573. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Lu, Y.; Xie, J.; Fei, Y.; Zheng, G.; Wang, Z.; Liu, J.; Lv, L.; Ling, Z.; Berglund, B.; et al. Probiotic Gastrointestinal Transit and Colonization After Oral Administration: A Long Journey. Front. Cell. Infect. Microbiol. 2021, 11, 609722. [Google Scholar] [CrossRef] [PubMed]
- Del Pozo, J.L. Biofilm-related disease. Expert Rev. Anti-Infect. Ther. 2018, 16, 51–65. [Google Scholar] [CrossRef]
- Sonnenborn, U. Escherichia coli strain Nissle 1917-from bench to bedside and back: History of a special Escherichia coli strain with probiotic properties. FEMS Microbiol. Lett. 2016, 363, fnw212. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xu, J.; Shi, T.; Zhang, Y.; Chen, F.; Yang, C.; Guo, X.; Liu, G.; Shao, D.; Leong, K.W.; et al. Probiotic-Inspired Nanomedicine Restores Intestinal Homeostasis in Colitis by Regulating Redox Balance, Immune Responses, and the Gut Microbiome. Adv. Mater. 2023, 35, e2207890. [Google Scholar] [CrossRef] [PubMed]
- Cukrowska, B.; LodInová-ZádnIková, R.; Enders, C.; Sonnenborn, U.; Schulze, J.; Tlaskalová-Hogenová, H. Specific proliferative and antibody responses of premature infants to intestinal colonization with nonpathogenic probiotic E. coli strain Nissle 1917. Scand. J. Immunol. 2002, 55, 204–209. [Google Scholar] [CrossRef]
- Lasaro, M.A.; Salinger, N.; Zhang, J.; Wang, Y.; Zhong, Z.; Goulian, M.; Zhu, J. F1C fimbriae play an important role in biofilm formation and intestinal colonization by the Escherichia coli commensal strain Nissle 1917. Appl. Environ. Microbiol. 2009, 75, 246–251. [Google Scholar] [CrossRef]
- Ventolini, G. Vaginal Lactobacillus: Biofilm formation in vivo—Clinical implications. Int. J. Women’s Health 2015, 7, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Rieu, A.; Aoudia, N.; Jego, G.; Chluba, J.; Yousfi, N.; Briandet, R.; Deschamps, J.; Gasquet, B.; Monedero, V.; Garrido, C.; et al. The biofilm mode of life boosts the anti-inflammatory properties of Lactobacillus. Cell. Microbiol. 2014, 16, 1836–1853. [Google Scholar] [CrossRef] [PubMed]
- Cheow, W.S.; Kiew, T.Y.; Hadinoto, K. Controlled release of Lactobacillus rhamnosus biofilm probiotics from alginate-locust bean gum microcapsules. Carbohydr. Polym. 2014, 103, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Glasenapp, Y.; Cattò, C.; Villa, F.; Saracchi, M.; Cappitelli, F.; Papenbrock, J. Promoting Beneficial and Inhibiting Undesirable Biofilm Formation with Mangrove Extracts. Int. J. Mol. Sci. 2019, 20, 3549. [Google Scholar] [CrossRef] [PubMed]
- Al Azzaz, J.; Al Tarraf, A.; Heumann, A.; Da Silva Barreira, D.; Laurent, J.; Assifaoui, A.; Rieu, A.; Guzzo, J.; Lapaquette, P. Resveratrol Favors Adhesion and Biofilm Formation of Lacticaseibacillus paracasei subsp. paracasei Strain ATCC334. Int. J. Mol. Sci. 2020, 21, 5423. [Google Scholar] [CrossRef] [PubMed]
- Vikram, A.; Jayaprakasha, G.; Jesudhasan, P.; Pillai, S.; Patil, B. Suppression of bacterial cell-cell signalling, biofilm formation and type III secretion system by citrus flavonoids. J. Appl. Microbiol. 2010, 109, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Dzubak, P.; Hajduch, M.; Vydra, D.; Hustova, A.; Kvasnica, M.; Biedermann, D.; Markova, L.; Urban, M.; Sarek, J. Pharmacological activities of natural triterpenoids and their therapeutic implications. Nat. Prod. Rep. 2006, 23, 394–411. [Google Scholar] [CrossRef]
- Shanmugam, M.K.; Dai, X.; Kumar, A.P.; Tan, B.K.; Sethi, G.; Bishayee, A. Oleanolic acid and its synthetic derivatives for the prevention and therapy of cancer: Preclinical and clinical evidence. Cancer Lett. 2014, 346, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Sheng, H.; Sun, H. Synthesis, biology and clinical significance of pentacyclic triterpenes: A multi-target approach to prevention and treatment of metabolic and vascular diseases. Nat. Prod. Rep. 2011, 28, 543–593. [Google Scholar] [CrossRef]
- Zhou, L.; Ding, Y.; Chen, W.; Zhang, P.; Chen, Y.; Lv, X. The in vitro study of ursolic acid and oleanolic acid inhibiting cariogenic microorganisms as well as biofilm. Oral Dis. 2013, 19, 494–500. [Google Scholar] [CrossRef]
- Kurek, A.; Markowska, K.; Grudniak, A.M.; Janiszowska, W.; Wolska, K.I. The effect of oleanolic and ursolic acids on the hemolytic properties and biofilm formation of Listeria monocytogenes. Pol. J. Microbiol. 2014, 63, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Marini, E.; Di Giulio, M.; Magi, G.; Di Lodovico, S.; Cimarelli, M.E.; Brenciani, A.; Nostro, A.; Cellini, L.; Facinelli, B. Curcumin, an antibiotic resistance breaker against a multiresistant clinical isolate of Mycobacterium abscessus. Phytother. Res. 2018, 32, 488–495. [Google Scholar] [CrossRef]
- Kulshrestha, S.; Khan, S.; Hasan, S.; Khan, M.E.; Misba, L.; Khan, A.U. Calcium fluoride nanoparticles induced suppression of Streptococcus mutans biofilm: An in vitro and in vivo approach. Appl. Microbiol. Biotechnol. 2016, 100, 1901–1914. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.K.; Wang, S.; Huang, Y.; Zhou, X.; Xu, H.H.K.; Ren, B.; Peng, X.; Xiao, Y.; Li, M.; Cheng, L. Starvation Survival and Biofilm Formation under Subminimum Inhibitory Concentration of QAMs. BioMed Res. Int. 2021, 2021, 8461245. [Google Scholar] [CrossRef]
- Upadhyay, A.; Upadhyaya, I.; Kollanoor-Johny, A.; Venkitanarayanan, K. Antibiofilm effect of plant derived antimicrobials on Listeria monocytogenes. Food Microbiol. 2013, 36, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-S.; García-Contreras, R.; Wood, T.K. YcfR (BhsA) influences Escherichia coli biofilm formation through stress response and surface hydrophobicity. J. Bacteriol. 2007, 189, 3051–3062. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, J.P.; de Azerêdo, G.A.; de Araújo Torres, R.; da Silva Vasconcelos, M.A.; da Conceição, M.L.; de Souza, E.L. Synergies of carvacrol and 1,8-cineole to inhibit bacteria associated with minimally processed vegetables. Int. J. Food Microbiol. 2012, 154, 145–151. [Google Scholar] [CrossRef]
- Kharga, K.; Dhar, I.; Kashyap, S.; Sengupta, S.; Kumar, D.; Kumar, L. Zingerone inhibits biofilm formation and enhances antibiotic efficacy against Salmonella biofilm. World J. Microbiol. Biotechnol. 2023, 39, 268. [Google Scholar] [CrossRef] [PubMed]
- Kumar, L.; Chhibber, S.; Kumar, R.; Kumar, M.; Harjai, K. Zingerone silences quorum sensing and attenuates virulence of Pseudomonas aeruginosa. Fitoterapia 2015, 102, 84–95. [Google Scholar] [CrossRef]
- Reichhardt, C.; Jacobson, A.N.; Maher, M.C.; Uang, J.; McCrate, O.A.; Eckart, M.; Cegelski, L. Congo Red Interactions with Curli-Producing E. coli and Native Curli Amyloid Fibers. PLoS ONE 2015, 10, e0140388. [Google Scholar] [CrossRef]
- Nazik, H.; Penner, J.C.; Ferreira, J.A.; Haagensen, J.A.; Cohen, K.; Spormann, A.M.; Martinez, M.; Chen, V.; Hsu, J.L.; Clemons, K.V.; et al. Effects of Iron Chelators on the Formation and Development of Aspergillus fumigatus Biofilm. Antimicrob. Agents Chemother. 2015, 59, 6514–6520. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente-Núñez, C.; Korolik, V.; Bains, M.; Nguyen, U.; Breidenstein, E.B.M.; Horsman, S.; Lewenza, S.; Burrows, L.; Hancock, R.E.W. Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob. Agents Chemother. 2012, 56, 2696–2704. [Google Scholar] [CrossRef] [PubMed]
- Köhler, T.; Curty, L.K.; Barja, F.; van Delden, C.; Pechère, J.-C. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 2000, 182, 5990–5996. [Google Scholar] [CrossRef] [PubMed]
- Overhage, J.; Lewenza, S.; Marr, A.K.; Hancock, R.E.W. Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 mini-Tn5-lux mutant library. J. Bacteriol. 2007, 189, 2164–2169. [Google Scholar] [CrossRef] [PubMed]
- Blomfield, I.C. The regulation of pap and type 1 fimbriation in Escherichia coli. Adv. Microb. Physiol. 2001, 45, 1–49. [Google Scholar] [PubMed]
- Boddicker, J.D.; Ledeboer, N.A.; Jagnow, J.; Jones, B.D.; Clegg, S. Differential binding to and biofilm formation on, HEp-2 cells by Salmonella enterica serovar Typhimurium is dependent upon allelic variation in the fimH gene of the fim gene cluster. Mol. Microbiol. 2002, 45, 1255–1265. [Google Scholar] [CrossRef] [PubMed]
- Hadjifrangiskou, M.; Hultgren, S.J. What does it take to stick around? Molecular insights into biofilm formation by uropathogenic Escherichia coli. Virulence 2012, 3, 231–233. [Google Scholar] [CrossRef]
- Otto, K.; Norbeck, J.; Larsson, T.; Karlsson, K.-A.; Hermansson, M. Adhesion of type 1-fimbriated Escherichia coli to abiotic surfaces leads to altered composition of outer membrane proteins. J. Bacteriol. 2001, 183, 2445–2453. [Google Scholar] [CrossRef] [PubMed]
- Hammer, N.D.; Schmidt, J.C.; Chapman, M.R. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc. Natl. Acad. Sci. USA 2007, 104, 12494–12499. [Google Scholar] [CrossRef]
- Jeffries, J.; Thongsomboon, W.; Visser, J.A.; Enriquez, K.; Yager, D.; Cegelski, L. Variation in the ratio of curli and phosphoethanolamine cellulose associated with biofilm architecture and properties. Biopolymers 2021, 112, e23395. [Google Scholar] [CrossRef]
- Brombacher, E.; Baratto, A.; Dorel, C.; Landini, P. Gene expression regulation by the Curli activator CsgD protein: Modulation of cellulose biosynthesis and control of negative determinants for microbial adhesion. J. Bacteriol. 2006, 188, 2027–2037. [Google Scholar] [CrossRef] [PubMed]
- Ogasawara, H.; Ishizuka, T.; Hotta, S.; Aoki, M.; Shimada, T.; Ishihama, A. Novel regulators of the csgD gene encoding the master regulator of biofilm formation in Escherichia coli K-12. Microbiology 2020, 166, 880–890. [Google Scholar] [CrossRef] [PubMed]
- Lacanna, E.; Bigosch, C.; Kaever, V.; Boehm, A.; Becker, A. Evidence for Escherichia coli Diguanylate Cyclase DgcZ Interlinking Surface Sensing and Adhesion via Multiple Regulatory Routes. J. Bacteriol. 2016, 198, 2524–2535. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Chen, S.; Li, L.; Cao, H.; Li, T.; Hu, M.; Liao, L.; Zhang, L.-H.; Xu, Z. Genome characterization of a uropathogenic Pseudomonas aeruginosa isolate PA_HN002 with cyclic di-GMP-dependent hyper-biofilm production. Front. Cell. Infect. Microbiol. 2022, 12, 956445. [Google Scholar] [CrossRef] [PubMed]
- Alves, J.R.; Pereira, A.C.M.; Souza, M.C.; Costa, S.B.; Pinto, A.S.; Mattos-Guaraldi, A.L.; Hirata-Júnior, R.; Rosa, A.C.P.; Rosa, L.M.B.O. Iron-limited condition modulates biofilm formation and interaction with human epithelial cells of enteroaggregative Escherichia coli (EAEC). J. Appl. Microbiol. 2010, 108, 246–255. [Google Scholar] [CrossRef]
- Lin, M.-H.; Shu, J.-C.; Huang, H.-Y.; Cheng, Y.-C. Involvement of iron in biofilm formation by Staphylococcus aureus. PLoS ONE 2012, 7, e34388. [Google Scholar] [CrossRef] [PubMed]
- Wiens, J.R.; Vasil, A.I.; Schurr, M.J.; Vasil, M.L. Iron-regulated expression of alginate production, mucoid phenotype, and biofilm formation by Pseudomonas aeruginosa. MBio 2014, 5, e01010-3. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; He, Y.; She, Q.; Larese-Casanova, P.; Li, P.; Chai, Y. Heterogeneity in respiratory electron transfer and adaptive iron utilization in a bacterial biofilm. Nat. Commun. 2019, 10, 3702. [Google Scholar] [CrossRef]
- Eijkelkamp, B.A.; Hassan, K.A.; Paulsen, I.T.; Brown, M.H. Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genom. 2011, 12, 126. [Google Scholar] [CrossRef]
- Matilla, M.A.; Ramos, J.L.; Duque, E.; Alché, J.d.D.; Espinosa-Urgel, M.; Ramos-González, M.I. Temperature and pyoverdine-mediated iron acquisition control surface motility of Pseudomonas putida. Environ. Microbiol. 2007, 9, 1842–1850. [Google Scholar] [CrossRef]
- Déziel, E.; Lépine, F.; Milot, S.; Villemur, R. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy) alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 2003, 149, 2005–2013. [Google Scholar] [CrossRef] [PubMed]
- McCarter, L.; Silverman, M. Iron regulation of swarmer cell differentiation of Vibrio parahaemolyticus. J. Bacteriol. 1989, 171, 731–736. [Google Scholar] [CrossRef] [PubMed]

| Gene ID | Gene | Product | Log2FC | p-Value |
|---|---|---|---|---|
| Iron uptake system | ||||
| RS09490 | irp2 | yersiniabactin biosynthetic protein | 1.2530 | 8.66 × 10−6 |
| RS09500 | ybtP | yersiniabactin ABC transporter ATP-binding/permease protein YbtP | 1.3492 | 3.75 × 10−2 |
| RS09505 | ybtQ | yersiniabactin ABC transporter ATP-binding/permease protein YbtQ | 1.1286 | 8.81 × 10−3 |
| RS17060 | entH | proofreading thioesterase in enterobactin biosynthesis | 1.2178 | 2.09 × 10−2 |
| RS20935 | fecR | ferric citrate regulator FecR | 1.1570 | 1.10 × 10−2 |
| RS11870 | dmsB | dimethyl sulfoxide reductase subunit B | 0.7426 | 1.54 × 10−2 |
| RS17130 | fepA | ferric enterobactin outer membrane transporter | 0.7580 | 1.15 × 10−2 |
| RS17095 | fepD | ferric enterobactin outer membrane transporter | 0.6541 | 2.15 × 10−2 |
| RS14160 | fhuE | ferric coprogen/ferric rhodotorulic acid outer membrane transporter | 0.8396 | 2.32 × 10−3 |
| RS16085 | fiu | iron catecholate outer membrane transporter Fiu | 0.7884 | 5.44 × 10−3 |
| RS09515 | ybtS | salicylate synthase Irp9 | 0.9727 | 1.39 × 10−2 |
| RS17135 | entD | phosphopantetheinyl transferase | 0.8360 | 2.92 × 10−3 |
| RS17075 | entE | 2,3-dihydroxybenzoate-AMP ligase | 0.7360 | 2.35 × 10−2 |
| RS09485 | irp1 | yersiniabactin polyketide synthase HMWP1 | 0.8076 | 2.83 × 10−3 |
| RS17115 | entF | activating enzyme | 0.8829 | 4.21 × 10−2 |
| Adaptability factors | ||||
| RS12115 | fimA | major type 1 subunit fimbrin | −1.1391 | 1.79 × 10−2 |
| RS11230 | ihfA | integration host factor subunit alpha | −1.0739 | 1.80 × 10−3 |
| RS14465 | csgE | curli assembly component CsgE | −1.0947 | 9.19 × 10−3 |
| RS14460 | csgD | transcriptional regulator | −0.9228 | 1.11 × 10−3 |
| RS18375 | ecpA | common pilus major subunit | −0.6306 | 2.49 × 10−2 |
| RS18370 | ecpR | transcriptional regulator for the ecp operon | −0.7083 | 9.96 × 10−3 |
| RS01085 | hdeD | acid-resistance membrane protein | −1.0482 | 6.51 × 10−4 |
| RS14310 | flgM | anti-sigma factor for FliA | −0.9128 | 2.32 × 10−2 |
| RS09885 | fliA | RNA polymerase sigma factor FliA | −0.9049 | 8.27 × 10−3 |
| RS18390 | ecpD | polymerized tip adhesin of ECP fibers | 0.6233 | 2.99 × 10−2 |
| RS01150 | slp | outer membrane lipoprotein | −1.4296 | 8.88 × 10−4 |
| RS13225 | hns | DNA-binding transcriptional dual regulator H-NS | −0.8428 | 1.99 × 10−3 |
| Two component system | ||||
| RS06395 | qseE/GlrK | two component system sensor histidine kinase QseE/GlrK | 0.7615 | 5.53 × 10−3 |
| RS13670 | ymgA | putative two-component system connector protein YmgA | 0.9766 | 1.06 × 10−2 |
| RS23010 | cpxP | inhibitor of the cpx response periplasmic adaptor protein | 0.8645 | 8.94 × 10−4 |
| RS01145 | dctR/yhiF | LuxR family repressor for dicarboxylate transport | −1.0504 | 2.56 × 10−2 |
| Stress | ||||
| RS19265 | degP | periplasmic serine endoprotease DegP | 0.5917 | 2.83 × 10−2 |
| c-di-GMP | ||||
| RS01915 | argD | N-acetylornithine aminotransferase/N-succinyldiaminopimelate aminotransferase | −1.0445 | 4.18 × 10−2 |
| RS11960 | dgcZ | diguanylate cyclase | 0.9421 | 4.09 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, D.; Liu, J.; Ran, L.; Yang, Z.; He, Y.; Yang, H.; Yu, Y.; Fu, L.; Zhu, M.; Chen, H. Oleanolic Acid Promotes the Formation of Probiotic Escherichia coli Nissle 1917 (EcN) Biofilm by Inhibiting Bacterial Motility. Microorganisms 2024, 12, 1097. https://doi.org/10.3390/microorganisms12061097
Liu D, Liu J, Ran L, Yang Z, He Y, Yang H, Yu Y, Fu L, Zhu M, Chen H. Oleanolic Acid Promotes the Formation of Probiotic Escherichia coli Nissle 1917 (EcN) Biofilm by Inhibiting Bacterial Motility. Microorganisms. 2024; 12(6):1097. https://doi.org/10.3390/microorganisms12061097
Chicago/Turabian StyleLiu, Dan, Jingjing Liu, Lei Ran, Zhuo Yang, Yuzhang He, Hongzao Yang, Yuandi Yu, Lizhi Fu, Maixun Zhu, and Hongwei Chen. 2024. "Oleanolic Acid Promotes the Formation of Probiotic Escherichia coli Nissle 1917 (EcN) Biofilm by Inhibiting Bacterial Motility" Microorganisms 12, no. 6: 1097. https://doi.org/10.3390/microorganisms12061097
APA StyleLiu, D., Liu, J., Ran, L., Yang, Z., He, Y., Yang, H., Yu, Y., Fu, L., Zhu, M., & Chen, H. (2024). Oleanolic Acid Promotes the Formation of Probiotic Escherichia coli Nissle 1917 (EcN) Biofilm by Inhibiting Bacterial Motility. Microorganisms, 12(6), 1097. https://doi.org/10.3390/microorganisms12061097






