Triglyceride-Catabolizing Lactiplantibacillus plantarum GBCC_F0227 Shows an Anti-Obesity Effect in a High-Fat-Diet-Induced C57BL/6 Mouse Obesity Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Identification of New Lactic Acid Bacteria
2.2. Measurement of Triglyceride in Culture Medium
2.3. Culture Conditions of L. plantarum GBCC_F0227
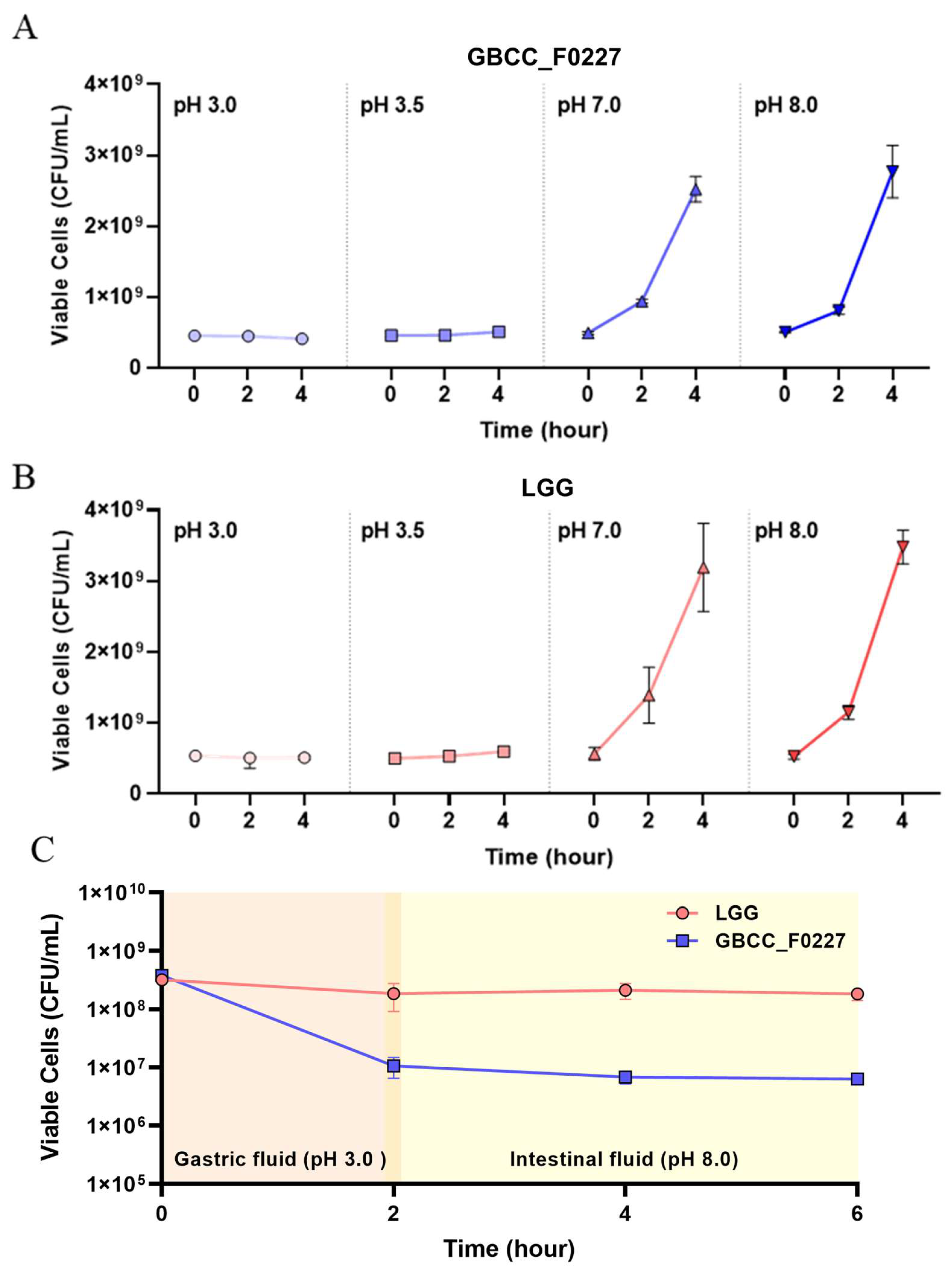
2.4. Acid Resistance Test
2.5. Cytotoxicity Assay
2.6. Whole-Genome Sequencing and Analysis
2.7. Identification of Bacterial Enzymes with Lipase Activity Using Comparative Genome Analysis
2.8. RNA Extraction and Quantitative RT-PCR (qRT-PCR) Analysis
2.9. HFD-Induced Obesity Mouse Model
2.10. Histological Quantification of Lipid Droplet Size in Epididymal Adipose Tissue
2.11. Statistical Analysis
3. Results
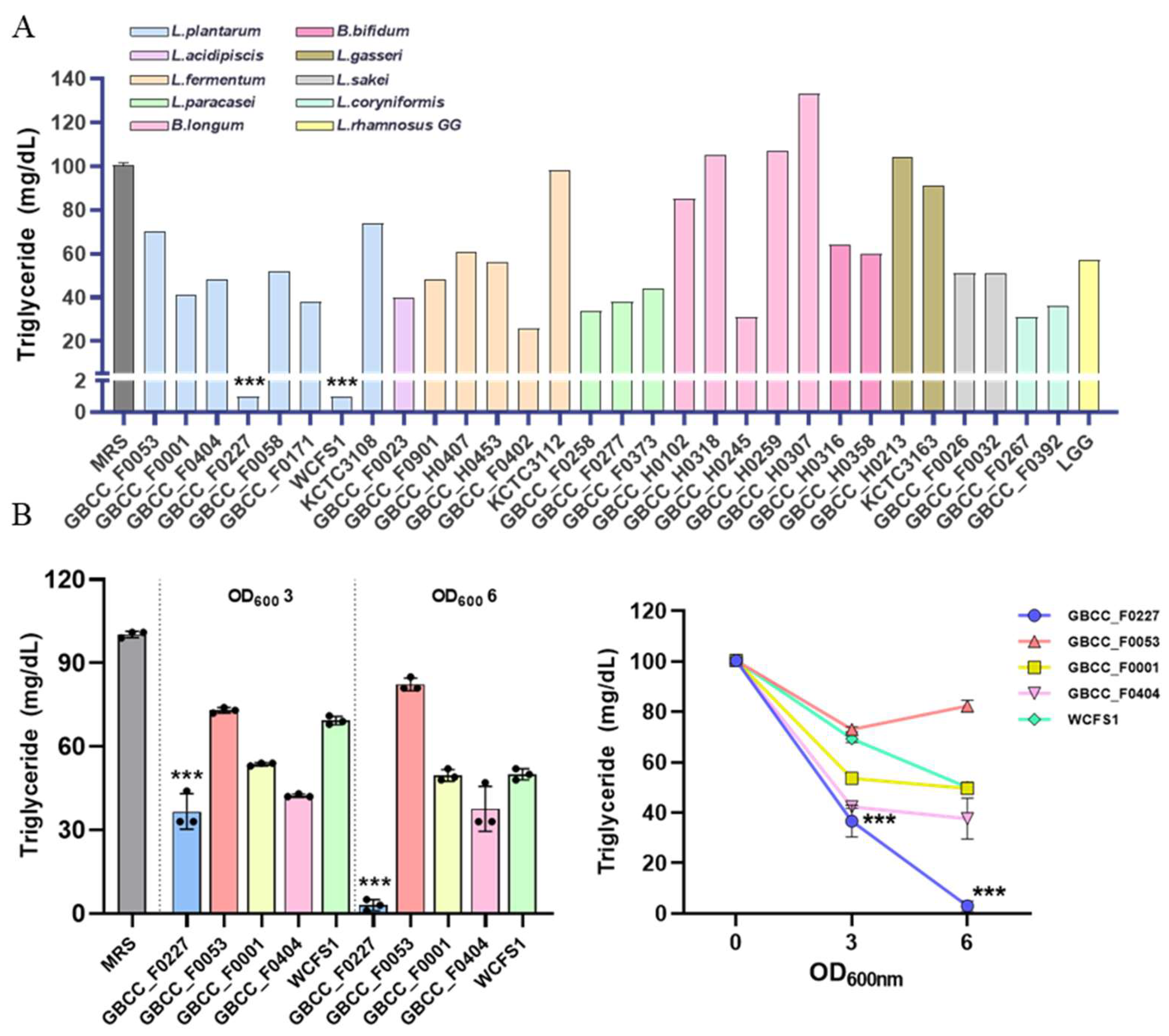
3.1. Discovery of L. plantarum GBCC_F0227, Which Effectively Catabolizes Triglycerides, through an In Vitro Screening Method
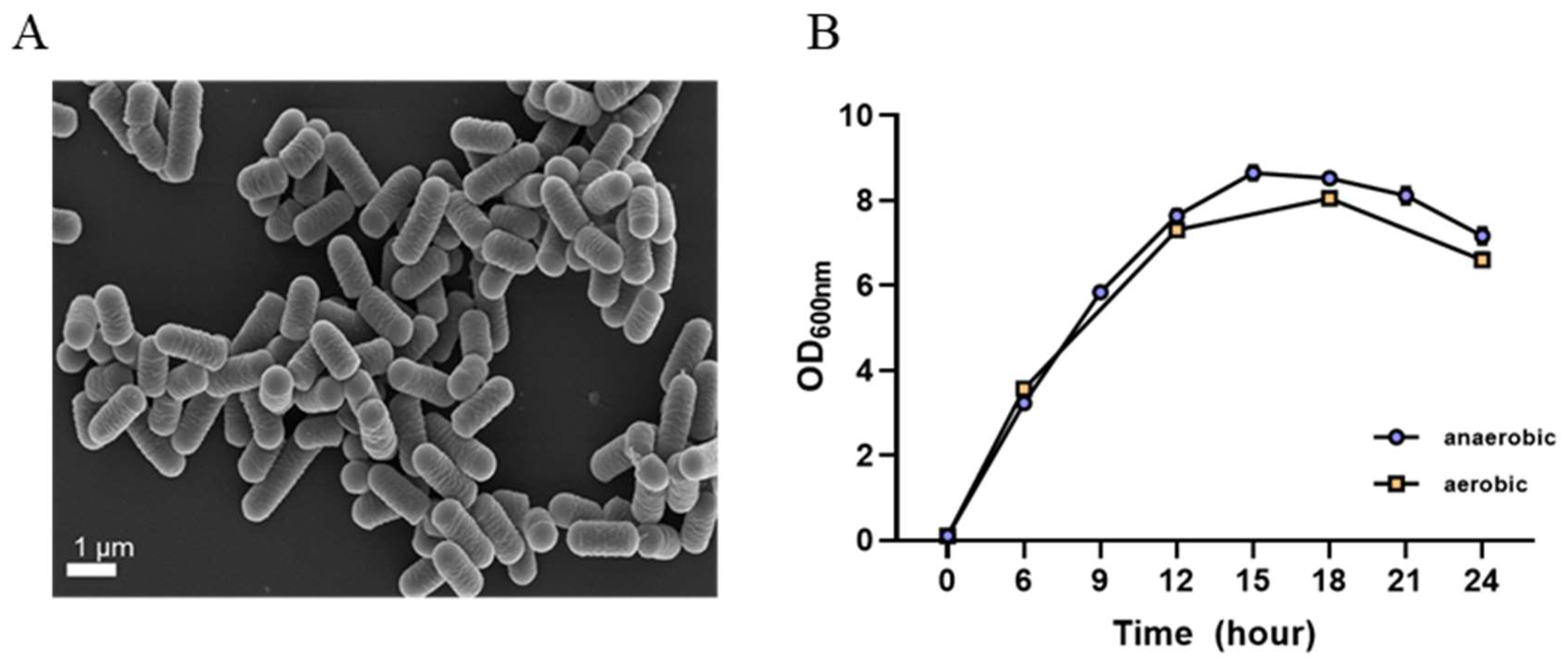
3.2. Morphological, Genetic, and Physiological Characteristics of L. plantarum GBCC_F0227
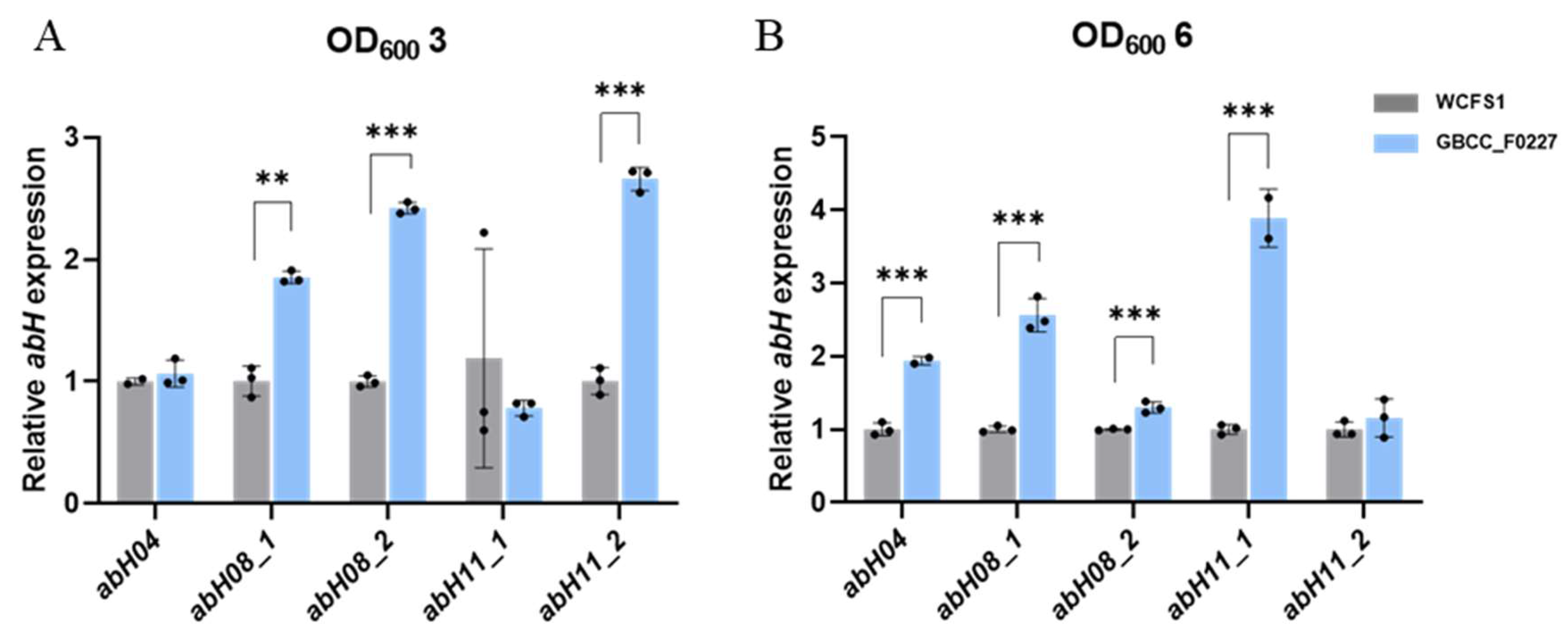
3.3. α/β Hydrolase Genes with Lipase Activity Are Highly Expressed in L. plantarum GBCC_F0227
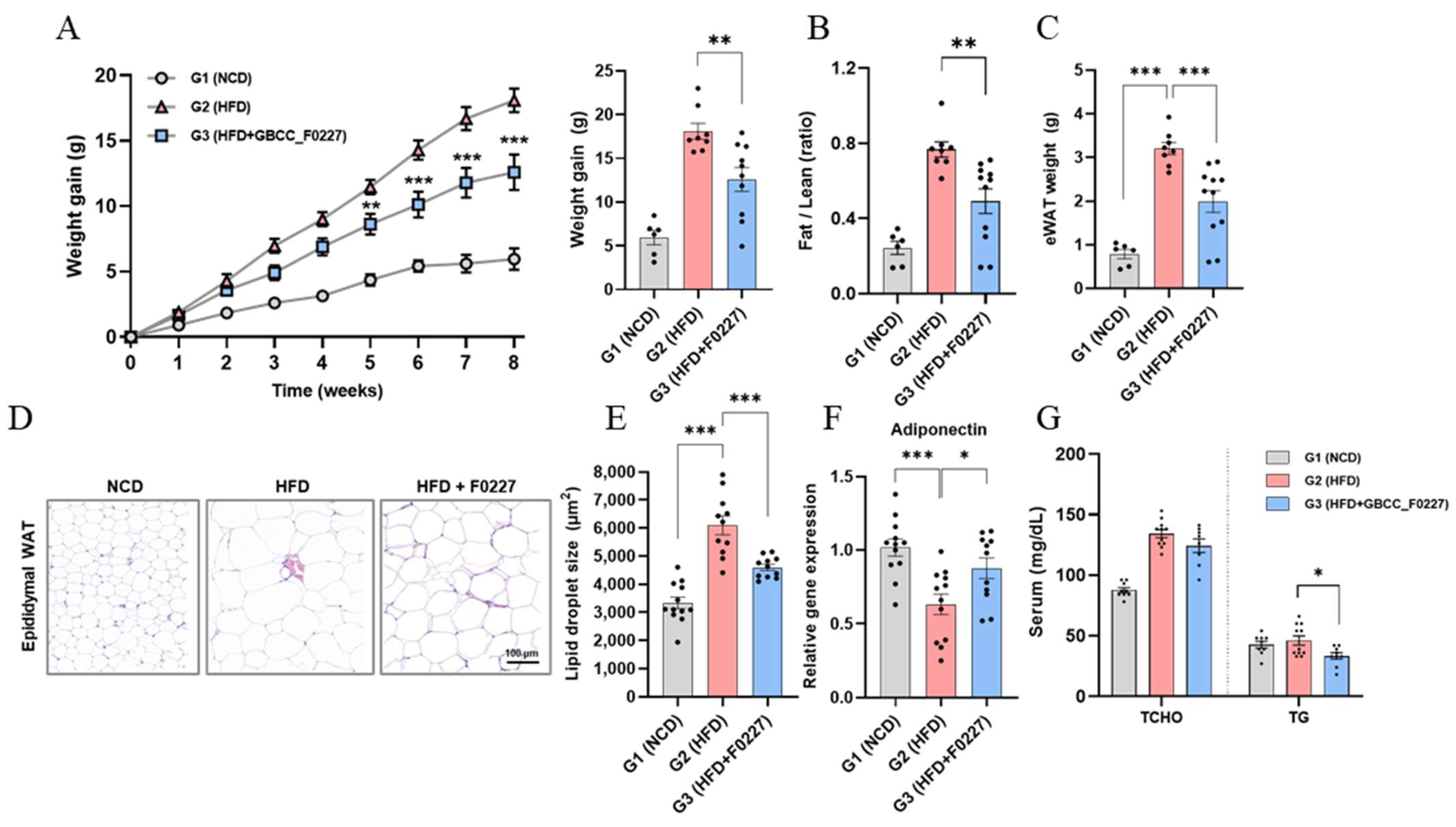
3.4. Anti-Obesity Effects of L. plantarum GBCC_F0227 in HFD-Induced Mouse Obesity Model
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Mraz, M.; Haluzik, M. The role of adipose tissue immune cells in obesity and low-grade inflammation. J. Endocrinol. 2014, 222, R113–R127. [Google Scholar] [CrossRef] [PubMed]
- Winer, S.; Chan, Y.; Paltser, G.; Truong, D.; Tsui, H.; Bahrami, J.; Dorfman, R.; Wang, Y.; Zielenski, J.; Mastronardi, F.; et al. Normalization of obesity-associated insulin resistance through immunotherapy. Nat. Med. 2009, 15, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Feuerer, M.; Herrero, L.; Cipolletta, D.; Naaz, A.; Wong, J.; Nayer, A.; Lee, J.; Goldfine, A.B.; Benoist, C.; Shoelson, S.; et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat. Med. 2009, 15, 930–939. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, S.; Manabe, I.; Nagasaki, M.; Eto, K.; Yamashita, H.; Ohsugi, M.; Otsu, M.; Hara, K.; Ueki, K.; Sugiura, S.; et al. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat. Med. 2009, 15, 914–920. [Google Scholar] [CrossRef] [PubMed]
- Lumeng, C.N.; Bodzin, J.L.; Saltiel, A.R. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Investig. 2007, 117, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Weisberg, S.P.; McCann, D.; Desai, M.; Rosenbaum, M.; Leibel, R.L.; Ferrante, A.W., Jr. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Investig. 2003, 112, 1796–1808. [Google Scholar] [CrossRef] [PubMed]
- Rohm, T.V.; Meier, D.T.; Olefsky, J.M.; Donath, M.Y. Inflammation in obesity, diabetes, and related disorders. Immunity 2022, 55, 31–55. [Google Scholar] [CrossRef] [PubMed]
- Kirichenko, T.V.; Markina, Y.V.; Bogatyreva, A.I.; Tolstik, T.V.; Varaeva, Y.R.; Starodubova, A.V. The Role of Adipokines in Inflammatory Mechanisms of Obesity. Int. J. Mol. Sci. 2022, 23, 14982. [Google Scholar] [CrossRef]
- Jung, U.J.; Choi, M.S. Obesity and its metabolic complications: The role of adipokines and the relationship between obesity, inflammation, insulin resistance, dyslipidemia and nonalcoholic fatty liver disease. Int. J. Mol. Sci. 2014, 15, 6184–6223. [Google Scholar] [CrossRef]
- Achari, A.E.; Jain, S.K. Adiponectin, a Therapeutic Target for Obesity, Diabetes, and Endothelial Dysfunction. Int. J. Mol. Sci. 2017, 18, 1321. [Google Scholar] [CrossRef]
- Yamauchi, T.; Kamon, J.; Ito, Y.; Tsuchida, A.; Yokomizo, T.; Kita, S.; Sugiyama, T.; Miyagishi, M.; Hara, K.; Tsunoda, M.; et al. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature 2003, 423, 762–769. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, T.; Yamauchi, T.; Kubota, N.; Hara, K.; Ueki, K.; Tobe, K. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J. Clin. Investig. 2006, 116, 1784–1792. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef] [PubMed]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.-M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.W.; Parkhill, J. Fighting Obesity with Bacteria. Science 2013, 341, 1069–1070. [Google Scholar] [CrossRef] [PubMed]
- Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Cheng, J.; Duncan, A.E.; Kau, A.L.; Griffin, N.W.; Lombard, V.; Henrissat, B.; Bain, J.R.; et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013, 341, 1241214. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef]
- Duan, Y.; Zeng, L.; Zheng, C.; Song, B.; Li, F.; Kong, X.; Xu, K. Inflammatory Links Between High Fat Diets and Diseases. Front. Immunol. 2018, 9, 2649. [Google Scholar] [CrossRef]
- Di Vincenzo, F.; Del Gaudio, A.; Petito, V.; Lopetuso, L.R.; Scaldaferri, F. Gut microbiota, intestinal permeability, and systemic inflammation: A narrative review. Intern. Emerg. Med. 2024, 19, 275–293. [Google Scholar] [CrossRef]
- Bervoets, L.; Van Hoorenbeeck, K.; Kortleven, I.; Van Noten, C.; Hens, N.; Vael, C.; Goossens, H.; Desager, K.N.; Vankerckhoven, V. Differences in gut microbiota composition between obese and lean children: A cross-sectional study. Gut Pathog. 2013, 5, 10. [Google Scholar] [CrossRef]
- Karlsson, C.L.; Onnerfalt, J.; Xu, J.; Molin, G.; Ahrne, S.; Thorngren-Jerneck, K. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity 2012, 20, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Kobyliak, N.; Conte, C.; Cammarota, G.; Haley, A.P.; Styriak, I.; Gaspar, L.; Fusek, J.; Rodrigo, L.; Kruzliak, P. Probiotics in prevention and treatment of obesity: A critical view. Nutr. Metab. 2016, 13, 14. [Google Scholar] [CrossRef]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, Q.; Pan, Y.; Chen, S.; Zhao, Y.; Hu, Y. New insights into the role of dietary triglyceride absorption in obesity and metabolic diseases. Front. Pharmacol. 2023, 14, 1097835. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Pacific Biosciences. 2020. Available online: https://www.pacb.com/support/software-downloads/ (accessed on 2 August 2022).
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Wu, C.H.; Apweiler, R.; Bairoch, A.; Natale, D.A.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; et al. The Universal Protein Resource (UniProt): An expanding universe of protein information. Nucleic Acids Res. 2006, 34, D187–D191. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef] [PubMed]
- Gardner, P.P.; Daub, J.; Tate, J.G.; Nawrocki, E.P.; Kolbe, D.L.; Lindgreen, S.; Wilkinson, A.C.; Finn, R.D.; Griffiths-Jones, S.; Eddy, S.R.; et al. Rfam: Updates to the RNA families database. Nucleic Acids Res. 2009, 37, D136–D140. [Google Scholar] [CrossRef] [PubMed]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rossello-Mora, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Stoeckert, C.J., Jr.; Roos, D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Fischer, M.; Pleiss, J. The Lipase Engineering Database: A navigation and analysis tool for protein families. Nucleic Acids Res. 2003, 31, 319–321. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Ivanovic, N.; Minic, R.; Dimitrijevic, L.; Radojevic Skodric, S.; Zivkovic, I.; Djordjevic, B. Lactobacillus rhamnosus LA68 and Lactobacillus plantarum WCFS1 differently influence metabolic and immunological parameters in high fat diet-induced hypercholesterolemia and hepatic steatosis. Food Funct. 2015, 6, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Xavier-Santos, D.; Bedani, R.; Lima, E.D.; Saad, S.M.I. Impact of probiotics and prebiotics targeting metabolic syndrome. J. Funct. Foods 2020, 64, 103666. [Google Scholar] [CrossRef]
- Plovier, H.; Everard, A.; Druart, C.; Depommier, C.; Van Hul, M.; Geurts, L.; Chilloux, J.; Ottman, N.; Duparc, T.; Lichtenstein, L.; et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 2017, 23, 107–113. [Google Scholar] [CrossRef] [PubMed]

| Features | Values | % |
|---|---|---|
| No. of replicons | 9 | - |
| Genome size (bp) | 3,378,947 | 100.0% |
| DNA coding (bp) | 2,808,715 | 83.1% |
| DNA G+C (bp) | 1,498,741 | 44.4% |
| Total genes | 3227 | 100.0% |
| Protein-coding genes | 3073 | 95.2% |
| RNA genes | 85 | 2.6% |
| rRNA genes | 16 | 0.5% |
| tRNA genes | 66 | 2.1% |
| Pseudo genes | 69 | 2.1% |
| Genes with function prediction | 2600 | 80.6% |
| Genes assigned to COGs | 2325 | 72.1% |
| Genes with Pfam domains | 2441 | 75.6% |
| Genes with signal peptides | 168 | 5.2% |
| Genes with transmembrane helices | 860 | 26.7% |
| CRISPR regions | 0 | - |
| Strains | Species | GBCC_F0227 | DSM 20174 | DSM 16365 | DSM 10667 | DSM 20314 | TCF032-E4 | LMG 26013 | FI11369 |
|---|---|---|---|---|---|---|---|---|---|
| GBCC_F0227 | - | - | 99.0 | 95.1 | 85.9 | 79.6 | 77.1 | 75.9 | 75.4 |
| DSM 20174 | L. plantarum | 99.0 | - | 95.2 | 85.6 | 79.5 | 77.2 | 75.6 | 75.2 |
| DSM 16365 | L. argentoratensis | 94.9 | 95.0 | - | 85.3 | 79.9 | 77.0 | 76.4 | 76.4 |
| DSM 10667 | L. paraplantarum | 85.9 | 85.6 | 85.5 | - | 79.6 | 77.5 | 76.0 | 75.7 |
| DSM 20314 | L. pentosus | 79.5 | 79.4 | 79.8 | 79.6 | - | 76.9 | 76.1 | 75.7 |
| TCF032-E4 | L. herbarum | 77.1 | 77.1 | 76.9 | 77.5 | 76.7 | - | 75.9 | 75.5 |
| LMG 26013 | L. xiangfangensis | 76.1 | 75.6 | 76.4 | 76.2 | 76.3 | 76.0 | - | 79.7 |
| FI11369 | L. garii | 75.2 | 75.1 | 76.2 | 75.8 | 75.6 | 75.4 | 79.5 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Jeon, S.-G.; Kwak, M.-J.; Park, S.-J.; Hong, H.; Choi, S.-B.; Lee, J.-H.; Kim, S.-W.; Kim, A.-R.; Park, Y.-K.; et al. Triglyceride-Catabolizing Lactiplantibacillus plantarum GBCC_F0227 Shows an Anti-Obesity Effect in a High-Fat-Diet-Induced C57BL/6 Mouse Obesity Model. Microorganisms 2024, 12, 1086. https://doi.org/10.3390/microorganisms12061086
Kim J, Jeon S-G, Kwak M-J, Park S-J, Hong H, Choi S-B, Lee J-H, Kim S-W, Kim A-R, Park Y-K, et al. Triglyceride-Catabolizing Lactiplantibacillus plantarum GBCC_F0227 Shows an Anti-Obesity Effect in a High-Fat-Diet-Induced C57BL/6 Mouse Obesity Model. Microorganisms. 2024; 12(6):1086. https://doi.org/10.3390/microorganisms12061086
Chicago/Turabian StyleKim, Jinwook, Seong-Gak Jeon, Min-Jung Kwak, So-Jung Park, Heeji Hong, Seon-Bin Choi, Ji-Hyun Lee, So-Woo Kim, A-Ram Kim, Young-Kyu Park, and et al. 2024. "Triglyceride-Catabolizing Lactiplantibacillus plantarum GBCC_F0227 Shows an Anti-Obesity Effect in a High-Fat-Diet-Induced C57BL/6 Mouse Obesity Model" Microorganisms 12, no. 6: 1086. https://doi.org/10.3390/microorganisms12061086
APA StyleKim, J., Jeon, S.-G., Kwak, M.-J., Park, S.-J., Hong, H., Choi, S.-B., Lee, J.-H., Kim, S.-W., Kim, A.-R., Park, Y.-K., Kim, B. K., & Yang, B.-G. (2024). Triglyceride-Catabolizing Lactiplantibacillus plantarum GBCC_F0227 Shows an Anti-Obesity Effect in a High-Fat-Diet-Induced C57BL/6 Mouse Obesity Model. Microorganisms, 12(6), 1086. https://doi.org/10.3390/microorganisms12061086






