Abstract
Polycyclic aromatic hydrocarbons (PAHs) cause serious stress to biological health and the soil environment as persistent pollutants. Despite the wide use of biochar in promoting soil improvement, the mechanism of biochar removing soil PAHs through rhizosphere effect in the process of phytoremediation remain uncertain. In this study, the regulation of soil niche and microbial degradation strategies under plants and biochar were explored by analyzing the effects of plants and biochar on microbial community composition, soil metabolism and enzyme activity in the process of PAH degradation. The combination of plants and biochar significantly increased the removal of phenanthrene (6.10%), pyrene (11.50%), benzo[a]pyrene (106.02%) and PAHs (27.10%) when compared with natural attenuation, and significantly increased the removal of benzo[a]pyrene (34.51%) and PAHs (5.96%) when compared with phytoremediation. Compared with phytoremediation, the combination of plants and biochar significantly increased soil nutrient availability, enhanced soil enzyme activity (urease and catalase), improved soil microbial carbon metabolism and amino acid metabolism, thereby benefiting microbial resistance to PAH stress. In addition, the activity of soil enzymes (dehydrogenase, polyphenol oxidase and laccase) and the expression of genes involved in the degradation and microorganisms (streptomyces, curvularia, mortierella and acremonium) were up-regulated through the combined action of plants and biochar. In view of the aforementioned results, the combined application of plants and biochar can enhance the degradation of PAHs and alleviate the stress of PAH on soil microorganisms.
1. Introduction
Polycyclic aromatic hydrocarbons (PAHs) are aromatic compounds which widely distributed in soil and produced in large quantities by industrial activities that are hazardous to human beings and extremely difficult to degrade [1]. Previous studies have demonstrated that PAHs can diminish soil fertility and quality by impacting the nutrient cycle, including carbon and nitrogen [2]. The remediation of PAHs in soil largely depends on indigenous microorganisms [3], whereas PAHs have a negative effect on soil microorganisms by inhibiting soil metabolic processes [2]. This may adversely affect the removal of PAHs. Therefore, it is important to clarify the effects of different treatments on the PAHs adaptability of indigenous microorganisms.
Phytoremediation is an environmentally-friendly remediation method that accelerates the degradation of PAHs through rhizosphere processes. Nutrient cycling, physical and chemical properties of soil will be affected by roots to form a unique rhizosphere environment [4]. Furthermore, plants can shape rhizosphere microorganisms by actively or passively releasing root exudation that act as a carbon source or a signal for microorganisms, recruiting specific beneficial microorganisms and altering the composition of indigenous microorganisms [5,6]. Other studies have determined that plants also engage in competition with microorganisms with microorganisms for soil nutrients, thereby influencing the functionality of microorganisms [7]. This may explain why the degradation efficiency of PAH remains unchanged or decreases under phytoremediation. Therefore, exploring the effect of plant roots on soil microorganisms is very important for the removal of PAHs.
Soil additives have widely emerged as a treatment for degrading pollution in recent years [8]. Additives can significantly affect soil remediation by improving the soil environment due to their own characteristics, such as rich nutrient elements and special structure [9]. Among these, biochar has been found to enhance remediation efficiency by modulating soil microbial structure and improving microbial metabolism due to its abundant nutrient content [9,10]. Moreover, biochar, as a potential source of soil PAHs [11], may also reduce the bioavailability of PAHs [12]. Although the feasibility of the combined application of phytoremediation and biochar is still controversial, it has been proved that biochar can improve the reduced soil fertility and soil metabolism under stress [13,14]. In addition, biochar applied may affect the composition of root exudates and the root morphology, such as increasing the content of organic acids and root area [15]. The elevated concentration of organic acids could potentially facilitate the degradation of PAH [16]. Therefore, biochar may promote the rhizodegradation of PAH by changing the root exudation strategy. Hence, when biochar and plants combine for remediating PAH contaminated soil, they can mediate the function of soil microorganisms and establish intricate interactions with microorganisms during the microbial degradation process of PAH, and ultimately affect the bioremediation. However, adverse results have been observed in studies of biochar remediation for the remediation of PAH contamination [17]. This may be due to the fact that biochar not only stimulates microbial biodegradation but also competes for the nutrients needed for rhizosphere microbial growth while stimulating the growth of plant roots [18,19,20]. This shows that the complex interaction between plants and microorganisms will be affected by biochar and ultimately promote or inhibit the adaptation of soil microorganisms to PAHs stress. And the adaptation of microorganisms to PAHs will affect the removal of PAHs in soil. However, the existing studies have not systematically studied the PAHs degradation and adaptability of microorganisms. Therefore, it is urgent to fully clarify the effects of biochar on PAHs degradation, microorganisms and rhizosphere soil environment, thereby assess the adaptability and degradation of biochar and plants to microorganisms to PAHs.
The resistance of microorganisms to PAH stress can be clarified, and the role of microorganisms in PAH degradation can be revealed by analysis of soil enzyme activity, microbial structure and function combined with soil metabolism. Buchloe dactyloides is a highly tolerant herbaceous plant [21]. In this study, we conducted 60-day experiment on potted plants in greenhouse to clarify the mechanism of soil remediation for PAH pollution and the effects on soil adaptability by combining B. dactyloides and biochar. The following hypotheses were addressed: (1) The combination of B. dactyloides roots and biochar stimulate microorganisms involved in PAH degradation, including changing the structure and functional patterns of microorganisms. (2) B. dactyloides roots and biochar can improve soil health and metabolism. This study may provide guidance for the remediation of PAH contaminated soil by plants combined with biochar.
2. Materials and Methods
2.1. Pot Experiment
The soil was collected at depths of 0–20 cm from an agricultural field in Beijing, China (40°0′27″ N, 116°15′22″ E). The properties of soil and biochar were detailed in the Tables S1 and S2. The PAHs contaminated soil containing phenanthrene (Phe), pyrene (Pyr) and benzo(a)pyrene (Bap), with or without 1% of biochar addition, was prepared. Preparation of PAHs-contaminated soil was performed as previously reported [14].
Three B. dactyloides were transferred to a pot full of 0.7 kg soil and placed under natural light at a temperature of 25–35 °C. The following four treatments were utilized: PAH contaminated soil (N); PAH contaminated soil with biochar (B); PAH contaminated soil with B. dactyloides (P); PAH contaminated soil with B. dactyloides and biochar (PB). The field capacity of soil was maintained at 60% by regularly weighing the pots and adding distilled water. The position of the pot changes randomly every week. After 60 days of incubation, the loose soil attached to the roots was removed by shaking roots vigorously, and the rhizosphere soil was carefully collected with a sterilized brush. Additionally, non-rhizosphere soil was also collected. The soil samples were sieved (2-mm mesh). Soil was stored in liquid nitrogen for further analysis. The soil properties after treatment were detailed in the Table S3.
2.2. Determination of PAHs
Freeze-dried soil (Christ-Alpha 1–4 LD plus) was subjected to ultrasonic extraction with a 30 mL mixture of n-hexane and acetone (2:1, v/v). The contents of Phe, Pyr and Bap in soil were quantitatively analyzed by the internal standard method [22]. Determination of PAHs content by Agilent HP 7890 gas chromatograph combined with Agilent HP 5975C inert mass selective detector (7890/5975C).
2.3. Determination of Enzyme Activity
The soil polyphenol oxidase (PPO) activity was measured using the pyrogallol colorimetric method. Soil dehydrogenases (DHA) activity was measured as previously reported [23]. Soil laccase activity was determined by measuring the oxidation of ABTS. The soil catalase (CAT) activity was measured using a soil catalase activity assay kit (Solarbio, Beijing, China). The soil urease activity was determined by using the indophenol blue colorimetry method.
2.4. High-Throughput Sequencing and Quantitative Polymerase Chain Reaction (qPCR)
The DNA extraction, Illumina sequencing and qPCR were carried out with reference to previous reports [22]. Soil DNA was extracted from 0.5 g of each soil sample using the E.Z.N.A. Soil DNA Kit (Omega Biotek, Norcross, GA, USA) according to the manufacturer’s protocol. 16S rRNA and ITS genes of distinct regions (16S V3–V4, ITS1) were amplified using specific primers with the barcode. The primer pairs and standard curve of 16S and ITS qPCR genes were shown in Supporting Information (Table S4, Figure S1).
2.5. Soil Metabolite Assay
Soil metabolites were extracted and determination was performed as previously reported [24,25,26]. The LC analysis was performed on a Vanquish UHPLC System (Thermo Fisher Scientific, Waltham, MA, USA). Chromatography was carried out with an ACQUITY UPLC ® HSS T3 (150 × 2.1 mm, 1.8 µm) (Waters, Milford, MA, USA). The column maintained at 40 °C. The flow rate and injection volume were set at 0.25 mL/min and 2 μL, respectively. For LC-ESI (+)-MS analysis, the mobile phases consisted of (C) 0.1% formic acid in acetonitrile (v/v) and (D) 0.1% formic acid in water (v/v). Separation was conducted under the following gradient: 0~1 min, 2% C; 1~9 min, 2~50% C; 9~12 min, 50~98% C; 12~13.5 min, 98% C; 13.5~14 min, 98~2% C; 14~20 min, 2% C. For LC-ESI (−)-MS analysis, the analytes was carried out with (A) acetonitrile and (B) ammonium formate (5 mM). Separation was conducted under the following gradient: 0~1 min, 2%A; 1~9 min, 2~50%A; 9~12 min, 50~98%A; 12~13.5 min, 98%A; 13.5~14 min, 98~2%A; 14~17 min, 2%A.
Mass spectrometric detection of metabolites was performed on Orbitrap Exploris 120 with ESI ion source. Simultaneous MS1 and MS/MS (Full MS-ddMS2 mode, data-dependent MS/MS) acquisition was used. The parameters were as follows: sheath gas pressure, 30 arb; aux gas flow, 10 arb; spray voltage, 3.50 kV and −2.50 kV for ESI(+) and ESI(−), respectively; capillary temperature, 325 °C; MS1 range, m/z 100–1000; MS1 resolving power, 60,000 FWHM; number of data dependant scans per cycle, 4; MS/MS resolving power, 15,000 FWHM; normalized collision energy, 30%; dynamic exclusion time, automatic.
2.6. Date Analysis
The normality of the datasets was tested using SPSS 22.0 (SPSS Inc., Chicago, IL, USA). The variables were analyzed using two-way analysis of variance, and using the statistical package IBM SPSS Statistics software (SPSS 22.0). The co-occurrence network, non-metric multidimensional scaling (NMDS), PICRUSt (KEGG, http://www.kegg.jp/, accessed on 17 April 2023), mantel test and metabolomics date analysis were measured using a modified method [2,22]. Linear discriminant analysis effect size (LEfSe) was utilized to identify significant microbial responders (LDA > 3.0). Adonis, Anosim, environmental niche width, Variance Partitioning Analysis (VPA) and Envfit analysis were conducted using the retatix R packages. Envfit analysis was used to determine the correlation between environment, microbial communities and KEGG.
3. Results
3.1. Degradation Rate of PAHs and Enzymatic Activity
There were significant differences in the removal rate of PAHs under different treatments (Figure 1A–C). The degradation efficiencies of three kinds of PAHs in the B treatment were lower than N treatment. The degradation rate of PAHs in the rhizosphere was significantly higher than that in the N treatment. The combined application of B. dactyloides and biochar brought the highest removal rate of Bap and total PAHs. The activities of urease and DHA under N and PB treatments were higher than those under other treatments. The activity of PPO was the highest in rhizosphere soil containing biochar. Under P treatment, the activities of laccase and CAT were lowest.
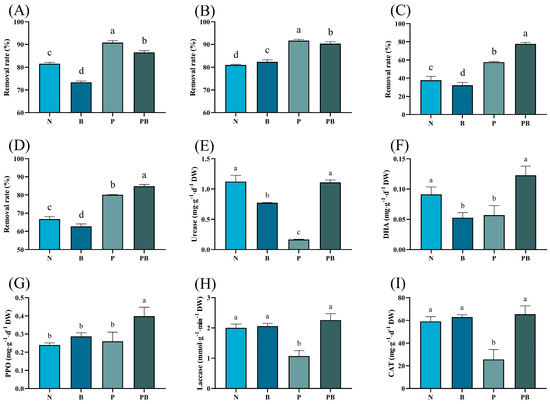
Figure 1.
The removal rate of Phenanthrene (A), Pyrene (B), Benzo[a]pyrene (C) and total PAHs (D) in different treatment. Effect of different bioremediation treatments on soil enzyme activities, including urease (E), DHA (F), PPO (G), laccase (H) and CAT (I). N: natural attenuation; B: biochar remediation; P: phytoremediation; PB: plant−biochar remediation. Averages ± SE were listed (n = 3). Different letters indicate that values are significantly different (p < 0.05).
3.2. Shifts in Microbial Community and qPCR
The chao1 index of microbial community and the shannon index of fungal community in rhizosphere soil were significantly higher than those in non-rhizosphere soil (Figure 2A–D). This indicated that the abundance and diversity of the rhizosphere microbial community were higher. NMDS showed that different treatments affected the similarity of microbial communities (Figure 2E,F). Lefse analysis showed that there were significant differences in the biomarkers of microbial groups under different treatments. Bacillus.g, Novosphingobium.g, Paenibacillus.g and Pseudomonas.g were the main bacterial participants in PAH degradation under N treatment, while Streptococcus.g and Massilia.g were the main bacterial participants in PAH degradation under B treatment. Moreover, Methylibium.g, Devosia.g and Hydrogenophaga.g were the main bacterial participants in the degradation of PAHs under P treatment, while Pseudoxanthomonas.g, Streptomyces.g, Sphingomonas.g and Ensifer.g were the main participants in the degradation of PAHs under PB treatment (Figure 3A). Mortierella.g, Phoma.g, Aspergillus.g and Trichocladium.g were the main fungal participants of PAH degradation under N treatment, while Chaetomium.g was the main fungal participant of PAH degradation under P treatment. In addition, Curvularia.g, Cladosporium.g and Acremonium.g were the main fungal participants of PAH degradation under PB treatment (Figure 3B). In the PB group, the bacterial biomass was the highest, and the fungal biomass was higher than that of N and B treatments (Figure 3C).
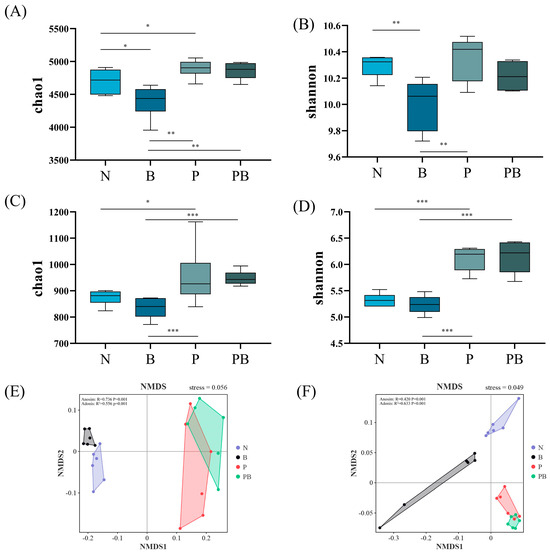
Figure 2.
Chao1 and Shannon indices of the bacterial (A,B) and fungal (C,D). NMDS of the bacterial (E) and fungal (F). N: natural attenuation; B: biochar remediation; P: phytoremediation; PB: plant−biochar remediation. The significance of the test was analyzed by Adonis and Anosim. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
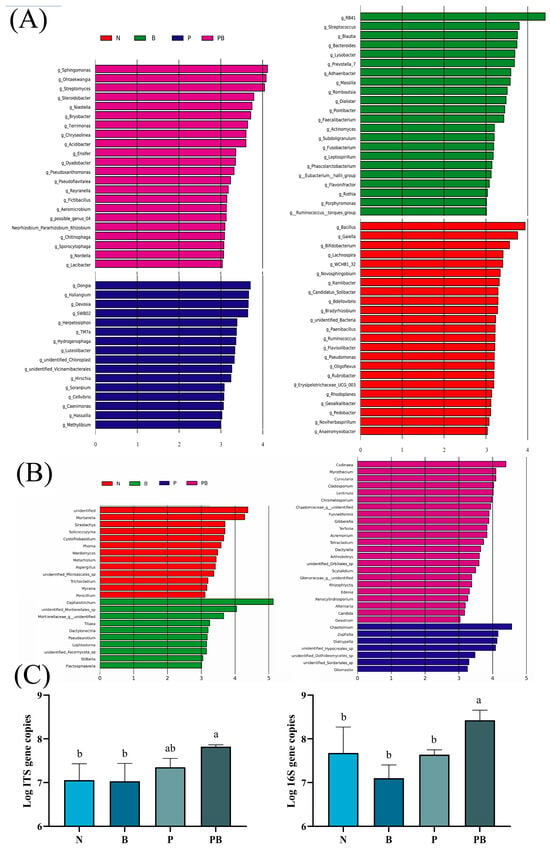
Figure 3.
Chicken breed and line-specific biomarkers. LEfSe analysis shows differentially abundant genera of bacteria (A) and fungi (B) as biomarkers determined using Kruskal-Wallis test (p < 0.05) and Wilcoxon test (p < 0.05) with LDA > 3.0. Gene copy numbers of 16S and ITS in different treatments (C). N: natural attenuation; B: biochar remediation; P: phytoremediation; PB: plant−biochar remediation. Averages ± SE were listed (n = 3). Different letters indicate that values are significantly different (p < 0.05).
3.3. Environmental Factors and Function of Microbial Community
Mantel test and Spearman models were used to screen the soil factors related to PAH degradation and PAH degrading microorganisms (Figure 4A–C). TN, TK, AK, AN and NO3− may be the main factors potentially involved in the degradation of PAHs. Among them, TK, AN and NO3− were significantly correlated with microorganisms. There were significant differences in niche breadths of soil microbial bacteria and fungi communities under different treatments (Figure 4D). The niche breadth of rhizosphere bacteria under biochar conditions was lower than P treatment, which indicated that the bacterial community tended to be specialized species. PICRUSt was used to predict the biodegradation intensity of genes involved in PAH degradation (Figure 5A). The expression levels of PAH degradation gene in PB treatment was significantly higher than that in other treatments. Bacterial functions in amino acid, carbohydrate, terpene, and flavonoid metabolism, as well as exogenous biodegradation and metabolism were significantly improved under PB treatments (Figure 5B). And soil microflora, soil metabolism and soil environment were significantly related to microbial function (Figure 6B). Finally, VPA showed that the difference in PAH removal rate was mainly attributed to soil microflora, soil metabolism, soil environment and microbial function (Figure 6A).
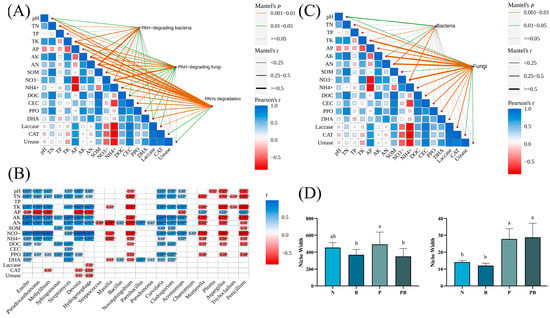
Figure 4.
(A) Mantel tests between PAHs and soil property. (B) Correlation coefficients among soil property and PAH-degrading microorganism. (C) Mantel tests between microorganism and soil property. (D) The environmental niche width of bacterial (left) and fungal (right) under different treatments. N: natural attenuation; B: biochar remediation; P: phytoremediation; PB: plant−biochar remediation. Averages ± SE were listed (n = 6). Different letters indicate that values are significantly different (p < 0.05). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
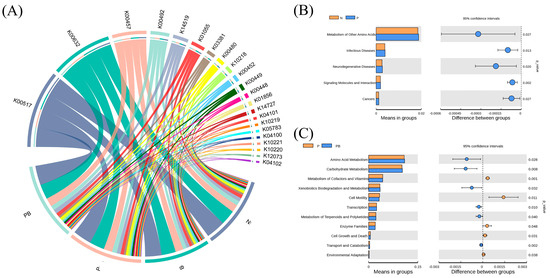
Figure 5.
(A) Alters in function profiles of microorganisms under different groups. Predicted relative abundance of bacterial metabolic functions in the N vs. P treatment (B) and P vs. PB treatment (C). N: natural attenuation; B: biochar remediation; P: phytoremediation; PB: plant−biochar remediation. Averages ± SE were listed (n = 6).
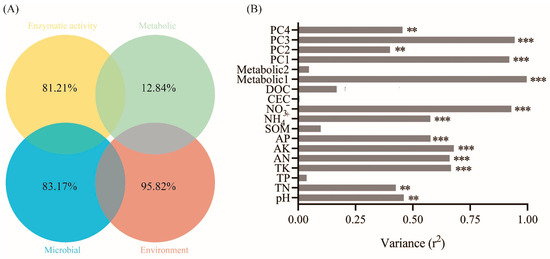
Figure 6.
(A) Venn showing the results of VPA (adjusted R2 value) for PAH degradation explained by enzymatic activity, metabolic, microbial and environmental properties categories. (B) Bacterial function profiles of KEGG (PAHs degradation genes) explained by metabolic, metabolic and environmental variables based on the Envfit analysis. **, p < 0.01; ***, p < 0.001.
3.4. Soil Metabolism and Co-Occurrence Network Analysis
The differential metabolite pathway showed that the combined application of biochar and plants significantly improved the main metabolic processes in PAH contaminated soil. Compared with P treatment, the amino acid metabolism, secondary metabolite biosynthesis, terpenoid and flavonoid metabolism, carbohydrate metabolism, and lipid metabolism of the bacterial community under PB treatment exhibited significant improvements. Furthermore, the secondary metabolite biosynthesis and amino acid metabolism of the bacterial community under P treatment were significantly improved compared with N treatment (Figure 7A). Compared with P treatment, the amino acid metabolism, secondary metabolite biosynthesis, carbohydrate metabolism and lipid metabolism of fungal community under PB treatment were significantly improved. The secondary metabolite biosynthesis and amino acid metabolism of fungal community under P treatment were significantly improved compared with N treatment (Figure 7B).
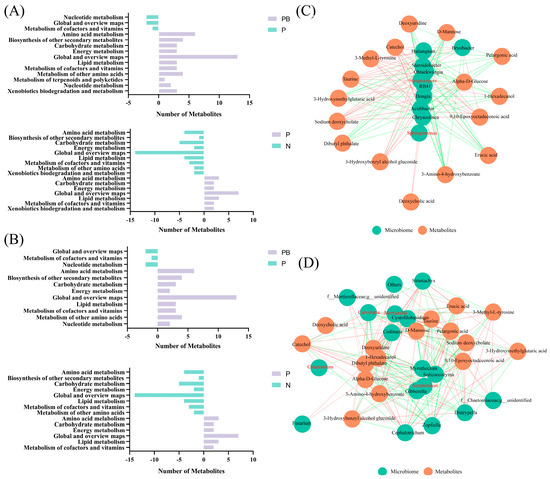
Figure 7.
Differential metabolite-based metabolic pathways in bacteria (A) and fungi (B) under different treatments. Network of co−occurring different bacteria (C) and fungi (D) with different metabolites. The red line is a positive correlation and the green is a negative correlation. The thickness of the line represents the correlation coefficient. N: natural attenuation; P: phytoremediation; PB: plant−biochar remediation.
For the soil bacterial community, Sphingomonas.g, Streptomyces.g, Bacillus.g, which may be involved in the degradation of PAHs, are the core bacteria. Theobromine, 3-Hydroxybenzyl alcohol glucoside and 9, and 10-Epoxyoctadecenoic acid are the metabolites with significant positive correlation with Sphingomonas.g, Streptomyces.g (Figure 7C). Acremonium.g, Mortierella.g, Chaetomium.g, Curvularia.g are fungi involved in PAH degradation and constitute the core fungal community. Catechol, Gingerol, Quercetin, 3-Hydroxybenzyl alcohol glucoside and 9, 10-Epoxyoctadecenoic acid are the metabolites with significant positive correlation with Mortierella.g, Curvularia.g, Chaetomium.g (Figure 7D).
4. Discussion
4.1. The Effects of Biochar and Plant Roots on PAH Contaminated Soil and Microorganisms
In this study, the application of biochar increased the removal rate of Pyr, while still decreased the removal of Phe and Bap, which is different from the results of other studies [14]. This discrepancy may be attributed to variations in the physical and chemical properties of biochar. The difference in removal of Phe and other PAHs by biochar may be explained as the variation in bioavailability caused by the number of benzene rings and the bond angle of ring [17]. P treatment and PB treatment significantly increased the removal rate of PAHs, which indicated that phytoremediation was an attractive approach to remove PAHs from soil. PAH-degrading enzyme activity is generally considered to reflect the ability of PAH degradation [27]. In this study, the activity of DHA, PPO, laccase involved in PAH degradation was the highest in PB treatment, and they were significantly related to the degradation rate of PAH (Figure 4A). The removal rate of Bap increased significantly under PB treatment may be attributed to the increase in PPO activity contributing to the degradation of high molecular weight PAH [28,29]. Moreover, the activities of urease and CAT increased under PB treatment, reflecting the enhancement of soil nitrogen use efficiency and the improvement of soil health. The decrease in CAT, DHA, laccase, and urease activities in the rhizosphere soil may be attributed to the competition between B. dactyloides and indigenous microorganisms for nutrients such as carbon and nitrogen, and the combined treatment reversed this negative effect (Table S3). Our investigation revealed a substantial increase in microbial biomass in PAH contaminated soil as a result of the synergistic effect of B. dactyloides and biochar (Figure 3C). Thus, B. dactyloides and biochar increased microbial metabolic activity, enhanced microbial PAH degradation ability and alleviated PAH stress.
The structural composition of microorganisms is considered to be one of the main drivers of PAH dissipation [30,31]. Pseudoxanthomonas.g, Streptomyces.g, Sphingomonas.g, Ensifer.g, Methylibium.g, Devosia.g, Hydrogenophaga.g, Bacillus.g, Paenibacillus.g, Pseudomonas.g, Curvularia.g, Cladosporium.g, Acremonium.g and Chaetomium.g have been identified as potential participants in the degradation of PAH. We found that biochar and B. dactyloides roots induced soil microbial community reconstruction (Figure 2E,F). The difference between different treatments may be due to soil properties. TN, pH, TK, AK, AN, NO3-, SOM and DOC were identified as soil factors related to the change of microbial community structure and PAH degradation (Figure 4A). In this study, biochar and B. dactyloides increased soil nutrient content, while the increased nutrient content increased the bioavailability of PAHs, and may promote the biodegradation of PAHs through co-metabolic pathway. Pseudoxanthomonas.g, Sphingomonas.g, Streptomyces.g and Ensifer.g were significantly enriched as PAH degrading bacteria in rhizosphere soil with the presence of biochar. Biochar provided protection for Sphingomonas.g that can utilize PAH as carbon source [32], thus promoting the degradation of PAH under the combined application of plants and biochar. The enrichment of Pseudoxanthomonas.g may be attributed to the enrichment of soil nutrients subsequent to biochar application [33], potentially leading to the promotion of NH4+-N transformation and nitrogen fixation along with Ensifer.g (Table S3) [34,35]. The increase of soil PAH removal rate could be ascribed to root exudation promoting the degradation of soil PAH by Streptomyces.g [36]. Devosia.g, Hydrogenophaga.g, and Methylibium.g mainly contributed to the degradation of low molecular weight PAH [37,38,39], which supported the significantly increased Phe and Pyr removal in the rhizosphere of B. dactyloides under P treatment. The biodegradation intensity of PAH is related to the abundance and composition of genes. Dioxygenase, dehydrogenase, and hydroxylase genes are directly involved in the oxidative degradation of PAH [40,41]. The proportions of functional genes related to dioxygenase, dehydrogenase and hydroxylase in different treatments was consistent with the degradation of PAHs (Figure 5A). Consistent with previous studies [10,22], this suggests that key bacteria and functional genes played a pivotal role in soil PAH degradation, and this may explain the increase in removal rate of PB treatments.
4.2. The Effects of Biochar and Plant Roots on Soil Carbon Metabolism Associated with PAH Degradation
Microbial metabolism often reflects soil microbial community and function in response to stress. As an organic pollutant, PAH can inhibit the soil microbial activity, alter the community structure, function and metabolic activities of microorganisms, ultimately affect the soil quality [2]. Plants and biochar usually have different strategies for regulating microbial metabolism. In this study, no significant improvement in metabolic activity of microorganisms under P treatment (Figure 7A,B). Biochar amendment significantly improved the microbial carbon metabolism and amino acid metabolism in rhizosphere soil (Figure 7A,B), and specialized soil niche (Figure 4D), which was beneficial for microorganisms to adapt to PAH stress. This may be due to the fact that biochar improved the soil environment and affects the recruitment of rhizosphere microorganisms by altering the composition of root exudation [42]. In addition, the increased organic acid metabolites can effectively improve the bioavailability and promote the biodegradation of PAH by releasing PAH bound in organic matter through modifications to the rhizosphere enhanced with biochar [43]. The improvement of microbial amino acid metabolism by B. dactyloides roots and biochar may also favor the expression of PAH degradation genes [44]. And the involvement of amino acid metabolism in microbial detoxification reflects the improvement of microbial tolerance [10]. Similarly, the combination of B. dactyloides roots and biochar improved PAH bioavailability by significantly increasing lipid metabolism. Intermediate metabolites produced by indigenous microorganisms involved in the degradation of PAH, especially high molecular weight PAH, are components of the soil carbon cycle [45]. Therefore, the promotion of microbial carbon metabolism is beneficial for the in-situ degradation of PAH.
Biochar promoted soil carbon metabolism and bioremediation by increasing the contents of soil organic carbon [46]. Some soil metabolites and root exudation participate in the process of PAH degradation through the co-metabolic pathway [47]. Therefore, biochar could affect the in-situ degradation of PAH by regulating the utilization of soil carbon resources and changing soil metabolites. Moreover, soil metabolism affects microbial function together with microbial structure and soil characteristics (Figure 6B). This shows that there are complex interactions among bacteria, fungi, soil metabolites and biodegradation. Metabolic intermediates (catechol, dibutyl phthalate and 3-amino-4-hydroxybenzoate), root exudates (deoxycholic acid and taurine), coexisted with microorganisms involved in PAH degradation (streptomyces, curvularia, mortierella and acremonium) (Figure 7C,D) [48]. This suggests that plants and microorganisms forming complex interactions under PAHs stress jointly participate in and influence the removal of PAHs from soil. And it indicates that these compounds may be involved in the degradation of PAHs as co-metabolic substrates. This suggests that these compounds may be involved in PAH degradation as co-metabolic substrates. In addition, VPA showed that the significant improvement of PAH removal rate under PB treatment was driven by soil enzyme activity, soil metabolism, microflora and soil characteristics. Our research shows that biochar and B. dactyloides as remediation methods for PAHs contaminated soil can significantly improve the removal rate of PAHs, soil environment and the adaptability of microorganisms to PAHs.
5. Conclusions
B. dactyloides and biochar improved the removal rate of PAH. B. dactyloides and biochar increased soil enzyme activity, affected soil environment, regulated soil metabolism, changed the structure and function of soil microorganisms, in which the changes of soil environment, the structural composition of microorganisms and the expression of functional genes were considered to be the main driving forces of PAH removal. In addition, the microbial activity was increased, and the microbial carbon metabolism and amino acid metabolism were improved to cope with PAH stress under the combined action of B. dactyloides and biochar. These findings are of great significance for elucidating the underlying mechanism of this bioremediation strategy.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms12050968/s1. Table S1: Properties of soil in this study. Table S2: Main physiochemical characteristics and PAHs concentrations of woody biomass biochar pyrolyzed at 400 °C. Table S3: Properties of soil in this study. Table S4 Primers used for qPCR. Figure S1: Standard curves of 16S (a) and ITS (b).
Author Contributions
Y.W.: Conceptualization, Data curation, Methodology, Visualization, Writing—original draft; A.L.: Writing—review & editing; B.Z.: Writing—review & editing; Y.Q.: Resources; X.L.: Supervision, Funding acquisition, Writing—review & editing; Z.S.: Supervision, Project administration, Writing—review & editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Fundamental Research Funds of Chinese Academy of Forestry [grant numbers CAFYBB2022XA002] and Open Competition Project to Select the Best Candidates of the National Forestry and Grassland Administration “Breeding of Excellent Grass Varieties” (202201).
Data Availability Statement
We will upload the data to the platform and provide web pages before the article is published.
Acknowledgments
We would like to thank all colleagues and friends who have contributed to this study.
Conflicts of Interest
No conflict of interest exists in the submission of this manuscript, and the manuscript is approved by all authors for publication. I would like to declare on behalf of my co-authors that the work described was original research that has not been previously published, in whole or in part, and it is not under consideration by any other journal, in whole or in part.
References
- Thacharodi, A.; Hassan, S.; Singh, T.; Mandal, R.; Chinnadurai, J.; Khan, H.A.; Hussain, M.A.; Brindhadevi, K.; Pugazhendhi, A. Bioremediation of Polycyclic Aromatic Hydrocarbons: An Updated Microbiological Review. Chemosphere 2023, 328, 138498. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Qu, C.; Bian, Y.; Gu, C.; Jiang, X.; Song, Y. New Insights into the Responses of Soil Microorganisms to Polycyclic Aromatic Hydrocarbon Stress by Combining Enzyme Activity and Sequencing Analysis with Metabolomics. Environ. Pollut. 2019, 255, 113312. [Google Scholar] [CrossRef]
- Singha, L.P.; Pandey, P. Rhizosphere Assisted Bioengineering Approaches for the Mitigation of Petroleum Hydrocarbons Contamination in Soil. Crit. Rev. Biotechnol. 2021, 41, 749–766. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, M.; Tateno, R. Rhizosphere Effects on Soil Extracellular Enzymatic Activity and Microbial Abundance during the Low-Temperature Dormant Season in a Northern Hardwood Forest. Rhizosphere-Neth. 2022, 21, 100465. [Google Scholar] [CrossRef]
- Haichar, F.E.Z.; Santaella, C.; Heulin, T.; Achouak, W. Root Exudates Mediated Interactions Belowground. Soil Biol. Biochem. 2014, 77, 69–80. [Google Scholar] [CrossRef]
- Zhong, Y.; Xun, W.; Wang, X.; Tian, S.; Zhang, Y.; Li, D.; Zhou, Y.; Qin, Y.; Zhang, B.; Zhao, G.; et al. Root-Secreted Bitter Triterpene Modulates the Rhizosphere Microbiota to Improve Plant Fitness. Nat. Plants 2022, 8, 887–896. [Google Scholar] [CrossRef]
- Qi, D.; Feng, F.; Lu, C.; Fu, Y. C:N:P Stoichiometry of Different Soil Components after the Transition of Temperate Primary Coniferous and Broad-Leaved Mixed Forests to Secondary Forests. Soil Tillage Res. 2022, 216, 105260. [Google Scholar] [CrossRef]
- Hamid, Y.; Tang, L.; Hussain, B.; Usman, M.; Lin, Q.; Rashid, M.S.; He, Z.; Yang, X. Organic Soil Additives for the Remediation of Cadmium Contaminated Soils and Their Impact on the Soil-Plant System: A Review. Sci. Total Environ. 2020, 707, 136121. [Google Scholar] [CrossRef]
- Ji, M.; Wang, X.; Usman, M.; Liu, F.; Dan, Y.; Zhou, L.; Campanaro, S.; Luo, G.; Sang, W. Effects of Different Feedstocks-Based Biochar on Soil Remediation: A Review. Environ. Pollut. 2022, 294, 118655. [Google Scholar] [CrossRef]
- Li, X.; Yao, S.; Bian, Y.; Jiang, X.; Song, Y. The Combination of Biochar and Plant Roots Improves Soil Bacterial Adaptation to PAH Stress: Insights from Soil Enzymes, Microbiome, and Metabolome. J. Hazard. Mater. 2020, 400, 123227. [Google Scholar] [CrossRef]
- Quilliam, R.S.; Rangecroft, S.; Emmett, B.A.; Deluca, T.H.; Jones, D.L. Is Biochar a Source or Sink for Polycyclic Aromatic Hydrocarbon (PAH) Compounds in Agricultural Soils? GCB Bioenergy 2013, 5, 96–103. [Google Scholar] [CrossRef]
- Hao, Z.; Wang, Q.; Yan, Z.; Jiang, H. Novel Magnetic Loofah Sponge Biochar Enhancing Microbial Responses for the Remediation of Polycyclic Aromatic Hydrocarbons-Contaminated Sediment. J. Hazard. Mater. 2021, 401, 123859. [Google Scholar] [CrossRef]
- Ali, S.; Rizwan, M.; Qayyum, M.F.; Ok, Y.S.; Ibrahim, M.; Riaz, M.; Arif, M.S.; Hafeez, F.; Al-Wabel, M.I.; Shahzad, A.N. Biochar Soil Amendment on Alleviation of Drought and Salt Stress in Plants: A Critical Review. Environ. Sci. Pollut. Res. 2017, 24, 12700–12712. [Google Scholar] [CrossRef]
- Li, X.; Song, Y.; Bian, Y.; Gu, C.; Yang, X.; Wang, F.; Jiang, X. Insights into the Mechanisms Underlying Efficient Rhizodegradation of PAHs in Biochar-Amended Soil: From Microbial Communities to Soil Metabolomics. Ecol. Indic. 2020, 144, 105995. [Google Scholar] [CrossRef] [PubMed]
- Bornø, M.L.; Müller-Stöver, D.S.; Liu, F. Biochar Modifies the Content of Primary Metabolites in the Rhizosphere of Well-Watered and Drought-Stressed Zea Mays L. (Maize). Biol. Fertil. Soils 2022, 58, 633–647. [Google Scholar] [CrossRef]
- Wang, Y.; Fang, L.; Lin, L.; Luan, T.; Tam, N.F.Y. Effects of Low Molecular-Weight Organic Acids and Dehydrogenase Activity in Rhizosphere Sediments of Mangrove Plants on Phytoremediation of Polycyclic Aromatic Hydrocarbons. Chemosphere 2014, 99, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Guo, X.; Zhu, Y.; Liu, X.; Han, Z.; Sun, K.; Ji, L.; He, Q.; Han, L. The Effects of Different Biochars on Microbial Quantity, Microbial Community Shift, Enzyme Activity, and Biodegradation of Polycyclic Aromatic Hydrocarbons in Soil. Geoderma 2018, 328, 100–108. [Google Scholar] [CrossRef]
- Kramer-Walter, K.R.; Laughlin, D.C. Root Nutrient Concentration and Biomass Allocation Are More Plastic than Morphological Traits in Response to Nutrient Limitation. Plant Soil 2017, 416, 539–550. [Google Scholar] [CrossRef]
- Song, X.; Razavi, B.S.; Ludwig, B.; Zamanian, K.; Zang, H.; Kuzyakov, Y.; Dippold, M.A.; Gunina, A. Combined Biochar and Nitrogen Application Stimulates Enzyme Activity and Root Plasticity. Sci. Total Environ. 2020, 735, 139393. [Google Scholar] [CrossRef]
- Wan, H.; Liu, X.; Shi, Q.; Chen, Y.; Jiang, M.; Zhang, J.; Cui, B.; Hou, J.; Wei, Z.; Hossain, M.A.; et al. Biochar Amendment Alters Root Morphology of Maize Plant: Its Implications in Enhancing Nutrient Uptake and Shoot Growth under Reduced Irrigation Regimes. Front. Plant Sci. 2023, 14, 1122742. [Google Scholar] [CrossRef]
- Zhu, H.; Gao, Y.; Li, D. Germination of Grass Species in Soil Affected by Crude Oil Contamination. Int. J. Phytoremediat. 2018, 20, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Li, X.; Liu, J.; Cheng, Y.; Zou, J.; Zhai, F.; Sun, Z.; Han, L. Soil Microbial Community Succession and Interactions during Combined Plant/White-Rot Fungus Remediation of Polycyclic Aromatic Hydrocarbons. Sci. Total Environ. 2021, 752, 142224. [Google Scholar] [CrossRef] [PubMed]
- Garcia, C.; Hernandez, T.; Costa, F. Potential Use of Dehydrogenase Activity as an Index of Microbial Activity in Degraded Soils. Commun. Soil Sci. Plant Anal. 1997, 28, 123–134. [Google Scholar] [CrossRef]
- Zelena, E.; Dunn, W.B.; Broadhurst, D.; Francis-McIntyre, S.; Carroll, K.M.; Begley, P.; O’Hagan, S.; Knowles, J.D.; Halsall, A.; HUSERMET Consortium; et al. Development of a Robust and Repeatable UPLC−MS Method for the Long-Term Metabolomic Study of Human Serum. Anal. Chem. 2009, 81, 1357–1364. [Google Scholar] [CrossRef] [PubMed]
- Want, E.J.; Masson, P.; Michopoulos, F.; Wilson, I.D.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J.K. Global Metabolic Profiling of Animal and Human Tissues via UPLC-MS. Nat. Protoc. 2013, 8, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Vasilev, N.; Boccard, J.; Lang, G.; Grömping, U.; Fischer, R.; Goepfert, S.; Rudaz, S.; Schillberg, S. Structured Plant Metabolomics for the Simultaneous Exploration of Multiple Factors. Sci. Rep. 2016, 6, 37390. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Liu, R.; Zhou, Y.; Li, N.; Hou, L.; Ma, Q.; Gao, B. Fire Phoenix Facilitates Phytoremediation of PAH-Cd Co-Contaminated Soil through Promotion of Beneficial Rhizosphere Bacterial Communities. Environ. Int. 2020, 136, 105421. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Yang, B.; Song, Z.; Wang, H.; He, F.; Han, X. Wheat Straw Biochar Amendments on the Removal of Polycyclic Aromatic Hydrocarbons (PAHs) in Contaminated Soil. Ecotoxicol. Environ. Saf. 2016, 130, 248–255. [Google Scholar] [CrossRef]
- Shi, W.; Zhang, X.; Jia, H.; Feng, S.; Yang, Z.; Zhao, O.; Li, Y. Effective Remediation of Aged HMW-PAHs Polluted Agricultural Soil by the Combination of Fusarium sp. and Smooth Bromegrass (Bromus inermis Leyss.). J. Integr. Agric. 2017, 16, 199–209. [Google Scholar] [CrossRef]
- Wang, X.; Chen, J.; Wang, Y.; Li, Y.; Chen, S.; Zhang, Q. Promoting Resuscitation of the VBNC Bacteria and Enrichment of Naphthalene-Degrading Bacteria from Oil-Contaminated Soils with the Rpf4 from Rhodococcus Erythropolis KB1. Geomicrobiol. J. 2022, 39, 916–924. [Google Scholar] [CrossRef]
- Lin, X.; Qiao, B.; Chang, R.; Li, Y.; Zheng, W.; He, Z.; Tian, Y. Characterization of Two Keystone Taxa, Sulfur-Oxidizing, and Nitrate-Reducing Bacteria, by Tracking Their Role Transitions in the Benzo[a]Pyrene Degradative Microbiome. Microbiome 2023, 11, 139. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Niu, X.; Zhang, N.; Li, T. Effect of Biochar-Immobilized Sphingomonas Sp. PJ2 on Bioremediation of PAHs and Bacterial Community Composition in Saline Soil. Chemosphere 2021, 279, 130427. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Tang, H.; Wang, Y.Z.; Yang, C.; Gao, M.-T.; Tsang, Y.F.; Li, J. Effect of Dissolved Solids Released from Biochar on Soil Microbial Metabolism. Environ. Sci. Process. Impacts 2022, 598–608. [Google Scholar] [CrossRef] [PubMed]
- Mei, J.; Li, B.; Su, L.; Zhou, X.; Duan, E. Effects of Potassium Persulfate on Nitrogen Loss and Microbial Community during Cow Manure and Corn Straw Composting. Bioresour. Technol. 2022, 363, 127919. [Google Scholar] [CrossRef] [PubMed]
- Shakoor, N.; Hussain, M.; Adeel, M.; Azeem, I.; Ahmad, M.A.; Zain, M.; Zhang, P.; Li, Y.; Quanlong, W.; Horton, R.; et al. Lithium-Induced Alterations in Soybean Nodulation and Nitrogen Fixation through Multifunctional Mechanisms. Sci. Total Environ. 2023, 904, 166438. [Google Scholar] [CrossRef] [PubMed]
- Baoune, H.; Aparicio, J.D.; Acuña, A.; El Hadj-khelil, A.O.; Sanchez, L.; Polti, M.A.; Alvarez, A. Effectiveness of the Zea Mays-Streptomyces Association for the Phytoremediation of Petroleum Hydrocarbons Impacted Soils. Ecotoxicol. Environ. Saf. 2019, 184, 109591. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, Q.; Xie, S. Stable Isotope Probing Identifies Anthracene Degraders under Methanogenic Conditions. Biodegradation 2012, 23, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Singha, L.P.; Pandey, P. Rhizobacterial Community of Jatropha Curcas Associated with Pyrene Biodegradation by Consortium of PAH-Degrading Bacteria. Appl. Soil Ecol. 2020, 155, 103685. [Google Scholar] [CrossRef]
- Wang, X.; Teng, Y.; Wang, X.; Li, X.; Luo, Y. Microbial Diversity Drives Pyrene Dissipation in Soil. Sci. Total Environ. 2022, 819, 153082. [Google Scholar] [CrossRef]
- Zhang, X.-X.; Cheng, S.-P.; Zhu, C.-J.; Sun, S.-L. Microbial PAH-Degradation in Soil: Degradation Pathways and Contributing Factors1 1Project Supported by the National High Technology Research and Development Program (863 Program) of China (No. 2001AA214191). Pedosphere 2006, 16, 555–565. [Google Scholar] [CrossRef]
- Li, X.; Song, Y.; Wang, F.; Bian, Y.; Jiang, X. Combined Effects of Maize Straw Biochar and Oxalic Acid on the Dissipation of Polycyclic Aromatic Hydrocarbons and Microbial Community Structures in Soil: A Mechanistic Study. J. Hazard. Mater. 2018, 364, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, A.; Dennis, P.G.; Forstner, C.; Raghavendra, A.K.H.; Mathesius, U.; Richardson, A.E.; Delhaize, E.; Gilliham, M.; Watt, M.; Ryan, P.R. Manipulating Exudate Composition from Root Apices Shapes the Microbiome throughout the Root System. Plant Physiol. 2021, 187, 2279–2295. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Hu, X.; Zhou, Z.; Zhang, W.; Wang, Y.; Sun, B. Phytoavailability and Mechanism of Bound PAH Residues in Filed Contaminated Soils. Environ. Pollut. 2017, 222, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Badejo, A.C.; Choi, C.-W.; Badejo, A.O.; Shin, K.-H.; Hyun, J.-H.; Lee, Y.-G.; Kim, S.; Park, K.-S.; Kim, S.H.; Jung, K.H.; et al. A Global Proteome Study of Mycobacterium Gilvum PYR-GCK Grown on Pyrene and Glucose Reveals the Activation of Glyoxylate, Shikimate and Gluconeogenetic Pathways through the Central Carbon Metabolism Highway. Biodegradation 2013, 24, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Ressler, B.P.; Kneifel, H.; Winter, J. Bioavailability of Polycyclic Aromatic Hydrocarbons and Formation of Humic Acid-like Residues during Bacterial PAH Degradation. Appl. Microbiol. Biotechnol. 1999, 85–91. [Google Scholar] [CrossRef]
- Wang, S.; Guo, S. Effects of Soil Organic Carbon Metabolism on Electro-Bioremediation of Petroleum-Contaminated Soil. J. Hazard. Mater. 2023, 459, 132180. [Google Scholar] [CrossRef] [PubMed]
- Techer, D.; D’Innocenzo, M.; Laval-Gilly, P.; Henry, S.; Bennasroune, A.; Martinez-Chois, C.; Falla, J. Assessment of Miscanthus×giganteus Secondary Root Metabolites for the Biostimulation of PAH-Utilizing Soil Bacteria. Appl. Soil Ecol. 2012, 62, 142–146. [Google Scholar] [CrossRef]
- Zhao, W.; Li, S.; Dong, L.; Wang, P.; Lu, X.; Zhang, X.; Su, Z.; Guo, Q.; Ma, P. Effects of Different Crop Rotations on the Incidence of Cotton Verticillium Wilt and Structure and Function of the Rhizospheric Microbial Community. Plant Soil 2023, 485, 457–474. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).