Abstract
Antimicrobial resistance is recognised as one of the top threats healthcare is bound to face in the future. There have been various attempts to preserve the efficacy of existing antimicrobials, develop new and efficient antimicrobials, manage infections with multi-drug resistant strains, and improve patient outcomes, resulting in a growing mass of routinely available data, including electronic health records and microbiological information that can be employed to develop individualised antimicrobial stewardship. Machine learning methods have been developed to predict antimicrobial resistance from whole-genome sequencing data, forecast medication susceptibility, recognise epidemic patterns for surveillance purposes, or propose new antibacterial treatments and accelerate scientific discovery. Unfortunately, there is an evident gap between the number of machine learning applications in science and the effective implementation of these systems. This narrative review highlights some of the outstanding opportunities that machine learning offers when applied in research related to antimicrobial resistance. In the future, machine learning tools may prove to be superbugs’ kryptonite. This review aims to provide an overview of available publications to aid researchers that are looking to expand their work with new approaches and to acquaint them with the current application of machine learning techniques in this field.
1. Introduction
Antimicrobial resistance is recognised as one of the top threats healthcare is bound to face in the future. There have been various attempts to preserve the efficacy of existing antimicrobials, develop new and efficient antimicrobials, manage infections with multi-drug resistant strains, and improve patient outcomes, resulting in a growing mass of routinely available data including electronic health records and microbiological information that can be employed to develop individualised antimicrobial stewardship [1].
As science has evolved, there has been an overlap between fields, and researchers are seeing more benefits from utilising recent advancements in computational methods, machine learning, and artificial intelligence (AI) development. These technologies are rapidly being incorporated across various disciplines and almost every research area to accelerate scientific discoveries. As they allow the optimal utilisation of large amounts of data, for example, clinical and laboratory data, and can support evidence-based decision making, they substantially save time on research [1,2,3,4].
High-quality machine learning models rely on comprehensive, structured, well-curated datasets [5]. When adequately designed and trained, models can extract meaningful rules and develop predictive tools; they learn objectively and, more often than not, make more accurate predictions compared with those seen in everyday practice [1]. Ultimately, the goal is to develop a system that autonomously improves its performance and accuracy without programming [6]. Such systems would substantially contribute to easing the burden of antimicrobial resistance [7].
A recent study revealed a substantial increase in publications and research on the use of machine learning in the field of antimicrobial resistance [8]. These methods have broad applications in microbiology, from diagnostics, drug and vaccine discovery and applications in epidemiology [1,3,9].
In biomedical research, in the past ten years, broad application of artificial intelligence, deep learning, and machine learning methods can be seen across the field. This narrative review highlights some of the remarkable opportunities that machine learning offers when applied in research related to antimicrobial resistance, including genome analysis for prediction of resistant strains, susceptibility testing, drug discovery, and potential clinical applications. Due to the exponential number of publications on the topic of machine learning applications in the field of antimicrobial resistance, this review aims to provide an overview rather than an in-depth analysis of available publications to aid researchers that are looking to expand their work with new approaches and to acquaint them with the current application of machine learning techniques in this field.
2. Genome Analysis for Prediction of Resistant Strains and Susceptibility Testing
Whole-genome sequence data and susceptibility profiles of different microorganisms can be used to train models to predict resistance in bacteria [10,11,12]. Machine learning has been widely utilised in predicting antimicrobial resistance genes and translating genetic mechanisms [13,14,15,16,17,18,19,20,21]. Machine learning based on protein sequences successfully predicted antimicrobial resistance genes in Gram-negative bacteria with over 90% accuracy [22,23]. Researchers investigating biomarkers for Pseudomonas aeruginosa’s carbapenem resistance implemented a learning model using a gradient boosting decision tree (GBDT) algorithm to screen for important glycan structures associated with resistant strains [24]. A machine learning model was successfully implemented in research for the classification of resistant genomes in Pseudomonas aeruginosa. According to the authors, the machine learning classifier had more predictive power for resistance to individual carbapenems than the broader mechanistic categories, as it included even the uncharacterised mutations found on the genes associated with these mechanisms. Furthermore, it identified several specific features relevant to predicting resistance, although these were not present in greater frequency among the resistant isolates [25]. Resistance prediction models of Pseudomonas aeruginosa for imipenem and meropenem were constructed using machine learning techniques paired with next-generation sequencing through matching resistance genotypes with phenotypes. Such models can be used to directly predict carbapenem resistance in clinical samples [26]. Machine learning was utilised to determine whether Elizabethkingia bruuniana and Elizabethkingia meningoseptica strains were resistant or susceptible to different antimicrobials [27]. Using 25 whole-genome sequences, researchers developed a genome-based machine-learning approach for discrimination between vancomycin-intermediate and vancomycin-susceptible Staphylococcus aureus [28]. Furthermore, PRAP, the Pan-Resistome Analysis Pipeline, has successfully analysed the genome and identified antimicrobial resistance genes of Salmonella enterica isolates [29].
Machine learning can improve the prediction of the antimicrobial resistance phenotype from genomic data [30]. Efforts are being made to make the models more generalisable as training a distinct model for each antimicrobial and species for predicting resistance may miss some valuable opportunities [31]. When integrating machine learning into research, antimicrobial resistance genes are often identified that have never previously been associated with resistance in any microbe [32,33]. Machine learning techniques can analyse large datasets much faster than traditional statistical methods. This is particularly relevant in genomics, where whole genome sequencing data is used to predict antibiotic resistance. Machine learning can swiftly identify patterns or resistance genes that would take humans much longer to find. Moreover, machine learning models, once trained, can predict antibiotic resistance from genomic data almost instantly. This is much faster than culture-based methods, which can take days [30,31,32,33].
Moreover, successfully developed novel machine learning models for predicting resistance of Escherichia coli to colistin have identified some previously unknown colistin-resistant biomarkers [34]. Supervised machine learning classifiers identified known and unknown resistance-associated mutations and genes related to resistance to 28 antimicrobials in Escherichia coli and Salmonella enterica [35]. Machine learning can be utilised to successfully identify known and potential new antimicrobial genes in Neisseria gonorrhoeae [36]. DeepARG is an example of a model developed for predicting antimicrobial resistance based on metagenomic data [37] (Table 1). These techniques have allowed the prediction of antimicrobial resistance based on genome sequences [38,39].
A reservoir of resistance genes may come from previously unknown sources. Other than identifying known resistance genes, machine learning algorithms may be utilised to identify novel yet unrecognised resistance genes [40]. Artificial intelligence has the potential to predict the presence and spread of antimicrobial resistance genes within and across populations [4]. Machine learning was successfully implemented for the identification of antimicrobial resistance genes retrieved from strains on the International Space Station’s environmental surfaces, overcoming traditional cut-offs based on high similarity in DNA sequencing and expanding the catalogue of antimicrobial resistance genes, creating opportunity for alternatives to the current gather-and-return sampling model used for the International Space Station [41].
Many machine learning systems have been developed for the field of clinical microbiology, a number of them for antimicrobial susceptibility testing [4,42]. There is an opportunity for utilisation of machine learning methods in the design and development of sequence-based diagnostics that also predict antimicrobial susceptibility, hence enabling tailored treatment [4,43]. These methods have been utilised to develop tools that predict resistance to a specific antimicrobial, i.e., rifampicin [44] (Table 1). A multi-component, microscopy-based approach that included a deep learning method for automated image classification and prediction of antimicrobial minimal inhibitory concentrations was proposed [45]. Minimum inhibitory concentrations of ciprofloxacin against Escherichia coli were successfully predicted based on different mutations [46] (Table 1). There has been much research on machine learning for the prediction of minimum inhibitory concentrations and the determination of resistant strains of different pathogens [47] (Table 1). This can be further developed to predict multi-drug resistance in pathogens [48] (Table 1). Machine learning techniques were adopted to support research into bacterial resistance to a panel of antimicrobials using whole-genome sequence data of Pseudomonas aeruginosa, with more than 95% accuracy [49] (Table 1). Furthermore, a similar system was used for the identification of methicillin resistance of Staphylococcus aureus, with an accuracy of 87.6%, sensitivity of 91.8%, and specificity of 83.3% [50] (Table 1). In other research, machine learning models were able to predict the minimum inhibitory concentrations of ten different antimicrobial agents for Staphylococcus aureus [51] (Table 1). Machine learning models have been utilised to predict minimum inhibitory concentrations of cefixime, ciprofloxacin, and azithromycin against Neisseria gonorrhoeae [52].
Whole-genome sequencing data have been successfully used to train models to predict minimum inhibitory concentrations of different antimicrobials against different pathogens, for example, Klebsiella pneumoniae [53]. Machine learning based on whole-genome sequencing was used to predict the resistance of four antimicrobials, ciprofloxacin, ceftazidime, cefotaxime, and gentamicin, to Escherichia coli [54,55].
Integrating traditional machine learning with deep learning models was used to predict minimum inhibitory concentrations of 15 antimicrobials against Salmonella [56]. Another study predicted minimum inhibitory concentrations of 15 antimicrobials for nontyphoidal Salmonella [39]. With the application of a machine learning feature-selection approach on a Salmonella enterica pan-genome, researchers could predict minimum inhibitory concentration values with very high accuracy [57]. Researchers utilised three machine learning algorithms to estimate minimum inhibitory concentrations of 13 antimicrobials against Acinetobacter baumannii [58] (Table 1). Furthermore, using multi-branch CNN and Attention model, a deep learning method outperformed traditional machine learning methods when applied in the prediction of the minimum inhibitory concentrations of peptides with antimicrobial activity against Escherichia coli [59] (Table 1). However, when several different machine learning models were designed for the prediction of resistance or susceptibility to ciprofloxacin, ceftazidime, and meropenem, some achieved lower accuracy depending on the input data used for training [60] (Table 1). Several available datasets have been used in the training of machine learning models [61]. Most antimicrobial resistance-related machines are not yet ready for implementation in real-life settings. Implementing these methods would only be feasible with further improvement in accuracy and a reduced financial cost, especially for pathogens that take longer to culture [62]. Models that accurately predict antimicrobial resistance phenotypes from genotypes are growing and being further optimised [63,64]. With more significant amounts of data, the performance of machine learning models improves [65]. A deep transfer learning model was developed to overcome the known pitfalls of machine learning when operating with limited datasets. This model achieved accurate and robust antimicrobial resistance predictions based on small, imbalanced datasets [66].
Raman spectroscopy was enhanced with machine learning techniques for identification and antimicrobial susceptibility testing [67]. When surface-enhanced Raman spectroscopy was coupled with machine learning methods, it detected virulence and carbapenem resistance in Klebsiella pneumoniae [68,69] (Table 1). Furthermore, it was successfully employed to differentiate resistant and susceptible strains of Escherichia coli [70]. Rapid identification of methicillin-resistant Staphylococcus aureus is possible when surface-enhanced Raman spectroscopy is coupled with deep learning methods [71].
Coupled with matrix-assisted laser desorption/ionisation and time-of-flight mass spectrometry (MALDI-TOF MS), machine learning has successfully been implemented for antimicrobial susceptibility testing and prediction of antimicrobial resistance in pathogens [72,73]. Examples include identifying antimicrobial resistance to benzylpenicillin or multidrug Staphylococcus aureus isolates and development of high-performance classifiers for ciprofloxacin and tetracycline-resistant Campylobacter jejuni and Campylobacter coli isolates [74,75]. Methods have been developed to distinguish vancomycin-intermediate Staphylococcus aureus from vancomycin-susceptible Staphylococcus aureus [76,77]. A convolutional neural network model was developed to rapidly predict vancomycin-resistant Enterococcus faecium in clinical samples after analysis of the matrix-assisted laser desorption ionisation time-of-flight mass spectrometry peaks’ pattern [78]. MALDI-TOF MS spectra coupled with a machine learning algorithm rapidly predicted the susceptibility of Enterococcus faecium to vancomycin with a mean accuracy of 0.78 [79].
Artificial intelligence and machine learning have been implemented to diagnose microorganisms and predict susceptibility to antimicrobial agents [80]. Researchers in China investigating the Pseudomonas aeruginosa ST316 sublineage causing ear infections used the K-mer machine learning approach to generate predictive models and identify biomarkers of resistance to gentamicin, fosfomycin, and cefoperazone–sulbactam [81]. A machine learning approach using mutations in Neisseria gonorrhoeae was proven helpful in predicting ceftriaxone susceptibility and decreased susceptibility. From seven investigated models, the random forest classified model showed the highest performance [82]. Machine learning was utilised for differentiating Enterobacter cloacae complex species [83].
Machine learning algorithms were implemented while developing antimicrobial resistance prediction models for Escherichia coli [84,85]. These algorithms were successfully utilised to enhance methods for discrimination between piperacillin/tazobactam-resistant and susceptible Escherichia coli isolates [86]. Furthermore, deep learning of single-cell subcellular phenotypes was utilised for determining antimicrobial susceptibility in Escherichia coli for ciprofloxacin, gentamicin, rifampicin, and co-amoxiclav [87].
Machine learning techniques have been successfully applied to distinguish the methicillin-resistant form of methicillin-sensitive Staphylococcus aureus [88,89]. A deep residual learning framework was trained for the classification of bacteria samples. The model predicted Staphylococcus aureus, Klebsiella pneumoniae, and Bacillus subtilis classes with 100% sensitivity. Such attempts may enable bacterial identification, avoiding cell culturing [90]. Furthermore, researchers have developed a mNGS-based machine learning model for rapid antimicrobial susceptibility testing of Acinetobacter baumannii that may come to take the place of time-consuming culture-based procedures [91]. This may be particularly beneficial for pathogens that require long culturing, such as Mycobacterium tuberculosis [92].
Machine learning classifiers can predict resistant and susceptible phenotypes of Mycobacterium tuberculosis, Klebsiella pneumoniae, and Salmonella enterica using bacterial genome sequence data [93]. There are several applications for machine learning in predicting drug resistance of Mycobacterium tuberculosis from sequence data [92,94,95,96,97,98,99,100,101]. The deep graph learning method has been used to predict drug resistance in tuberculosis with a sensitivity of over 90% for isoniazid, ethambutol, and pyrazinamide [102]. Incorporating different models into research, potential new candidate mutations in Mycobacterium tuberculosis not previously reported in the literature, which can be related to resistance, have been identified [103]. Convolutional neural network applied in the analysis of the Mycobacterium tuberculosis genome identified 18 sites not previously associated with antimicrobial resistance [104]. Some tools have been made available to interested researchers. The TuBerculosis Drug Resistance Optimal Prediction tool allows users to input sequencing data for the prediction of resistance in Mycobacterium tuberculosis [105]. The Translational Genomics platform for Tuberculosis, GenTB, is a publicly available online tool designed to predict drug resistance in Mycobacterium tuberculosis from genomic data [106].
These methods can be used to predict pyrazinamide resistance of Mycobacterium tuberculosis and to pinpoint genes and mutations responsible for the resistance [107,108]. A pipeline for developing machine learning models of mycobacterium tuberculosis drug resistance prediction was made publicly available [109]. Moreover, a metabolic allele classifier was developed to predict antimicrobial resistance phenotypes of Mycobacterium tuberculosis [110]. Overall, there have been many applications in drug resistance profiling of tuberculosis [111]. In drug resistance profiling of tuberculosis, some models have achieved 99% accuracy [112]. Machine learning classifiers were used to predict resistance mutations related to rifampicin as an alternative rapid method [113].
Machine learning techniques have been proposed for the prediction of antimicrobial resistance of Klebsiella pneumoniae [114]. These models coupled with logistic regression have been successfully used for identification of strong resistance predictors in Klebsiella pneumoniae. Such models may guide diagnosis and treatment selection in clinical practice [115]. Analysis of genomic data with the implementation of machine learning has allowed predictoin of resistance to polymyxins in Klebsiella pneumoniae [116].
Machine learning technologies can be implemented for rapid differentiation of resistant Mycobacterium abscessus complex subspecies from macrolide-susceptible subspecies [117]. These technologies have been used with other methods to investigate mechanisms of resistance in Pneumocystis jirovecii [118]. Furthermore, machine learning models were utilised to analyse the drug resistance of Candida auris [119].
Machine learning algorithms can identify risk and clinical predictors of multidrug-resistant Enterobacterales infections in persons infected with HIV [120]. Moreover, different machine learning approaches have been utilised to predict the drug resistance of HIV [121,122,123,124,125,126,127,128,129,130,131,132]. For example, models were implemented to predict HIV-1 protease resistance [133]. A web application, SHIVA, that can determine drug resistance in HIV has been developed [134]. Furthermore, the freely available web service HVR, based on a validated machine learning model, predicts human immunodeficiency virus type 1 drug resistance to nucleoside and non-nucleoside reverse transcriptase and protease inhibitors [135]. Furthermore, research has demonstrated that deep learning networks are able to differentiate viruses in clinical samples and that the addition of deep learning to single-particle fluorescence microscopy can rapidly detect and classify viruses [136]. There have been efforts to predict antiviral drug resistance of the Influenza virus with machine learning [137]. In research incorporating statistical analysis, a machine learning algorithm was used for predictive analysis of virulence genes to explore further the ones potentially related to specific antimicrobial resistance phenotypes [138].

Table 1.
Summary of selected studies showing the efficacy of machine learning in AMR prediction.
Table 1.
Summary of selected studies showing the efficacy of machine learning in AMR prediction.
| Study | Pathogen | Algorithm | Target | Result |
|---|---|---|---|---|
| Aytan-Aktug et al. [10] | Mycobacterium tuberculosis, Escherichia coli, Salmonella enterica, Staphylococcus aureus | Neural network | Multiple AMR profile prediction | AUC 0.90–0.95 on test dataset |
| Chowdhury et al. [22] | Acinetobacter, Klebsiella, Campylobacter, Salmonella, and Escherichia | SVM | AMR gene classification | >90% accuracy |
| Pesesky et al. [23] | Gram-negative Bacilli | Logistic regression | Antibiotic resistance prediction | Agreement of 90.8% to the phenotypic ASTs |
| Dang et al. [24] | P. aeruginosa | GBT | Predicting glycopatterns for carbapenem resistance | AUC 0.95 |
| Liu et al. [25] | P. aeruginosa | Neural network | Predicting imipenem and carbapenem resistance | AUC 0.906 and 0.925 |
| Tian et al. [34] | E. coli | Lasso regression | Predicting colistin resistance | AUC 0.902 on validation dataset |
| Shi et al. [36] | Neisseria gonorrhoeae | DNP-AAP | Predicting AMR | AUC 0.97–0.99 |
| Arango-Argoty et al. [37] | Antibiotic resistance genes from multiple pathogens | DeepARG | Predicting ARG | Precision (>0.97), recall (>0.90) |
| Portelli et al. [44] | M. tuberculosis | Linear classifiers, decision trees, ensemble classifiers | Predicting rifampicin resistance | Sensitivity 92.2%, specificity 83.6% |
| Pataki et al. [46] | E. coli | Random forest | Predicting ciprofloxacin minimum inhibitory concentration | AUC 0.99 |
| Valizdeh et al. [47] | C. jejuni, S. enterica, N. gonorrhoeae and K. pneumoniae | XgBoost | Predicting AMR | Accuracy 0.95–0.97 |
| Ren et al. [48] | E. coli | Random forest | Multi-drug resistance prediction | F score 0.93 ± 0.04 |
| Noman et al. [49] | P. aeruginosa | Random forest | Predicting AMR | Accuracy > 0.97 |
| Jeon et al. [50] | S. aureus | AMRQuest | Presumptive identification of MRSA | Sensitivity of 91.8%, Specificity of 83.3%, Accuracy of 87.6% |
| Wang et al. [51] | S. aureus | XgBoost, random forest, SVM | Identification of MRSA | Category agreement > 85% and >90% (one-two fold dilution) |
| Ayoola et al. [56] | Salmonella spp. | Genome feature extractor pipeline (combining random forest and MLP) | Predicting MIC | Accuracy > 96% |
| Gao et al. [58] | A. baumani | Random forest, SVM, and XgBoost | Predicting MIC | Average essential agreement 90.90% (95% CI, 89.03–92.77%) |
| Yan et al. [59] | E. coli | MBC-Attention | Predicting AMR | PCC of 0.775 and a root mean squared error (RMSE) of 0.533 (log μM) |
| Yasir et al. [60] | P. aeruginosa | Random forest, and nine other classifiers | Predicting AMR | Accuracy 0.73 |
| Ren et al. [66] | E. coli | CNN with transfer learning | Predicting AMR | Validation AUC 0.72–0.93 |
| Lu et al. [68] | K. pneumoniae | CNN on Raman spectroscopy | Predicting AMR | AUC 0.97 |
SVM—support vector machine, AMR—antimicrobial resistance, ASTs—antibiotic susceptibility tests, AUC—area under the ROC curve, DNP-AAP—(deep neural pursuit—average activation potential), DeepARG—deep learning antibiotic resistance gene, XgBoost—Extreme Gradient Boosting machine, MLP—multi-layered perceptron (neural network), MIC—minimal inhibitory concentration, MBC-Attention—a combination of multi-layered convolutional networks and attention mechanism, PCC—Pearson correlation coefficient, RMSE—root mean squared error, CNN—convolutional neural network.
3. Drug Discovery
Machine learning may find great application in rapid antimicrobial resistance predictions, and it may enhance the discovery of novel antimicrobials [66] (Figure 1). In research on antimicrobial peptides, support vector machines, random forest machine learning algorithms, deep neural networks, and convolutional neural networks have found application [139]. Previously developed machine learning models were used to screen a chemo library and identify potential drug candidates for known therapeutic targets [140]. Traditional machine learning may be used to discover antimicrobial peptides in large-scale natural known peptide libraries, while artificial neural networks may predict peptide activity against pathogens and identify highly active peptides. Furthermore, models may be used for design by optimisation and in de novo design of antimicrobial peptides [141]. Machine learning models may be used as aids in screening during the early stages of designing peptide-based antimicrobials [142,143,144,145,146,147,148,149,150]. Machine learning algorithms allow rapid in silico screening and classification of antimicrobial peptides for investigating activity against specific species, providing savings and replacing or enhancing time-consuming conventional methods [151]. These techniques can be used to screen libraries of compounds to identify drug candidates. This approach was used to discover beta-lactamase CMY-10 inhibitors against Enterobacteriaceae [152].
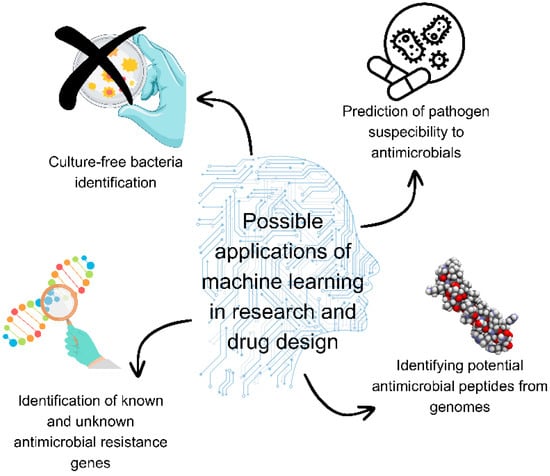
Figure 1.
Possible applications of machine learning in research and drug design. Figure was made using Canva.
Machine learning techniques have been widely applied in research to develop and enhance antimicrobials [153]. A machine learning model, CalcAMP, can predict the antimicrobial activity of peptides against both Gram-positive and Gram-negative bacteria [154]. An antimicrobial peptide classification model, AMP-BERT, was developed as a tool for pre-screening for antimicrobial drug candidates that may promote the discovery process of antimicrobial drugs [155]. Deep-AmPEP30 is a prediction tool to identify short-length antimicrobial peptides from genomic sequences [156]. However, antimicrobial peptides can be identified from various organisms [157]. Machine learning methods were implemented as tools to identify possible embedded antimicrobial peptides from different proteomes [158]. Deep learning models can be implemented for predicting antimicrobial peptides in peptide sequences, such as AMPlify, which identified four antimicrobial peptides in the bullfrog genome [159,160].
To overcome the fact that computational approaches are usually unable to predict the functional activities of antimicrobial peptides, researchers recently developed a deep learning approach, iAMPCN, that improved the prediction of antimicrobial peptides’ functional activities [161]. A proposed model of NIRBMMDA was developed for the prediction of potential microbe–drug association and has shown promising results. For example, 17 of the top 20 predicted microbes for ciprofloxacin were identified and confirmed in published literature. However, such models’ performance depends on the dataset used to train the model [162]. Furthermore, machine learning was utilised in research into the structure–activity relationship of New Delhi metallo-beta-lactamase-1 inhibitors, setting a path for further drug discovery and drug optimisation [163].
Machine learning techniques can be incorporated into all stages of drug discovery, including predicting biological responses. Machine learning has been utilised to guide decision making during the de novo design and differentiation of molecules with desired properties. Models have been applied to accelerate the discovery and design of drug candidates against different multidrug-resistant microbes. They may significantly cut the conventional cost and screening efforts required for the process of screening for new drug candidates [164].
Several tools have been developed for identification of lead compounds against Mycobacterium tuberculosis, for prediction of the potential toxicity of a compound of interest, and for classifying drug resistance in isolates and linking specific mutations with resistance [165]. Machine learning techniques have been utilised to predict the antitubercular effects of drugs [166]. Researchers have developed a computational model (termed iAMAP-SCM) that can identify and characterise peptides with antimalarial activity using just sequence information [167]. Furthermore, machine learning techniques have been utilised to design novel peptide sequences with expected antibacterial activity against Escherichia coli [168]. A similar method was employed to identify an antimicrobial peptide active against Staphylococcus epidermidis [169].
Available machine-learning algorithms have been used for assessing the theoretical function of newly designed antimicrobial peptides [170]. In research, deep learning and computational methods were utilised to create and prioritise de novo antimicrobial peptides, which showed no development of resistance in vitro [171]. Moreover, chemically modified peptides can be analysed to predict their potential antimicrobial activity [172].
Computational tools have been used for possible target identification, namely, quorum-sensing peptides [173]. Machine learning algorithms may be utilised to predict molecular properties of novel efflux pump inhibitors, as it is now essential to screen for therapeutic targets capable of restoring the effectiveness of known antimicrobials [174]. Furthermore, models were used in quantitative structure–activity relationship (QSAR) modelling of LpxC inhibitors to predict the inhibitory activity and identify the best model [175].
An interpretable machine learning algorithm, InterPred, can associate a bioactive molecule with its bioactive moiety that has a new mechanism of action and can further guide the selection of candidate molecules in drug repurposing and enable prioritisation of entities with novel mechanisms of action [176]. Moreover, the development of the novel antimicrobial halicin that has demonstrated in vitro and in vivo efficacy against the high-priority pathogen Acinetobacter baumannii was supported with machine learning models [4]. This drug was repurposed, as originally it was being developed as an anti-diabetic agent; however, an AI-supported analysis showed it has a bactericidal effect after screening via the Drug Repurposing Hub [2].
PmxPred was utilised as a predictive tool and may be useful in developing next-generation polymyxins as it accelerated identification of polymyxin analogues active against Gram-negative bacteria [177].
Machine learning models were utilised to predict peptides with biofilm inhibition activity [178]. Such models have been developed to mine peptide databases, classify peptides that may have potential antibiofilm activities, and identify the characteristics of current antibiofilm peptides. For example, the Extreme Gradient Boosting model has predicted peptides with an over 98% accuracy [179]. A machine learning multi-technique consensus workflow was developed to predict the protein targets in molecules with confirmed inhibitory activity against biofilm formation by Pseudomonas aeruginosa [180].
Machine learning techniques can also be used to predict the toxicity of antimicrobial peptides, which can present a notable challenge in their clinical implementation [181,182]. Today, there is a freely available microbial strain-specific antimicrobial peptide predictor that is based on published research and can make predictions of general antimicrobial, antibacterial, antifungal, antiviral, and haemolytic activity of peptides [183].
Machine learning techniques have been utilised to discover bacteriocins [184]. The peptides bacteriocins, produced by bacteria in their metabolic process during ribosome synthesis, are an appealing substitute for conventional antimicrobials, as they are highly specific. Researchers presented a bacteriocin prediction pipeline based on machine learning and developed an application called BPAGS so that users can test different protein sequences for bacteriocin prediction without prior programming knowledge [185]. BaPreS achieves a prediction accuracy of 95.54% for testing protein sequences. It was developed as a prediction tool to discover new highly dissimilar bacteriocins that may be further developed into highly effective antimicrobial drugs [186].
Machine learning techniques allow the in silico screening of compounds that may yield antimicrobial effects and then guide further in vivo tests [187]. Supervised learning techniques (like random forest and neural networks) were employed to identify bacterial cell wall lyases as candidates with antibacterial properties [188]. The models were trained using large datasets comprising chemical properties and biological activity data of known antibacterial agents, enhancing their ability to identify promising candidates effectively [188].
DefPred has been developed for the classification and identification of defensins, a group of antimicrobial peptides [189].
Machine learning methods have enabled fast identification of potential bacteriophages [190], and have been implemented to predict phage virion proteins with 83% accuracy, 82% sensitivity, and 89% specificity [191]. Furthermore, a machine learning tool was used for the targeted identification of phage depolymerase, which is likely to be a powerful weapon against antimicrobial-resistant bacteria [192].
Machine learning tools for the design of antimicrobial peptides are being further optimised to achieve higher precision and selectivity for resistant targets [193]. They are an essential tool for scientists to identify critical properties of structures [177]. AI accelerates the timeline of research through efficient analysis of enormous datasets and supports innovating strategies that reduce costs and accelerate the development of new drugs. The introduction of AI may necessitate only 2 years for the development of new antimicrobials. New antimicrobials can outperform existing compounds as they can evade resistance, possess anti-biofilm activities, and have new modes of action. Furthermore, AI can make estimations on further efficacy, resistance trends, and adverse reactions, thus prioritising the most promising compounds [164,194].
4. Potential Clinical Applications
Inappropriate treatment inevitably leads to unfavourable outcomes and is rooted in an inability to rapidly identify patients who are at risk of infection with the antimicrobial-resistant pathogen [1]. Machine learning models based on electronic health records have been used to predict bacterial infections and optimal antimicrobial treatment in hospitalised patients [195]. These algorithms have seen application in predicting risks of developing multi-drug resistant infection with Gram-negative bacteria to guide treatment choice and predict patients’ outcomes [196]. However, model training is crucial. There have been various attempts to implement models in clinical settings. One example was predicting perirectal colonisation with carbapenem-resistant Enterobacteriaceae (CRE) and other carbapenem-resistant organisms at hospital unit admission. However, the developed decision tree models did not achieve satisfactory results [197].
As machine learning is mathematically oriented, it does not depend on prior knowledge of resistant strains and has proven valuable in predicting antimicrobial resistance. As such, it may assist in informed drug decisions [198]. Adequately trained artificial intelligence successfully predicted the probability of antimicrobial resistance in patients, taking patients’ characteristics, admission data, historical drug treatments, and culture test results into account [199]. Another model was developed for the estimation of patients’ length of stay, mortality, and outcomes on any given day should the antimicrobial treatment be continued or stopped. In the mentioned study, results suggested that stopping antimicrobials earlier may be associated with shortening patients’ hospitalisation, indicating savings for healthcare. Such a model may be used as a proxy for treatment optimisation [200]. Algorithms were developed for the optimal dose of meropenem and polymyxin B against carbapenem-resistant Acinetobacter baumannii based on in vitro data [201].
A review from 2020 covering this topic identified 60 different machine learning clinical decision support systems developed for diagnosis or prediction of infection, prediction of treatment response or antimicrobial resistance, choice of antimicrobial or antiretroviral therapy, or early detection or stratification of sepsis. Some questions arose in the research regarding ideal machine learning systems for aiding clinical decision in reducing risk of antimicrobial resistance. The researchers concluded that none of the systems included data on local antimicrobial resistance rate. Moreover, they used somewhat fewer data than a clinician would [202]. Systems to aid diagnosis and distinguish bacterial from viral infection can greatly reduce unnecessary antimicrobials in dominantly viral infections. However, due to the imperfections of these systems, different patient variables that may be taken into consideration, different learning techniques, etc., they may currently be considered as support for clinical decision rather than an alternative. The development of such aids for clinical practice requires open access to databases for training these systems. Furthermore, great implementation in the future may be expected only should they be made financially accessible [202].
Machine learning models developed to predict antimicrobial susceptibility based on electronic health data have demonstrated they can optimise antimicrobial treatment and reduce the use of second-line antimicrobials [203]. The literature reports that machine learning-based algorithms in clinical settings can help reduce prescription of unnecessary antimicrobials by up to 40% [204]. Furthermore, machine learning methods successfully predicted whether patients were infected with extended-spectrum β-Lactamase–producing bacteria, with over 90% positive and negative predictive value [205]. Researchers have utilised machine learning-based clinical decision support systems in efforts to standardise decisions on when to switch patients from IV to oral antimicrobials [206]. As machine learning and artificial neural network models have been utilised for the detection of carbapenem-resistant Klebsiella pneumoniae, they may serve as a screening tool in clinical practice for rapid identification [207]. Moreover, approaches have been made combining biochemical markers and microbiology susceptibility tests to predict infection risk. Such analyses can easily be incorporated into everyday clinical work [208].
Different algorithms assessing antimicrobial susceptibility have been tested to improve empirical treatments in intensive care unit centres and avoid time-consuming conventional analyses [209,210]. Other than susceptibility testing, several other clinical applications of machine learning algorithms are described in the literature. Machine-learning algorithms applied directly to clinical samples may be utilised to accurately define effective antimicrobial therapy [211]. In antimicrobial stewardship, machine learning was employed for the identification of risk factors associated with the development of ventilated hospital-acquired pneumonia and mortality. Such models may enable early detection of at-risk patients and guide treatment decisions [212]. Moreover, machine learning models have been developed to help clinicians determine when an antimicrobial is unnecessary in the treatment of uncomplicated upper respiratory tract infections in the emergency department [213].
Machine learning models based on electronic health records can be used to aid clinicians in predicting antimicrobial-resistant urinary tract infections [214]. Attempts have been made to predict antimicrobial resistance based on the personal clinical history of patients [215]. Dsaas is a machine learning model developed to help clinicians identify patients at risk of contracting a multidrug-resistant urinary tract infection [216]. Furthermore, attempts have been made to identify biomarkers relevant for accurate point-of-care diagnosis of urinary tract infections, using machine learning models to examine large quantities of data [217]. Moreover, point-of-care testing of antimicrobial susceptibility of Escherichia coli in urine coupled with machine learning algorithms dramatically shortens the time needed for analysis [218]. Machine learning analysis of urinary tract and wound infections could predict treatment-induced resistance. Such algorithms can make personalised treatment recommendations [219]. Algorithms have been developed to predict antimicrobial resistance in uropathogens, lowering the probability of using ineffective antimicrobials in the emergency department by 20%. As such, they may aid in empirical treatment choices [220].
Research has demonstrated that AI models can promote the early prediction of urine tract infections and secondary bloodstream infections. In practice, such models can reduce the risk of delayed introduction of antimicrobial treatment in patients with nonspecific symptoms [221]. A supervised machine learning algorithm was adopted to diagnose bacterial infection based on routine blood parameters upon presentation to the hospital [222]. Machine learning to predict antimicrobial resistance in Escherichia coli has proven more efficient than laboratory testing. Researchers have developed a machine learning-based online tool that aids clinicians in predicting the resistance phenotype of Escherichia coli and accelerates clinical decision-making [223]. In a study by Luterbach et al., a machine-learning model was used to rank the variables predicting 30-day mortality in patients with carbapenem-resistant Klebsiella pneumoniae bloodstream infections [224]. Researchers used machine learning to determine changes in risk factors for the prognosis of patients during different periods of the bloodstream infection timeline [225]. Other models may aid in distinguishing commensal strains from Escherichia coli, causing bloodstream infections and identifying genetic factors linked to pathogenicity [226]. Furthermore, a machine learning pipeline was established for blood culture outcome prediction that, upon clinical implementation, may reduce the number of unnecessary blood culture tests [227].
Machine learning algorithms predicted intensive care unit patients likely to be colonised with resistant pathogens, with sensitivity values above 75% and specificity values ranging from 59% to 83% for different pathogens [228]. Other models were trained to predict resistance to fluoroquinolones in patients with rifampicin-resistant tuberculosis. This may guide treatment selection for these patients. Such models need excellent specificity to avoid unnecessary treatment interventions and additional resistance to alternative treatments. The final model used in this research included information on the prevalence of resistance to fluoroquinolones in the region [229].
Machine learning classifiers were utilised for rapid triage for COVID-19 [230]. A similar approach was employed to differentiate biomarker combinations related to COVID-19 pneumonia [231]. Furthermore, during the COVID-19 pandemic, researchers developed machine learning algorithms that supported the diagnosis of secondary bacterial infection in hospitalised patients. Such interventions may help halt the unnecessary expenditure of antimicrobials in COVID-19 and similar indications [232].
Analysis of hospital data via implementing machine learning techniques and developing clinical decision support tools that predict antimicrobial resistance can serve as a proxy to clinicians in choosing the appropriate antimicrobial, with the ultimate aim of reducing the unnecessary use of antimicrobials [233]. Applying machine learning algorithms to patient data can help guide targeted empirical antimicrobial prescribing [234]. An AI-based, offline smartphone application has even been developed that enables disk-diffusion antibiogram analysis [235]. Integrating artificial intelligence into clinical practice can undoubtedly shorten the time of analysis. For example, deep learning methods have been utilised for the rapid detection of carbapenemase-producing Gram-negative bacteria in only 15 min as an enhanced version of the blue carba test [236]. Although the first applications of machine learning in medicine were seen in the 1980s and 1990s, there is an evident gap between the number of machine learning applications in science and the effective implementation of these systems [237,238]. Most of the proposed solutions aim to enhance, not replace, clinicians’ work, allowing the automatic management of vast amounts of data. However, ideal implementation relies on quality datasets for training as well as training of healthcare personnel to utilise these tools [239].
Limitations of Machine Learning in Clinical Application
One of the main limitations of machine learning in healthcare and microbiological research is the dependency on high-quality, large-scale datasets. The performance of machine learning models is directly tied to the quality and quantity of the data they are trained on. Issues such as missing data, inconsistent data collection practices, and limited access to diverse datasets can significantly impact model accuracy and generalisability [60]. Many machine learning models, especially deep learning algorithms, are often seen as “black boxes” due to their complexity. This lack of transparency can hinder their acceptance and trust among healthcare practitioners, who may be reluctant to rely on predictions that cannot be easily explained or understood [238].
There is also the issue of model generalisability and the potential for overfitting. Machine learning models can perform exceptionally well on the data they were trained on but may fail to generalise to new, unseen datasets. This is particularly problematic in microbiological research, where the genetic diversity of pathogens and the emergence of new resistance mechanisms can render models quickly outdated. Overfitting to specific datasets can also mislead research directions and clinical decisions. Additionally, during training, models can also learn certain “shortcuts” that lead them to superficially correct classification or prediction [240]. Shortcut learning occurs when a model, rather than comprehending the underlying biological or clinical characteristics that dictate antimicrobial resistance, instead identifies and leverages incidental patterns or anomalies present in the training data to make predictions. These incidental patterns may not be related to the actual biological processes or mechanisms of resistance but can appear statistically significant to the algorithm during the training phase. For example, if a dataset predominantly consists of resistant strains collected from a particular geographic location or a specific laboratory, the model might erroneously associate resistance with location-specific or laboratory-specific metadata rather than the genetic or phenotypic traits that confer resistance. As a result, while the model may perform well on similar datasets, its ability to generalise and accurately predict resistance in strains from different contexts or environments is compromised. This reliance on non-generalisable, dataset-specific quirks rather than true, causative features of resistance can significantly limit the practical application and reliability of machine learning models in predicting antimicrobial resistance across diverse bacterial species and settings [241].
To mitigate shortcut learning, it is crucial to employ diverse and well-curated datasets that accurately reflect the complexity of microbial resistance mechanisms, alongside employing model training strategies that emphasise the learning of biologically relevant features. Additionally, rigorous validation on independent datasets from varied sources is necessary to ensure that the models have genuinely learned to identify the hallmarks of antimicrobial resistance and can make robust predictions across different contexts. Unfortunately, most published research lacks validation [238]. Not all research reports measure performance. Hence, legislation may come in place to set standards for machine learning systems that may be fully implemented in clinical work. As such, they may be regulated as medical devices and may have to include obligatory variables or at least several prespecified patient variables and pass rigorous testing before implementation to reduce possible errors should some clinicians blindly obey the system. Controlled randomised clinical trials evaluating patients’ outcomes are needed to confirm the validity and usefulness of machine learning systems in clinical settings. Furthermore, implementation of such systems may not result in savings for healthcare systems as other staff are needed for maintenance, and they should not be expected to replace clinicians. However, their implementation may result in a decrease in medication errors and other complications arising from suboptimal treatment choices, leading to savings in the long term [202].
5. Alternative Applications in the Medical Field
AI- or machine learning-enabled devices are seeing wide application in the medical field (Figure 2). Machine learning models can analyse data on antimicrobial resistance and antimicrobial use to identify potential hotspots of resistance and emerging resistance patterns [242]. Models were successfully utilised for mapping antimicrobial resistance threats in marine habitats with more than 75% accuracy [243]. Machine learning may establish itself in antimicrobial stewardship in terms of predicting antimicrobial resistance using genomic and antimicrobial susceptibility testing data (i.e., to identify novel resistance mechanisms and predict resistance from incomplete data) and also as a forecast tool for future changes in resistance rates based on historical prevalence data [6]. Machine learning methods can predict future trends in antimicrobial resistance and expenditure based on current data [244]. Moreover, using machine learning, researchers attempted to predict the future proportions of resistant strains using prior resistance information [245]. Causal machine learning was used to identify critical interventions for the reduction of antimicrobial resistance and outlined that quality of governance and immunisation strategies are vital [246]. In research, machine learning methods have been applied to connect dietary, physiological, and lifestyle features of healthy adults with antimicrobial resistance, suggesting that individuals that had more varied diets, richer in fibre and limited in animal protein, had lower abundances of antimicrobial resistance genes [247].
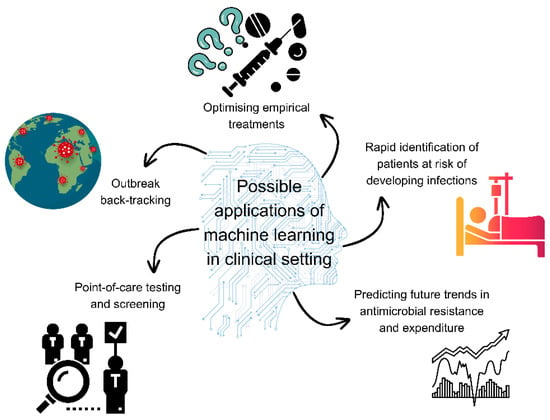
Figure 2.
Possible applications of machine learning in the clinical setting. Figure was made using Canva.
Machine learning tools can be used in epidemiology to back-track transmission of outbreaks [248,249]. This technique has been applied in identifying the geographical sources of new infections during public health outbreak investigations. A model was trained and implemented to identify and trace Salmonella enteritidis infections using whole-genome sequencing data [250]. Furthermore, implementation of machine learning to support logistic regression analysis in outbreak investigation revealed that patients receiving antimicrobials and those older than 55 years were at higher risk for colonisation with vancomycin-resistant enterococci. This was not apparent from the logistic regression [251].
Machine learning methods are well established in drug design, and they can also guide the design of pathogen-resistant coatings naturally resistant to biofilm formation [252]. Furthermore, machine learning methods have been utilised to predict the investigation of pathogen attachment to coating polymers for biomedical devices, thus guiding decisions towards the development of devices with less potential risk of complications [253].
6. Conclusions
Machine learning methods have been developed to predict antimicrobial resistance from whole-genome sequencing data, forecast medication susceptibility, recognise epidemic patterns for surveillance purposes, propose new antibacterial treatments, and accelerate scientific discovery. Unfortunately, there is an evident gap between the number of machine learning applications in science and the effective implementation of these systems. With more significant amounts of data, the performance of machine learning models improves. High-quality data is a prerequisite for accurate machine-learning based systems; hence, further development requires unrestricted access for all interested researchers.
Possible applications of artificial intelligence-based systems in healthcare are endless. In the way that digitalisation, Google, PubMed, and other search engines have revolutionised libraries and shortened the time needed for accessing and searching data, artificial intelligence can shorten the time needed for complex analyses and much more. In the way that the World Wide Web has facilitated communication and access to information, novel AI-based systems will enable people to speak the language of programming and allow everyone to program according to their specific needs. Machine learning facilitates the optimal use of data for evidence-based decision making. Algorithms can learn objectively and often outperform decisions observed in everyday practice. Machine learning should be able to significantly improve research efficiency, allowing scientists to focus on more complex scientific matters. In the not-so-distant future, training in machine learning may prove to be as essential to researchers as training in statistical analysis, although authors speculate that currently, classical statistical analysis is essential for determining causality [254]. Although the golden era for antimicrobial discovery is far behind us, using AI in the field of antimicrobial resistance may postpone the post-antimicrobial era. Undoubtedly, today is an exciting time to live in. We are looking into a future in which an AI-based system will become highly integrated in biomedical research.
Author Contributions
D.R., A.S.P., D.L., J.B. (Josipa Bukic), D.M. (Darko Modun), J.V., M.V. and M.G.: conceptualisation, original draft preparation, and review of literature; M.K., D.M. (Dinko Martinovic) and J.B. (Josko Bozic): reviewing, editing, and visualisation. All authors contributed to the final draft of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Sakagianni, A.; Koufopoulou, C.; Feretzakis, G.; Kalles, D.; Verykios, V.S.; Myrianthefs, P.; Fildisis, G. Using Machine Learning to Predict Antimicrobial Resistance—A Literature Review. Antibiotics 2023, 12, 452. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.Y.; Yu, D.; Fan, M.M.; Zhang, X.; Jin, Z.Y.; Tang, C.; Liu, X.F. Antimicrobial resistance crisis: Could artificial intelligence be the solution? Mil. Med. Res. 2024, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Goodswen, S.J.; Barratt, J.L.N.; Kennedy, P.J.; Kaufer, A.; Calarco, L.; Ellis, J.T. Machine learning and applications in microbiology. FEMS Microbiol. Rev. 2021, 45, fuab015. [Google Scholar] [CrossRef] [PubMed]
- Behling, A.H.; Wilson, B.C.; Ho, D.; Virta, M.; O’Sullivan, J.M.; Vatanen, T. Addressing antibiotic resistance: Computational answers to a biological problem? Curr. Opin. Microbiol. 2023, 74, 102305. [Google Scholar] [CrossRef]
- VanOeffelen, M.; Nguyen, M.; Aytan-Aktug, D.; Brettin, T.; Dietrich, E.M.; Kenyon, R.W.; Machi, D.; Mao, C.; Olson, R.; Pusch, G.D.; et al. A genomic data resource for predicting antimicrobial resistance from laboratory-derived antimicrobial susceptibility phenotypes. Brief. Bioinform. 2021, 22, bbab313. [Google Scholar] [CrossRef]
- Wheeler, N.E.; Price, V.; Cunningham-Oakes, E.; Tsang, K.K.; Nunn, J.G.; Midega, J.T.; Anjum, M.F.; Wade, M.J.; Feasey, N.A.; Peacock, S.J.; et al. Innovations in genomic antimicrobial resistance surveillance. Lancet Microbe 2023, 4, e1063–e1070. [Google Scholar] [CrossRef]
- Caioni, G.; Benedetti, E.; Perugini, M.; Amorena, M.; Merola, C. Personal Care Products as a Contributing Factor to Antimicrobial Resistance: Current State and Novel Approach to Investigation. Antibiotics 2023, 12, 724. [Google Scholar] [CrossRef]
- Farhat, F.; Athar, M.T.; Ahmad, S.; Madsen, D.O.; Sohail, S.S. Antimicrobial resistance and machine learning: Past, present, and future. Front. Microbiol. 2023, 14, 1179312. [Google Scholar] [CrossRef]
- Anahtar, M.N.; Yang, J.H.; Kanjilal, S. Applications of Machine Learning to the Problem of Antimicrobial Resistance: An Emerging Model for Translational Research. J. Clin. Microbiol. 2021, 59, e0126020. [Google Scholar] [CrossRef]
- Aytan-Aktug, D.; Clausen, P.; Bortolaia, V.; Aarestrup, F.M.; Lund, O. Prediction of Acquired Antimicrobial Resistance for Multiple Bacterial Species Using Neural Networks. mSystems 2020, 5, e00774-19. [Google Scholar] [CrossRef]
- Bhattacharyya, R.P.; Bandyopadhyay, N.; Ma, P.; Son, S.S.; Liu, J.; He, L.L.; Wu, L.; Khafizov, R.; Boykin, R.; Cerqueira, G.C.; et al. Simultaneous detection of genotype and phenotype enables rapid and accurate antibiotic susceptibility determination. Nat. Med. 2019, 25, 1858–1864. [Google Scholar] [CrossRef]
- Hicks, A.L.; Wheeler, N.; Sanchez-Buso, L.; Rakeman, J.L.; Harris, S.R.; Grad, Y.H. Evaluation of parameters affecting performance and reliability of machine learning-based antibiotic susceptibility testing from whole genome sequencing data. PLoS Comput. Biol. 2019, 15, e1007349. [Google Scholar] [CrossRef]
- Imchen, M.; Moopantakath, J.; Kumavath, R.; Barh, D.; Tiwari, S.; Ghosh, P.; Azevedo, V. Current Trends in Experimental and Computational Approaches to Combat Antimicrobial Resistance. Front. Genet. 2020, 11, 563975. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lin, J.; Hu, Y.; Zhou, J. PARMAP: A Pan-Genome-Based Computational Framework for Predicting Antimicrobial Resistance. Front. Microbiol. 2020, 11, 578795. [Google Scholar] [CrossRef] [PubMed]
- Majek, P.; Luftinger, L.; Beisken, S.; Rattei, T.; Materna, A. Genome-Wide Mutation Scoring for Machine-Learning-Based Antimicrobial Resistance Prediction. Int. J. Mol. Sci. 2021, 22, 13049. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.; Olson, R.; Shukla, M.; VanOeffelen, M.; Davis, J.J. Predicting antimicrobial resistance using conserved genes. PLoS Comput. Biol. 2020, 16, e1008319. [Google Scholar] [CrossRef] [PubMed]
- Van Camp, P.J.; Haslam, D.B.; Porollo, A. Prediction of Antimicrobial Resistance in Gram-Negative Bacteria from Whole-Genome Sequencing Data. Front. Microbiol. 2020, 11, 1013. [Google Scholar] [CrossRef] [PubMed]
- Khaledi, A.; Weimann, A.; Schniederjans, M.; Asgari, E.; Kuo, T.H.; Oliver, A.; Cabot, G.; Kola, A.; Gastmeier, P.; Hogardt, M.; et al. Predicting antimicrobial resistance in Pseudomonas aeruginosa with machine learning-enabled molecular diagnostics. EMBO Mol. Med. 2020, 12, e10264. [Google Scholar] [CrossRef] [PubMed]
- Boolchandani, M.; D’Souza, A.W.; Dantas, G. Sequencing-based methods and resources to study antimicrobial resistance. Nat. Rev. Genet. 2019, 20, 356–370. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.J.; Boisvert, S.; Brettin, T.; Kenyon, R.W.; Mao, C.; Olson, R.; Overbeek, R.; Santerre, J.; Shukla, M.; Wattam, A.R.; et al. Antimicrobial Resistance Prediction in PATRIC and RAST. Sci. Rep. 2016, 6, 27930. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Greenberg, D.E.; Pifer, R.; Jiang, S.; Xiao, G.; Shelburne, S.A.; Koh, A.; Xie, Y.; Zhan, X. VAMPr: VAriant Mapping and Prediction of antibiotic resistance via explainable features and machine learning. PLoS Comput. Biol. 2020, 16, e1007511. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, A.S.; Call, D.R.; Broschat, S.L. Antimicrobial Resistance Prediction for Gram-Negative Bacteria via Game Theory-Based Feature Evaluation. Sci. Rep. 2019, 9, 14487. [Google Scholar] [CrossRef] [PubMed]
- Pesesky, M.W.; Hussain, T.; Wallace, M.; Patel, S.; Andleeb, S.; Burnham, C.D.; Dantas, G. Evaluation of Machine Learning and Rules-Based Approaches for Predicting Antimicrobial Resistance Profiles in Gram-negative Bacilli from Whole Genome Sequence Data. Front. Microbiol. 2016, 7, 1887. [Google Scholar] [CrossRef] [PubMed]
- Dang, J.; Shu, J.; Wang, R.; Yu, H.; Chen, Z.; Yan, W.; Zhao, B.; Ding, L.; Wang, Y.; Hu, H.; et al. The glycopatterns of Pseudomonas aeruginosa as a potential biomarker for its carbapenem resistance. Microbiol. Spectr. 2023, 11, e0200123. [Google Scholar] [CrossRef] [PubMed]
- Stanton, R.A.; Campbell, D.; McAllister, G.A.; Breaker, E.; Adamczyk, M.; Daniels, J.B.; Lutgring, J.D.; Karlsson, M.; Schutz, K.; Jacob, J.T.; et al. Whole-Genome Sequencing Reveals Diversity of Carbapenem-Resistant Pseudomonas aeruginosa Collected through CDC’s Emerging Infections Program, United States, 2016–2018. Antimicrob. Agents Chemother. 2022, 66, e0049622. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Gao, J.; Liu, X.F.; Rao, G.; Luo, J.; Han, P.; Hu, W.; Zhang, Z.; Zhao, Q.; Han, L.; et al. Direct prediction of carbapenem resistance in Pseudomonas aeruginosa by whole genome sequencing and metagenomic sequencing. J. Clin. Microbiol. 2023, 61, e0061723. [Google Scholar] [CrossRef] [PubMed]
- Naidenov, B.; Lim, A.; Willyerd, K.; Torres, N.J.; Johnson, W.L.; Hwang, H.J.; Hoyt, P.; Gustafson, J.E.; Chen, C. Pan-Genomic and Polymorphic Driven Prediction of Antibiotic Resistance in Elizabethkingia. Front. Microbiol. 2019, 10, 1446. [Google Scholar] [CrossRef] [PubMed]
- Rishishwar, L.; Petit, R.A., 3rd; Kraft, C.S.; Jordan, I.K. Genome sequence-based discriminator for vancomycin-intermediate Staphylococcus aureus. J. Bacteriol. 2014, 196, 940–948. [Google Scholar] [CrossRef]
- He, Y.; Zhou, X.; Chen, Z.; Deng, X.; Gehring, A.; Ou, H.; Zhang, L.; Shi, X. PRAP: Pan Resistome analysis pipeline. BMC Bioinform. 2020, 21, 20. [Google Scholar] [CrossRef]
- Dillon, L.; Dimonaco, N.J.; Creevey, C.J. Accessory genes define species-specific routes to antibiotic resistance. Life Sci. Alliance 2024, 7, e202302420. [Google Scholar] [CrossRef]
- Visona, G.; Duroux, D.; Miranda, L.; Sukei, E.; Li, Y.; Borgwardt, K.; Oliver, C. Multimodal learning in clinical proteomics: Enhancing antimicrobial resistance prediction models with chemical information. Bioinformatics 2023, 39, btad717. [Google Scholar] [CrossRef] [PubMed]
- Youn, J.; Rai, N.; Tagkopoulos, I. Knowledge integration and decision support for accelerated discovery of antibiotic resistance genes. Nat. Commun. 2022, 13, 2360. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.R.; Wu, Y.W. Enhancing predictions of antimicrobial resistance of pathogens by expanding the potential resistance gene repertoire using a pan-genome-based feature selection approach. BMC Bioinform. 2022, 23, 131. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zhang, D.; Chen, F.; Rao, G.; Zhang, Y. Machine learning-based colistin resistance marker screening and phenotype prediction in Escherichia coli from whole genome sequencing data. J. Infect. 2024, 88, 191–193. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.; Zhang, X.; Maciel-Guerra, A.; Babaarslan, K.; Dong, Y.; Wang, W.; Hu, Y.; Renney, D.; Liu, L.; Li, H.; et al. Convergence of resistance and evolutionary responses in Escherichia coli and Salmonella enterica co-inhabiting chicken farms in China. Nat. Commun. 2024, 15, 206. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Yan, Y.; Links, M.G.; Li, L.; Dillon, J.R.; Horsch, M.; Kusalik, A. Antimicrobial resistance genetic factor identification from whole-genome sequence data using deep feature selection. BMC Bioinform. 2019, 20, 535. [Google Scholar] [CrossRef] [PubMed]
- Arango-Argoty, G.; Garner, E.; Pruden, A.; Heath, L.S.; Vikesland, P.; Zhang, L. DeepARG: A deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome 2018, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Satola, S.W.; Read, T.D. Genome-Based Prediction of Bacterial Antibiotic Resistance. J. Clin. Microbiol. 2019, 57, e01405-18. [Google Scholar] [CrossRef] [PubMed]
- Monk, J.M. Predicting Antimicrobial Resistance and Associated Genomic Features from Whole-Genome Sequencing. J. Clin. Microbiol. 2019, 57, e01610-18. [Google Scholar] [CrossRef]
- Sunuwar, J.; Azad, R.K. Identification of Novel Antimicrobial Resistance Genes Using Machine Learning, Homology Modeling, and Molecular Docking. Microorganisms 2022, 10, 2102. [Google Scholar] [CrossRef]
- Madrigal, P.; Singh, N.K.; Wood, J.M.; Gaudioso, E.; Hernandez-Del-Olmo, F.; Mason, C.E.; Venkateswaran, K.; Beheshti, A. Machine learning algorithm to characterize antimicrobial resistance associated with the International Space Station surface microbiome. Microbiome 2022, 10, 134. [Google Scholar] [CrossRef] [PubMed]
- Peiffer-Smadja, N.; Delliere, S.; Rodriguez, C.; Birgand, G.; Lescure, F.X.; Fourati, S.; Ruppe, E. Machine learning in the clinical microbiology laboratory: Has the time come for routine practice? Clin. Microbiol. Infect. 2020, 26, 1300–1309. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.L.; Mortimer, T.D.; Grad, Y.H. Machine learning models for Neisseria gonorrhoeae antimicrobial susceptibility tests. Ann. N. Y. Acad. Sci. 2023, 1520, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Portelli, S.; Myung, Y.; Furnham, N.; Vedithi, S.C.; Pires, D.E.V.; Ascher, D.B. Prediction of rifampicin resistance beyond the RRDR using structure-based machine learning approaches. Sci. Rep. 2020, 10, 18120. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.P.; Richmond, D.L.; Brennan-Krohn, T.; Elliott, H.L.; Kirby, J.E. Development of MAST: A Microscopy-Based Antimicrobial Susceptibility Testing Platform. SLAS Technol. 2017, 22, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Pataki, B.A.; Matamoros, S.; van der Putten, B.C.L.; Remondini, D.; Giampieri, E.; Aytan-Aktug, D.; Hendriksen, R.S.; Lund, O.; Csabai, I.; Schultsz, C. Understanding and predicting ciprofloxacin minimum inhibitory concentration in Escherichia coli with machine learning. Sci. Rep. 2020, 10, 15026. [Google Scholar] [CrossRef] [PubMed]
- ValizadehAslani, T.; Zhao, Z.; Sokhansanj, B.A.; Rosen, G.L. Amino Acid k-mer Feature Extraction for Quantitative Antimicrobial Resistance (AMR) Prediction by Machine Learning and Model Interpretation for Biological Insights. Biology 2020, 9, 365. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Chakraborty, T.; Doijad, S.; Falgenhauer, L.; Falgenhauer, J.; Goesmann, A.; Schwengers, O.; Heider, D. Multi-label classification for multi-drug resistance prediction of Escherichia coli. Comput. Struct. Biotechnol. J. 2022, 20, 1264–1270. [Google Scholar] [CrossRef] [PubMed]
- Noman, S.M.; Zeeshan, M.; Arshad, J.; Deressa Amentie, M.; Shafiq, M.; Yuan, Y.; Zeng, M.; Li, X.; Xie, Q.; Jiao, X. Machine Learning Techniques for Antimicrobial Resistance Prediction of Pseudomonas aeruginosa from Whole Genome Sequence Data. Comput. Intell. Neurosci. 2023, 2023, 5236168. [Google Scholar] [CrossRef]
- Jeon, K.; Kim, J.M.; Rho, K.; Jung, S.H.; Park, H.S.; Kim, J.S. Performance of a Machine Learning-Based Methicillin Resistance of Staphylococcus aureus Identification System Using MALDI-TOF MS and Comparison of the Accuracy according to SCCmec Types. Microorganisms 2022, 10, 1903. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, C.; Yin, Y.; Chen, F.; Chen, H.; Wang, H. A Practical Approach for Predicting Antimicrobial Phenotype Resistance in Staphylococcus aureus Through Machine Learning Analysis of Genome Data. Front. Microbiol. 2022, 13, 841289. [Google Scholar] [CrossRef] [PubMed]
- Yasir, M.; Karim, A.M.; Malik, S.K.; Bajaffer, A.A.; Azhar, E.I. Prediction of antimicrobial minimal inhibitory concentrations for Neisseria gonorrhoeae using machine learning models. Saudi J. Biol. Sci. 2022, 29, 3687–3693. [Google Scholar] [CrossRef]
- Nguyen, M.; Brettin, T.; Long, S.W.; Musser, J.M.; Olsen, R.J.; Olson, R.; Shukla, M.; Stevens, R.L.; Xia, F.; Yoo, H.; et al. Developing an in silico minimum inhibitory concentration panel test for Klebsiella pneumoniae. Sci. Rep. 2018, 8, 421. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Chakraborty, T.; Doijad, S.; Falgenhauer, L.; Falgenhauer, J.; Goesmann, A.; Hauschild, A.C.; Schwengers, O.; Heider, D. Prediction of antimicrobial resistance based on whole-genome sequencing and machine learning. Bioinformatics 2022, 38, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Her, H.L.; Wu, Y.W. A pan-genome-based machine learning approach for predicting antimicrobial resistance activities of the Escherichia coli strains. Bioinformatics 2018, 34, i89–i95. [Google Scholar] [CrossRef] [PubMed]
- Ayoola, M.B.; Das, A.R.; Krishnan, B.S.; Smith, D.R.; Nanduri, B.; Ramkumar, M. Predicting Salmonella MIC and Deciphering Genomic Determinants of Antibiotic Resistance and Susceptibility. Microorganisms 2024, 12, 134. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.R.; Su, S.F.; Wu, Y.W. Using bacterial pan-genome-based feature selection approach to improve the prediction of minimum inhibitory concentration (MIC). Front. Genet. 2023, 14, 1054032. [Google Scholar] [CrossRef]
- Gao, Y.; Li, H.; Zhao, C.; Li, S.; Yin, G.; Wang, H. Machine learning and feature extraction for rapid antimicrobial resistance prediction of Acinetobacter baumannii from whole-genome sequencing data. Front. Microbiol. 2023, 14, 1320312. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhang, B.; Zhou, M.; Campbell-Valois, F.X.; Siu, S.W.I. A deep learning method for predicting the minimum inhibitory concentration of antimicrobial peptides against Escherichia coli using Multi-Branch-CNN and Attention. mSystems 2023, 8, e0034523. [Google Scholar] [CrossRef]
- Yasir, M.; Karim, A.M.; Malik, S.K.; Bajaffer, A.A.; Azhar, E.I. Application of Decision-Tree-Based Machine Learning Algorithms for Prediction of Antimicrobial Resistance. Antibiotics 2022, 11, 1593. [Google Scholar] [CrossRef]
- Sowers, A.; Wang, G.; Xing, M.; Li, B. Advances in Antimicrobial Peptide Discovery via Machine Learning and Delivery via Nanotechnology. Microorganisms 2023, 11, 1129. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.I.; Maguire, F.; Tsang, K.K.; Gouliouris, T.; Peacock, S.J.; McAllister, T.A.; McArthur, A.G.; Beiko, R.G. Machine Learning for Antimicrobial Resistance Prediction: Current Practice, Limitations, and Clinical Perspective. Clin. Microbiol. Rev. 2022, 35, e0017921. [Google Scholar] [CrossRef] [PubMed]
- Drouin, A.; Letarte, G.; Raymond, F.; Marchand, M.; Corbeil, J.; Laviolette, F. Interpretable genotype-to-phenotype classifiers with performance guarantees. Sci. Rep. 2019, 9, 4071. [Google Scholar] [CrossRef] [PubMed]
- Luftinger, L.; Majek, P.; Beisken, S.; Rattei, T.; Posch, A.E. Learning from Limited Data: Towards Best Practice Techniques for Antimicrobial Resistance Prediction from Whole Genome Sequencing Data. Front. Cell. Infect. Microbiol. 2021, 11, 610348. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, C.; Tsai, D.H.; Wu, I.C.; Boselli, E.; Hughes, C.; Padmanabhan, D.; Hsia, Y. Machine learning applications on neonatal sepsis treatment: A scoping review. BMC Infect. Dis. 2023, 23, 441. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Chakraborty, T.; Doijad, S.; Falgenhauer, L.; Falgenhauer, J.; Goesmann, A.; Schwengers, O.; Heider, D. Deep Transfer Learning Enables Robust Prediction of Antimicrobial Resistance for Novel Antibiotics. Antibiotics 2022, 11, 1611. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, B.L.; Christensen, J.B.; Rodenko, O.; Usenov, I.; Gronnemose, R.B.; Andersen, T.E.; Lassen, M. Accurate and fast identification of minimally prepared bacteria phenotypes using Raman spectroscopy assisted by machine learning. Sci. Rep. 2022, 12, 16436. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Chen, J.; Liu, C.; Zeng, Y.; Sun, Q.; Li, J.; Shen, Z.; Chen, S.; Zhang, R. Identification of antibiotic resistance and virulence-encoding factors in Klebsiella pneumoniae by Raman spectroscopy and deep learning. Microb. Biotechnol. 2022, 15, 1270–1280. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Tang, J.W.; Lyu, J.W.; Wang, J.J.; Pan, Y.C.; Shi, X.Y.; Liu, Q.H.; Zhang, X.; Gu, B.; Wang, L. Discrimination between Carbapenem-Resistant and Carbapenem-Sensitive Klebsiella pneumoniae Strains through Computational Analysis of Surface-Enhanced Raman Spectra: A Pilot Study. Microbiol. Spectr. 2022, 10, e0240921. [Google Scholar] [CrossRef]
- Nakar, A.; Pistiki, A.; Ryabchykov, O.; Bocklitz, T.; Rosch, P.; Popp, J. Detection of multi-resistant clinical strains of E. coli with Raman spectroscopy. Anal. Bioanal. Chem. 2022, 414, 1481–1492. [Google Scholar] [CrossRef]
- Ciloglu, F.U.; Caliskan, A.; Saridag, A.M.; Kilic, I.H.; Tokmakci, M.; Kahraman, M.; Aydin, O. Drug-resistant Staphylococcus aureus bacteria detection by combining surface-enhanced Raman spectroscopy (SERS) and deep learning techniques. Sci. Rep. 2021, 11, 18444. [Google Scholar] [CrossRef] [PubMed]
- Weis, C.V.; Jutzeler, C.R.; Borgwardt, K. Machine learning for microbial identification and antimicrobial susceptibility testing on MALDI-TOF mass spectra: A systematic review. Clin. Microbiol. Infect. 2020, 26, 1310–1317. [Google Scholar] [CrossRef] [PubMed]
- Weis, C.; Horn, M.; Rieck, B.; Cuenod, A.; Egli, A.; Borgwardt, K. Topological and kernel-based microbial phenotype prediction from MALDI-TOF mass spectra. Bioinformatics 2020, 36, i30–i38. [Google Scholar] [CrossRef] [PubMed]
- Esener, N.; Maciel-Guerra, A.; Giebel, K.; Lea, D.; Green, M.J.; Bradley, A.J.; Dottorini, T. Mass spectrometry and machine learning for the accurate diagnosis of benzylpenicillin and multidrug resistance of Staphylococcus aureus in bovine mastitis. PLoS Comput. Biol. 2021, 17, e1009108. [Google Scholar] [CrossRef] [PubMed]
- Feucherolles, M.; Nennig, M.; Becker, S.L.; Martiny, D.; Losch, S.; Penny, C.; Cauchie, H.M.; Ragimbeau, C. Combination of MALDI-TOF Mass Spectrometry and Machine Learning for Rapid Antimicrobial Resistance Screening: The Case of Campylobacter spp. Front. Microbiol. 2021, 12, 804484. [Google Scholar] [CrossRef] [PubMed]
- Mather, C.A.; Werth, B.J.; Sivagnanam, S.; SenGupta, D.J.; Butler-Wu, S.M. Rapid Detection of Vancomycin-Intermediate Staphylococcus aureus by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. J. Clin. Microbiol. 2016, 54, 883–890. [Google Scholar] [CrossRef]
- Asakura, K.; Azechi, T.; Sasano, H.; Matsui, H.; Hanaki, H.; Miyazaki, M.; Takata, T.; Sekine, M.; Takaku, T.; Ochiai, T.; et al. Rapid and easy detection of low-level resistance to vancomycin in methicillin-resistant Staphylococcus aureus by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. PLoS ONE 2018, 13, e0194212. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Y.; Hsieh, T.T.; Chung, C.R.; Chang, H.C.; Horng, J.T.; Lu, J.J.; Huang, J.H. Efficiently Predicting Vancomycin Resistance of Enterococcus faecium from MALDI-TOF MS Spectra Using a Deep Learning-Based Approach. Front. Microbiol. 2022, 13, 821233. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Y.; Chung, C.R.; Chen, C.J.; Lu, K.P.; Tseng, Y.J.; Chang, T.H.; Wu, M.H.; Huang, W.T.; Lin, T.W.; Liu, T.P.; et al. Clinically Applicable System for Rapidly Predicting Enterococcus faecium Susceptibility to Vancomycin. Microbiol. Spectr. 2021, 9, e0091321. [Google Scholar] [CrossRef]
- Mishra, A.; Khan, S.; Das, A.; Das, B.C. Evolution of Diagnostic and Forensic Microbiology in the Era of Artificial Intelligence. Cureus 2023, 15, e45738. [Google Scholar] [CrossRef]
- Sun, Z.; Yang, F.; Ji, J.; Cao, W.; Liu, C.; Ding, B.; Xu, X. Dissecting the genotypic features of a fluoroquinolone-resistant Pseudomonas aeruginosa ST316 sublineage causing ear infections in Shanghai, China. Microb. Genom. 2023, 9, mgen000989. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.M.; Lin, E.Y.; Klausner, J.D.; Adamson, P.C. Machine learning to predict ceftriaxone resistance using single nucleotide polymorphisms within a global database of Neisseria gonorrhoeae genomes. Microbiol. Spectr. 2023, 11, e0170323. [Google Scholar] [CrossRef] [PubMed]
- Candela, A.; Guerrero-Lopez, A.; Mateos, M.; Gomez-Asenjo, A.; Arroyo, M.J.; Hernandez-Garcia, M.; Del Campo, R.; Cercenado, E.; Cuenod, A.; Mendez, G.; et al. Automatic Discrimination of Species within the Enterobacter cloacae Complex Using Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry and Supervised Algorithms. J. Clin. Microbiol. 2023, 61, e0104922. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.R.; Wang, H.Y.; Yao, C.H.; Wu, L.C.; Lu, J.J.; Horng, J.T.; Lee, T.Y. Data-Driven Two-Stage Framework for Identification and Characterization of Different Antibiotic-Resistant Escherichia coli Isolates Based on Mass Spectrometry Data. Microbiol. Spectr. 2023, 11, e0347922. [Google Scholar] [CrossRef] [PubMed]
- Moradigaravand, D.; Palm, M.; Farewell, A.; Mustonen, V.; Warringer, J.; Parts, L. Prediction of antibiotic resistance in Escherichia coli from large-scale pan-genome data. PLoS Comput. Biol. 2018, 14, e1006258. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez Villodres, A.; Galvez Benitez, L.; Arroyo, M.J.; Mendez, G.; Mancera, L.; Vila Dominguez, A.; Lepe Jimenez, J.A.; Smani, Y. Ultrasensitive and rapid identification of ESRI developer- and piperacillin/tazobactam-resistant Escherichia coli by the MALDIpiptaz test. Emerg. Microbes Infect. 2022, 11, 2034–2044. [Google Scholar] [CrossRef] [PubMed]
- Zagajewski, A.; Turner, P.; Feehily, C.; El Sayyed, H.; Andersson, M.; Barrett, L.; Oakley, S.; Stracy, M.; Crook, D.; Nellaker, C.; et al. Deep learning and single-cell phenotyping for rapid antimicrobial susceptibility detection in Escherichia coli. Commun. Biol. 2023, 6, 1164. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Chen, M.; Lin, Q.; Ye, Y.; Fan, H.; Wen, K.; Zeng, J.; Huang, D.; Mo, W.; Lei, Y.; et al. Identification of Methicillin-Resistant Staphylococcus aureus from Methicillin-Sensitive Staphylococcus aureus and Molecular Characterization in Quanzhou, China. Front. Cell Dev. Biol. 2021, 9, 629681. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Parel, J.M.; Miller, D. Interactions between staphylococcal enterotoxins A and D and superantigen-like proteins 1 and 5 for predicting methicillin and multidrug resistance profiles among Staphylococcus aureus ocular isolates. PLoS ONE 2021, 16, e0254519. [Google Scholar] [CrossRef]
- Ahmad, A.; Hettiarachchi, R.; Khezri, A.; Singh Ahluwalia, B.; Wadduwage, D.N.; Ahmad, R. Highly sensitive quantitative phase microscopy and deep learning aided with whole genome sequencing for rapid detection of infection and antimicrobial resistance. Front. Microbiol. 2023, 14, 1154620. [Google Scholar] [CrossRef]
- Hu, X.; Zhao, Y.; Han, P.; Liu, S.; Liu, W.; Mai, C.; Deng, Q.; Ren, J.; Luo, J.; Chen, F.; et al. Novel Clinical mNGS-Based Machine Learning Model for Rapid Antimicrobial Susceptibility Testing of Acinetobacter baumannii. J. Clin. Microbiol. 2023, 61, e0180522. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.L.; Doddi, A.; Royer, J.; Freschi, L.; Schito, M.; Ezewudo, M.; Kohane, I.S.; Beam, A.; Farhat, M. Beyond multidrug resistance: Leveraging rare variants with machine and statistical learning models in Mycobacterium tuberculosis resistance prediction. eBioMedicine 2019, 43, 356–369. [Google Scholar] [CrossRef] [PubMed]
- Aytan-Aktug, D.; Nguyen, M.; Clausen, P.; Stevens, R.L.; Aarestrup, F.M.; Lund, O.; Davis, J.J. Predicting Antimicrobial Resistance Using Partial Genome Alignments. mSystems 2021, 6, e0018521. [Google Scholar] [CrossRef]
- Libiseller-Egger, J.; Wang, L.; Deelder, W.; Campino, S.; Clark, T.G.; Phelan, J.E. TB-ML-a framework for comparing machine learning approaches to predict drug resistance of Mycobacterium tuberculosis. Bioinform. Adv. 2023, 3, vbad040. [Google Scholar] [CrossRef] [PubMed]
- Deelder, W.; Napier, G.; Campino, S.; Palla, L.; Phelan, J.; Clark, T.G. A modified decision tree approach to improve the prediction and mutation discovery for drug resistance in Mycobacterium tuberculosis. BMC Genom. 2022, 23, 46. [Google Scholar] [CrossRef]
- Jamal, S.; Khubaib, M.; Gangwar, R.; Grover, S.; Grover, A.; Hasnain, S.E. Artificial Intelligence and Machine learning based prediction of resistant and susceptible mutations in Mycobacterium tuberculosis. Sci. Rep. 2020, 10, 5487. [Google Scholar] [CrossRef]
- Kavvas, E.S.; Catoiu, E.; Mih, N.; Yurkovich, J.T.; Seif, Y.; Dillon, N.; Heckmann, D.; Anand, A.; Yang, L.; Nizet, V.; et al. Machine learning and structural analysis of Mycobacterium tuberculosis pan-genome identifies genetic signatures of antibiotic resistance. Nat. Commun. 2018, 9, 4306. [Google Scholar] [CrossRef] [PubMed]
- Munir, A.; Kumar, N.; Ramalingam, S.B.; Tamilzhalagan, S.; Shanmugam, S.K.; Palaniappan, A.N.; Nair, D.; Priyadarshini, P.; Natarajan, M.; Tripathy, S.; et al. Identification and Characterization of Genetic Determinants of Isoniazid and Rifampicin Resistance in Mycobacterium tuberculosis in Southern India. Sci. Rep. 2019, 9, 10283. [Google Scholar] [CrossRef] [PubMed]
- Deelder, W.; Christakoudi, S.; Phelan, J.; Benavente, E.D.; Campino, S.; McNerney, R.; Palla, L.; Clark, T.G. Machine Learning Predicts Accurately Mycobacterium tuberculosis Drug Resistance from Whole Genome Sequencing Data. Front. Genet. 2019, 10, 922. [Google Scholar] [CrossRef]
- Yang, Y.; Niehaus, K.E.; Walker, T.M.; Iqbal, Z.; Walker, A.S.; Wilson, D.J.; Peto, T.E.A.; Crook, D.W.; Smith, E.G.; Zhu, T.; et al. Machine learning for classifying tuberculosis drug-resistance from DNA sequencing data. Bioinformatics 2018, 34, 1666–1671. [Google Scholar] [CrossRef]
- Yang, Y.; Walker, T.M.; Walker, A.S.; Wilson, D.J.; Peto, T.E.A.; Crook, D.W.; Shamout, F.; Zhu, T.; Clifton, D.A. DeepAMR for predicting co-occurrent resistance of Mycobacterium tuberculosis. Bioinformatics 2019, 35, 3240–3249. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Walker, T.M.; Kouchaki, S.; Wang, C.; Peto, T.E.A.; Crook, D.W.; Clifton, D.A. An end-to-end heterogeneous graph attention network for Mycobacterium tuberculosis drug-resistance prediction. Brief. Bioinform. 2021, 22, bbab299. [Google Scholar] [CrossRef] [PubMed]
- Yurtseven, A.; Buyanova, S.; Agrawal, A.A.; Bochkareva, O.O.; Kalinina, O.V. Machine learning and phylogenetic analysis allow for predicting antibiotic resistance in M. tuberculosis. BMC Microbiol. 2023, 23, 404. [Google Scholar] [CrossRef] [PubMed]
- Green, A.G.; Yoon, C.H.; Chen, M.L.; Ektefaie, Y.; Fina, M.; Freschi, L.; Groschel, M.I.; Kohane, I.; Beam, A.; Farhat, M. A convolutional neural network highlights mutations relevant to antimicrobial resistance in Mycobacterium tuberculosis. Nat. Commun. 2022, 13, 3817. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jiang, Z.; Liang, P.; Liu, Z.; Cai, H.; Sun, Q. TB-DROP: Deep learning-based drug resistance prediction of Mycobacterium tuberculosis utilizing whole genome mutations. BMC Genom. 2024, 25, 167. [Google Scholar] [CrossRef] [PubMed]
- Groschel, M.I.; Owens, M.; Freschi, L.; Vargas, R., Jr.; Marin, M.G.; Phelan, J.; Iqbal, Z.; Dixit, A.; Farhat, M.R. GenTB: A user-friendly genome-based predictor for tuberculosis resistance powered by machine learning. Genome Med. 2021, 13, 138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Teng, L.; Alterovitz, G. An explainable machine learning platform for pyrazinamide resistance prediction and genetic feature identification of Mycobacterium tuberculosis. J. Am. Med. Inform. Assoc. 2021, 28, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Karmakar, M.; Rodrigues, C.H.M.; Horan, K.; Denholm, J.T.; Ascher, D.B. Structure guided prediction of Pyrazinamide resistance mutations in pncA. Sci. Rep. 2020, 10, 1875. [Google Scholar] [CrossRef]
- Kuang, X.; Wang, F.; Hernandez, K.M.; Zhang, Z.; Grossman, R.L. Accurate and rapid prediction of tuberculosis drug resistance from genome sequence data using traditional machine learning algorithms and CNN. Sci. Rep. 2022, 12, 2427. [Google Scholar] [CrossRef]
- Kavvas, E.S.; Yang, L.; Monk, J.M.; Heckmann, D.; Palsson, B.O. A biochemically-interpretable machine learning classifier for microbial GWAS. Nat. Commun. 2020, 11, 2580. [Google Scholar] [CrossRef]
- Sharma, A.; Machado, E.; Lima, K.V.B.; Suffys, P.N.; Conceicao, E.C. Tuberculosis drug resistance profiling based on machine learning: A literature review. Braz. J. Infect. Dis. 2022, 26, 102332. [Google Scholar] [CrossRef]
- Muller, S.J.; Meraba, R.L.; Dlamini, G.S.; Mapiye, D.S. First-line drug resistance profiling of Mycobacterium tuberculosis: A machine learning approach. AMIA Annu. Symp. Proc. 2021, 2021, 891–899. [Google Scholar]
- Ning, Q.; Wang, D.; Cheng, F.; Zhong, Y.; Ding, Q.; You, J. Predicting rifampicin resistance mutations in bacterial RNA polymerase subunit beta based on majority consensus. BMC Bioinform. 2021, 22, 210. [Google Scholar] [CrossRef]
- Jaillard, M.; Palmieri, M.; van Belkum, A.; Mahe, P. Interpreting k-mer-based signatures for antibiotic resistance prediction. Gigascience 2020, 9, giaa110. [Google Scholar] [CrossRef]
- Russo, T.A.; Alvarado, C.L.; Davies, C.J.; Drayer, Z.J.; Carlino-MacDonald, U.; Hutson, A.; Luo, T.L.; Martin, M.J.; Corey, B.W.; Moser, K.A.; et al. Differentiation of hypervirulent and classical Klebsiella pneumoniae with acquired drug resistance. mBio 2024, 15, e0286723. [Google Scholar] [CrossRef] [PubMed]
- Macesic, N.; Bear Don’t Walk, O.I.; Pe’er, I.; Tatonetti, N.P.; Peleg, A.Y.; Uhlemann, A.C. Predicting Phenotypic Polymyxin Resistance in Klebsiella pneumoniae through Machine Learning Analysis of Genomic Data. mSystems 2020, 5, e00656-19. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Y.; Kuo, C.H.; Chung, C.R.; Lin, W.Y.; Wang, Y.C.; Lin, T.W.; Yu, J.R.; Lu, J.J.; Wu, T.S. Rapid and Accurate Discrimination of Mycobacterium abscessus Subspecies Based on Matrix-Assisted Laser Desorption Ionization-Time of Flight Spectrum and Machine Learning Algorithms. Biomedicines 2022, 11, 45. [Google Scholar] [CrossRef] [PubMed]
- Leidner, F.; Kurt Yilmaz, N.; Schiffer, C.A. Deciphering Antifungal Drug Resistance in Pneumocystis jirovecii DHFR with Molecular Dynamics and Machine Learning. J. Chem. Inf. Model. 2021, 61, 2537–2541. [Google Scholar] [CrossRef]
- Li, D.; Wang, Y.; Hu, W.; Chen, F.; Zhao, J.; Chen, X.; Han, L. Application of Machine Learning Classifier to Candida auris Drug Resistance Analysis. Front. Cell. Infect. Microbiol. 2021, 11, 742062. [Google Scholar] [CrossRef]
- Henderson, H.I.; Napravnik, S.; Kosorok, M.R.; Gower, E.W.; Kinlaw, A.C.; Aiello, A.E.; Williams, B.; Wohl, D.A.; van Duin, D. Predicting Risk of Multidrug-Resistant Enterobacterales Infections among People with HIV. Open Forum Infect. Dis. 2022, 9, ofac487. [Google Scholar] [CrossRef]
- Blassel, L.; Zhukova, A.; Villabona-Arenas, C.J.; Atkins, K.E.; Hue, S.; Gascuel, O. Drug resistance mutations in HIV: New bioinformatics approaches and challenges. Curr. Opin. Virol. 2021, 51, 56–64. [Google Scholar] [CrossRef]
- Qiu, J.; Tian, X.; Liu, J.; Qin, Y.; Zhu, J.; Xu, D.; Qiu, T. Revealing the Mutation Patterns of Drug-Resistant Reverse Transcriptase Variants of Human Immunodeficiency Virus through Proteochemometric Modeling. Biomolecules 2021, 11, 1302. [Google Scholar] [CrossRef]
- Blassel, L.; Tostevin, A.; Villabona-Arenas, C.J.; Peeters, M.; Hue, S.; Gascuel, O. Using machine learning and big data to explore the drug resistance landscape in HIV. PLoS Comput. Biol. 2021, 17, e1008873. [Google Scholar] [CrossRef] [PubMed]
- Ota, R.; So, K.; Tsuda, M.; Higuchi, Y.; Yamashita, F. Prediction of HIV drug resistance based on the 3D protein structure: Proposal of molecular field mapping. PLoS ONE 2021, 16, e0255693. [Google Scholar] [CrossRef]
- Matthew, A.N.; Leidner, F.; Lockbaum, G.J.; Henes, M.; Zephyr, J.; Hou, S.; Rao, D.N.; Timm, J.; Rusere, L.N.; Ragland, D.A.; et al. Drug Design Strategies to Avoid Resistance in Direct-Acting Antivirals and Beyond. Chem. Rev. 2021, 121, 3238–3270. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.; Freas, C.; Weber, I.T.; Harrison, R.W. Evolution of drug resistance in HIV protease. BMC Bioinform. 2020, 21, 497. [Google Scholar] [CrossRef] [PubMed]
- Steiner, M.C.; Gibson, K.M.; Crandall, K.A. Drug Resistance Prediction Using Deep Learning Techniques on HIV-1 Sequence Data. Viruses 2020, 12, 560. [Google Scholar] [CrossRef]
- Shen, C.; Yu, X.; Harrison, R.W.; Weber, I.T. Automated prediction of HIV drug resistance from genotype data. BMC Bioinform. 2016, 17 (Suppl. S8), 278. [Google Scholar] [CrossRef]
- Ramon, E.; Belanche-Munoz, L.; Perez-Enciso, M. HIV drug resistance prediction with weighted categorical kernel functions. BMC Bioinform. 2019, 20, 410. [Google Scholar] [CrossRef]
- Weber, I.T.; Harrison, R.W. Decoding HIV resistance: From genotype to therapy. Future Med. Chem. 2017, 9, 1529–1538. [Google Scholar] [CrossRef]
- Tarasova, O.; Biziukova, N.; Filimonov, D.; Poroikov, V. A Computational Approach for the Prediction of HIV Resistance Based on Amino Acid and Nucleotide Descriptors. Molecules 2018, 23, 2751. [Google Scholar] [CrossRef] [PubMed]
- Hepler, N.L.; Scheffler, K.; Weaver, S.; Murrell, B.; Richman, D.D.; Burton, D.R.; Poignard, P.; Smith, D.M.; Kosakovsky Pond, S.L. IDEPI: Rapid prediction of HIV-1 antibody epitopes and other phenotypic features from sequence data using a flexible machine learning platform. PLoS Comput. Biol. 2014, 10, e1003842. [Google Scholar] [CrossRef] [PubMed]
- Tunc, H.; Dogan, B.; Darendeli Kiraz, B.N.; Sari, M.; Durdagi, S.; Kotil, S. Prediction of HIV-1 protease resistance using genotypic, phenotypic, and molecular information with artificial neural networks. PeerJ 2023, 11, e14987. [Google Scholar] [CrossRef] [PubMed]
- Riemenschneider, M.; Hummel, T.; Heider, D. SHIVA—A web application for drug resistance and tropism testing in HIV. BMC Bioinform. 2016, 17, 314. [Google Scholar] [CrossRef] [PubMed]
- Paremskaia, A.I.; Rudik, A.V.; Filimonov, D.A.; Lagunin, A.A.; Poroikov, V.V.; Tarasova, O.A. Web Service for HIV Drug Resistance Prediction Based on Analysis of Amino Acid Substitutions in Main Drug Targets. Viruses 2023, 15, 2245. [Google Scholar] [CrossRef] [PubMed]
- Shiaelis, N.; Tometzki, A.; Peto, L.; McMahon, A.; Hepp, C.; Bickerton, E.; Favard, C.; Muriaux, D.; Andersson, M.; Oakley, S.; et al. Virus Detection and Identification in Minutes Using Single-Particle Imaging and Deep Learning. ACS Nano 2023, 17, 697–710. [Google Scholar] [CrossRef] [PubMed]
- Borkenhagen, L.K.; Allen, M.W.; Runstadler, J.A. Influenza virus genotype to phenotype predictions through machine learning: A systematic review. Emerg. Microbes Infect. 2021, 10, 1896–1907. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, M.; Kamoun, S.; Hkimi, C.; Ghedira, K.; Bejaoui, A.; Maaroufi, A. Relationships between Virulence Genes and Antibiotic Resistance Phenotypes/Genotypes in Campylobacter spp. Isolated from Layer Hens and Eggs in the North of Tunisia: Statistical and Computational Insights. Foods 2022, 11, 3554. [Google Scholar] [CrossRef] [PubMed]
- Mwangi, J.; Kamau, P.M.; Thuku, R.C.; Lai, R. Design methods for antimicrobial peptides with improved performance. Zool. Res. 2023, 44, 1095–1114. [Google Scholar] [CrossRef]
- Le, M.T.; Trinh, D.T.; Ngo, T.D.; Tran-Nguyen, V.K.; Nguyen, D.N.; Hoang, T.; Nguyen, H.M.; Do, T.G.; Mai, T.T.; Tran, T.D.; et al. Chalcone Derivatives as Potential Inhibitors of P-Glycoprotein and NorA: An In Silico and In Vitro Study. BioMed Res. Int. 2022, 2022, 9982453. [Google Scholar] [CrossRef]
- Yan, J.; Cai, J.; Zhang, B.; Wang, Y.; Wong, D.F.; Siu, S.W.I. Recent Progress in the Discovery and Design of Antimicrobial Peptides Using Traditional Machine Learning and Deep Learning. Antibiotics 2022, 11, 1451. [Google Scholar] [CrossRef]
- Ruiz-Blanco, Y.B.; Aguero-Chapin, G.; Romero-Molina, S.; Antunes, A.; Olari, L.R.; Spellerberg, B.; Munch, J.; Sanchez-Garcia, E. ABP-Finder: A Tool to Identify Antibacterial Peptides and the Gram-Staining Type of Targeted Bacteria. Antibiotics 2022, 11, 1708. [Google Scholar] [CrossRef]
- Ramazi, S.; Mohammadi, N.; Allahverdi, A.; Khalili, E.; Abdolmaleki, P. A review on antimicrobial peptides databases and the computational tools. Database 2022, 2022, baac011. [Google Scholar] [CrossRef]
- Wang, G.; Vaisman, I.I.; van Hoek, M.L. Machine Learning Prediction of Antimicrobial Peptides. In Computational Peptide Science; Methods in Molecular Biology; Humana: New York, NY, USA, 2022; Volume 2405, pp. 1–37. [Google Scholar] [CrossRef]
- Zakaryan, H.; Chilingaryan, G.; Arabyan, E.; Serobian, A.; Wang, G. Natural antimicrobial peptides as a source of new antiviral agents. J. Gen. Virol. 2021, 102, 001661. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Xu, J.; Yin, Y.; Quan, X.; Zhang, H. Antimicrobial peptide identification using multi-scale convolutional network. BMC Bioinform. 2019, 20, 730. [Google Scholar] [CrossRef]
- Lin, W.; Xu, D. Imbalanced multi-label learning for identifying antimicrobial peptides and their functional types. Bioinformatics 2016, 32, 3745–3752. [Google Scholar] [CrossRef]
- Veltri, D.; Kamath, U.; Shehu, A. Deep learning improves antimicrobial peptide recognition. Bioinformatics 2018, 34, 2740–2747. [Google Scholar] [CrossRef] [PubMed]
- Bin Hafeez, A.; Jiang, X.; Bergen, P.J.; Zhu, Y. Antimicrobial Peptides: An Update on Classifications and Databases. Int. J. Mol. Sci. 2021, 22, 11691. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, P.; Glossop, H.; Meikle, T.G.; Aburto-Medina, A.; Conn, C.E.; Sarojini, V.; Valery, C. Molecular engineering of antimicrobial peptides: Microbial targets, peptide motifs and translation opportunities. Biophys. Rev. 2021, 13, 35–69. [Google Scholar] [CrossRef]
- Redshaw, J.; Ting, D.S.J.; Brown, A.; Hirst, J.D.; Gartner, T. Krein support vector machine classification of antimicrobial peptides. Digit. Discov. 2023, 2, 502–511. [Google Scholar] [CrossRef]
- Parvaiz, N.; Ahmad, F.; Yu, W.; MacKerell, A.D., Jr.; Azam, S.S. Discovery of beta-lactamase CMY-10 inhibitors for combination therapy against multi-drug resistant Enterobacteriaceae. PLoS ONE 2021, 16, e0244967. [Google Scholar] [CrossRef] [PubMed]
- Melo, M.C.R.; Maasch, J.; de la Fuente-Nunez, C. Accelerating antibiotic discovery through artificial intelligence. Commun. Biol. 2021, 4, 1050. [Google Scholar] [CrossRef] [PubMed]
- Bournez, C.; Riool, M.; de Boer, L.; Cordfunke, R.A.; de Best, L.; van Leeuwen, R.; Drijfhout, J.W.; Zaat, S.A.J.; van Westen, G.J.P. CalcAMP: A New Machine Learning Model for the Accurate Prediction of Antimicrobial Activity of Peptides. Antibiotics 2023, 12, 725. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lee, S.; Lee, I.; Nam, H. AMP-BERT: Prediction of antimicrobial peptide function based on a BERT model. Protein Sci. 2023, 32, e4529. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Bhadra, P.; Li, A.; Sethiya, P.; Qin, L.; Tai, H.K.; Wong, K.H.; Siu, S.W.I. Deep-AmPEP30: Improve Short Antimicrobial Peptides Prediction with Deep Learning. Mol. Ther. Nucleic Acids 2020, 20, 882–894. [Google Scholar] [CrossRef]
- Chung, C.R.; Jhong, J.H.; Wang, Z.; Chen, S.; Wan, Y.; Horng, J.T.; Lee, T.Y. Characterization and Identification of Natural Antimicrobial Peptides on Different Organisms. Int. J. Mol. Sci. 2020, 21, 986. [Google Scholar] [CrossRef] [PubMed]
- Carballo, G.M.; Vazquez, K.G.; Garcia-Gonzalez, L.A.; Rio, G.D.; Brizuela, C.A. Embedded-AMP: A Multi-Thread Computational Method for the Systematic Identification of Antimicrobial Peptides Embedded in Proteome Sequences. Antibiotics 2023, 12, 139. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Warren, R.L.; Birol, I. Models and data of AMPlify: A deep learning tool for antimicrobial peptide prediction. BMC Res. Notes 2023, 16, 11. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Sutherland, D.; Hammond, S.A.; Yang, C.; Taho, F.; Bergman, L.; Houston, S.; Warren, R.L.; Wong, T.; Hoang, L.M.N.; et al. AMPlify: Attentive deep learning model for discovery of novel antimicrobial peptides effective against WHO priority pathogens. BMC Genom. 2022, 23, 77. [Google Scholar] [CrossRef]
- Xu, J.; Li, F.; Li, C.; Guo, X.; Landersdorfer, C.; Shen, H.H.; Peleg, A.Y.; Li, J.; Imoto, S.; Yao, J.; et al. iAMPCN: A deep-learning approach for identifying antimicrobial peptides and their functional activities. Brief. Bioinform. 2023, 24, bbad240. [Google Scholar] [CrossRef]
- Cheng, X.; Qu, J.; Song, S.; Bian, Z. Neighborhood-based inference and restricted Boltzmann machine for microbe and drug associations prediction. PeerJ 2022, 10, e13848. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Ahmad Malik, A.; Anuwongcharoen, N.; Eiamphungporn, W.; Nantasenamat, C.; Piacham, T. Towards combating antibiotic resistance by exploring the quantitative structure-activity relationship of NDM-1 inhibitors. EXCLI J. 2022, 21, 1331–1351. [Google Scholar] [CrossRef] [PubMed]
- Oselusi, S.O.; Dube, P.; Odugbemi, A.I.; Akinyede, K.A.; Ilori, T.L.; Egieyeh, E.; Sibuyi, N.R.; Meyer, M.; Madiehe, A.M.; Wyckoff, G.J.; et al. The role and potential of computer-aided drug discovery strategies in the discovery of novel antimicrobials. Comput. Biol. Med. 2024, 169, 107927. [Google Scholar] [CrossRef] [PubMed]
- Naidu, A.; Nayak, S.S.; Lulu, S.S.; Sundararajan, V. Advances in computational frameworks in the fight against TB: The way forward. Front. Pharmacol. 2023, 14, 1152915. [Google Scholar] [CrossRef] [PubMed]
- Kazakova, O.; Racoviceanu, R.; Petrova, A.; Mioc, M.; Militaru, A.; Udrescu, L.; Udrescu, M.; Voicu, A.; Cummings, J.; Robertson, G.; et al. New Investigations with Lupane Type A-Ring Azepane Triterpenoids for Antimycobacterial Drug Candidate Design. Int. J. Mol. Sci. 2021, 22, 12542. [Google Scholar] [CrossRef] [PubMed]
- Charoenkwan, P.; Schaduangrat, N.; Lio, P.; Moni, M.A.; Chumnanpuen, P.; Shoombuatong, W. iAMAP-SCM: A Novel Computational Tool for Large-Scale Identification of Antimalarial Peptides Using Estimated Propensity Scores of Dipeptides. ACS Omega 2022, 7, 41082–41095. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Garlick, S.; Zloh, M. Deep Learning for Novel Antimicrobial Peptide Design. Biomolecules 2021, 11, 471. [Google Scholar] [CrossRef] [PubMed]
- Boone, K.; Wisdom, C.; Camarda, K.; Spencer, P.; Tamerler, C. Combining genetic algorithm with machine learning strategies for designing potent antimicrobial peptides. BMC Bioinform. 2021, 22, 239. [Google Scholar] [CrossRef] [PubMed]
- Bobde, S.S.; Alsaab, F.M.; Wang, G.; Van Hoek, M.L. Ab initio Designed Antimicrobial Peptides against Gram-Negative Bacteria. Front. Microbiol. 2021, 12, 715246. [Google Scholar] [CrossRef]
- Pandi, A.; Adam, D.; Zare, A.; Trinh, V.T.; Schaefer, S.L.; Burt, M.; Klabunde, B.; Bobkova, E.; Kushwaha, M.; Foroughijabbari, Y.; et al. Cell-free biosynthesis combined with deep learning accelerates de novo-development of antimicrobial peptides. Nat. Commun. 2023, 14, 7197. [Google Scholar] [CrossRef]
- Agrawal, P.; Raghava, G.P.S. Prediction of Antimicrobial Potential of a Chemically Modified Peptide from Its Tertiary Structure. Front. Microbiol. 2018, 9, 2551. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Balaya, R.D.A.; Kanekar, S.; Raju, R.; Prasad, T.S.K.; Kandasamy, R.K. Computational tools for exploring peptide-membrane interactions in gram-positive bacteria. Comput. Struct. Biotechnol. J. 2023, 21, 1995–2008. [Google Scholar] [CrossRef] [PubMed]
- Gaurav, A.; Bakht, P.; Saini, M.; Pandey, S.; Pathania, R. Role of bacterial efflux pumps in antibiotic resistance, virulence, and strategies to discover novel efflux pump inhibitors. Microbiology 2023, 169, 001333. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Chong, L.C.; Nantasenamat, C.; Anuwongcharoen, N.; Piacham, T. Machine learning approaches to study the structure-activity relationships of LpxC inhibitors. EXCLI J. 2023, 22, 975–991. [Google Scholar] [CrossRef]
- Mongia, M.; Guler, M.; Mohimani, H. An interpretable machine learning approach to identify mechanism of action of antibiotics. Sci. Rep. 2022, 12, 10342. [Google Scholar] [CrossRef]
- Wang, X.; Patil, N.; Li, F.; Wang, Z.; Zhan, H.; Schmidt, D.; Thompson, P.; Guo, Y.; Landersdorfer, C.B.; Shen, H.H.; et al. PmxPred: A data-driven approach for the identification of active polymyxin analogues against gram-negative bacteria. Comput. Biol. Med. 2024, 168, 107681. [Google Scholar] [CrossRef] [PubMed]
- Imchen, M.; Anju, V.T.; Busi, S.; Mohan, M.S.; Subhaswaraj, P.; Dyavaiah, M.; Kumavath, R. Metagenomic insights into taxonomic, functional diversity and inhibitors of microbial biofilms. Microbiol. Res. 2022, 265, 127207. [Google Scholar] [CrossRef] [PubMed]
- Bose, B.; Downey, T.; Ramasubramanian, A.K.; Anastasiu, D.C. Identification of Distinct Characteristics of Antibiofilm Peptides and Prospection of Diverse Sources for Efficacious Sequences. Front. Microbiol. 2021, 12, 783284. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, J.; Magalhaes, R.P.; de la Oliva Roque, V.M.; Simoes, M.; Pratas, D.; Sousa, S.F. TargIDe: A machine-learning workflow for target identification of molecules with antibiofilm activity against Pseudomonas aeruginosa. J. Comput. Aided Mol. Des. 2023, 37, 265–278. [Google Scholar] [CrossRef]
- Khabbaz, H.; Karimi-Jafari, M.H.; Saboury, A.A.; BabaAli, B. Prediction of antimicrobial peptides toxicity based on their physico-chemical properties using machine learning techniques. BMC Bioinform. 2021, 22, 549. [Google Scholar] [CrossRef]
- Timmons, P.B.; Hewage, C.M. HAPPENN is a novel tool for hemolytic activity prediction for therapeutic peptides which employs neural networks. Sci. Rep. 2020, 10, 10869. [Google Scholar] [CrossRef] [PubMed]
- Vishnepolsky, B.; Grigolava, M.; Managadze, G.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M.; Pirtskhalava, M. Comparative analysis of machine learning algorithms on the microbial strain-specific AMP prediction. Brief. Bioinform. 2022, 23, bbac233. [Google Scholar] [CrossRef] [PubMed]
- Yount, N.Y.; Weaver, D.C.; de Anda, J.; Lee, E.Y.; Lee, M.W.; Wong, G.C.L.; Yeaman, M.R. Discovery of Novel Type II Bacteriocins Using a New High-Dimensional Bioinformatic Algorithm. Front. Immunol. 2020, 11, 1873. [Google Scholar] [CrossRef] [PubMed]
- Akhter, S.; Miller, J.H. BPAGS: A web application for bacteriocin prediction via feature evaluation using alternating decision tree, genetic algorithm, and linear support vector classifier. Front. Bioinform. 2023, 3, 1284705. [Google Scholar] [CrossRef] [PubMed]
- Akhter, S.; Miller, J.H. BaPreS: A software tool for predicting bacteriocins using an optimal set of features. BMC Bioinform. 2023, 24, 313. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Gonzalez, A.; Zanin, M.; Menasalvas-Ruiz, E. Public Health and Epidemiology Informatics: Can Artificial Intelligence Help Future Global Challenges? An Overview of Antimicrobial Resistance and Impact of Climate Change in Disease Epidemiology. Yearb. Med. Inform. 2019, 28, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.X.; Tang, H.; Li, W.C.; Wu, H.; Chen, W.; Ding, H.; Lin, H. Identification of Bacterial Cell Wall Lyases via Pseudo Amino Acid Composition. BioMed Res. Int. 2016, 2016, 1654623. [Google Scholar] [CrossRef] [PubMed]
- Kaur, D.; Patiyal, S.; Arora, C.; Singh, R.; Lodhi, G.; Raghava, G.P.S. In-Silico Tool for Predicting, Scanning, and Designing Defensins. Front. Immunol. 2021, 12, 780610. [Google Scholar] [CrossRef] [PubMed]
- Thung, T.Y.; White, M.E.; Dai, W.; Wilksch, J.J.; Bamert, R.S.; Rocker, A.; Stubenrauch, C.J.; Williams, D.; Huang, C.; Schittelhelm, R.; et al. Component Parts of Bacteriophage Virions Accurately Defined by a Machine-Learning Approach Built on Evolutionary Features. mSystems 2021, 6, e0024221. [Google Scholar] [CrossRef]
- Barman, R.K.; Chakrabarti, A.K.; Dutta, S. Prediction of Phage Virion Proteins Using Machine Learning Methods. Molecules 2023, 28, 2238. [Google Scholar] [CrossRef]
- Magill, D.J.; Skvortsov, T.A. DePolymerase Predictor (DePP): A machine learning tool for the targeted identification of phage depolymerases. BMC Bioinform. 2023, 24, 208. [Google Scholar] [CrossRef] [PubMed]
- Van Oort, C.M.; Ferrell, J.B.; Remington, J.M.; Wshah, S.; Li, J. AMPGAN v2: Machine Learning-Guided Design of Antimicrobial Peptides. J. Chem. Inf. Model. 2021, 61, 2198–2207. [Google Scholar] [CrossRef] [PubMed]
- Akinsulie, O.C.; Idris, I.; Aliyu, V.A.; Shahzad, S.; Banwo, O.G.; Ogunleye, S.C.; Olorunshola, M.; Okedoyin, D.O.; Ugwu, C.; Oladapo, I.P.; et al. The potential application of artificial intelligence in veterinary clinical practice and biomedical research. Front. Vet. Sci. 2024, 11, 1347550. [Google Scholar] [CrossRef] [PubMed]
- Eickelberg, G.; Sanchez-Pinto, L.N.; Luo, Y. Predictive modeling of bacterial infections and antibiotic therapy needs in critically ill adults. J. Biomed. Inform. 2020, 109, 103540. [Google Scholar] [CrossRef] [PubMed]
- Giacobbe, D.R.; Mora, S.; Giacomini, M.; Bassetti, M. Machine Learning and Multidrug-Resistant Gram-Negative Bacteria: An Interesting Combination for Current and Future Research. Antibiotics 2020, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Goodman, K.E.; Simner, P.J.; Klein, E.Y.; Kazmi, A.Q.; Gadala, A.; Toerper, M.F.; Levin, S.; Tamma, P.D.; Rock, C.; Cosgrove, S.E.; et al. Predicting probability of perirectal colonization with carbapenem-resistant Enterobacteriaceae (CRE) and other carbapenem-resistant organisms (CROs) at hospital unit admission. Infect. Control Hosp. Epidemiol. 2019, 40, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jia, C.; Li, H.; Yin, R.; Chen, J.; Li, Y.; Yue, M. Paving the way for precise diagnostics of antimicrobial resistant bacteria. Front. Mol. Biosci. 2022, 9, 976705. [Google Scholar] [CrossRef] [PubMed]
- Cavallaro, M.; Moran, E.; Collyer, B.; McCarthy, N.D.; Green, C.; Keeling, M.J. Informing antimicrobial stewardship with explainable AI. PLoS Digit. Health 2023, 2, e0000162. [Google Scholar] [CrossRef]
- Bolton, W.J.; Rawson, T.M.; Hernandez, B.; Wilson, R.; Antcliffe, D.; Georgiou, P.; Holmes, A.H. Machine learning and synthetic outcome estimation for individualised antimicrobial cessation. Front. Digit. Health 2022, 4, 997219. [Google Scholar] [CrossRef]
- Smith, N.M.; Lenhard, J.R.; Boissonneault, K.R.; Landersdorfer, C.B.; Bulitta, J.B.; Holden, P.N.; Forrest, A.; Nation, R.L.; Li, J.; Tsuji, B.T. Using machine learning to optimize antibiotic combinations: Dosing strategies for meropenem and polymyxin B against carbapenem-resistant Acinetobacter baumannii. Clin. Microbiol. Infect. 2020, 26, 1207–1213. [Google Scholar] [CrossRef]
- Peiffer-Smadja, N.; Rawson, T.M.; Ahmad, R.; Buchard, A.; Georgiou, P.; Lescure, F.X.; Birgand, G.; Holmes, A.H. Machine learning for clinical decision support in infectious diseases: A narrative review of current applications. Clin. Microbiol. Infect. 2020, 26, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Kanjilal, S.; Oberst, M.; Boominathan, S.; Zhou, H.; Hooper, D.C.; Sontag, D. A decision algorithm to promote outpatient antimicrobial stewardship for uncomplicated urinary tract infection. Sci. Transl. Med. 2020, 12, eaay5067. [Google Scholar] [CrossRef] [PubMed]
- Moran, E.; Robinson, E.; Green, C.; Keeling, M.; Collyer, B. Towards personalized guidelines: Using machine-learning algorithms to guide antimicrobial selection. J. Antimicrob. Chemother. 2020, 75, 2677–2680. [Google Scholar] [CrossRef] [PubMed]
- Goodman, K.E.; Lessler, J.; Cosgrove, S.E.; Harris, A.D.; Lautenbach, E.; Han, J.H.; Milstone, A.M.; Massey, C.J.; Tamma, P.D. A Clinical Decision Tree to Predict Whether a Bacteremic Patient Is Infected with an Extended-Spectrum beta-Lactamase-Producing Organism. Clin. Infect. Dis. 2016, 63, 896–903. [Google Scholar] [CrossRef] [PubMed]
- Bolton, W.J.; Wilson, R.; Gilchrist, M.; Georgiou, P.; Holmes, A.; Rawson, T.M. Personalising intravenous to oral antibiotic switch decision making through fair interpretable machine learning. Nat. Commun. 2024, 15, 506. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.M.; Tsao, M.F.; Chang, C.Y.; Lin, K.T.; Keller, J.J.; Lin, H.C. Rapid identification of carbapenem-resistant Klebsiella pneumoniae based on matrix-assisted laser desorption ionization time-of-flight mass spectrometry and an artificial neural network model. J. Biomed. Sci. 2023, 30, 25. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, B.; Herrero, P.; Rawson, T.M.; Moore, L.S.P.; Evans, B.; Toumazou, C.; Holmes, A.H.; Georgiou, P. Supervised learning for infection risk inference using pathology data. BMC Med. Inform. Decis. Mak. 2017, 17, 168. [Google Scholar] [CrossRef] [PubMed]
- Feretzakis, G.; Loupelis, E.; Sakagianni, A.; Kalles, D.; Martsoukou, M.; Lada, M.; Skarmoutsou, N.; Christopoulos, C.; Valakis, K.; Velentza, A.; et al. Using Machine Learning Techniques to Aid Empirical Antibiotic Therapy Decisions in the Intensive Care Unit of a General Hospital in Greece. Antibiotics 2020, 9, 50. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Aguero, S.; Mora-Jimenez, I.; Lerida-Garcia, J.; Alvarez-Rodriguez, J.; Soguero-Ruiz, C. Machine Learning Techniques to Identify Antimicrobial Resistance in the Intensive Care Unit. Entropy 2019, 21, 603. [Google Scholar] [CrossRef]
- Bauer, M.J.; Peri, A.M.; Luftinger, L.; Beisken, S.; Bergh, H.; Forde, B.M.; Buckley, C.; Cuddihy, T.; Tan, P.; Paterson, D.L.; et al. Optimized Method for Bacterial Nucleic Acid Extraction from Positive Blood Culture Broth for Whole-Genome Sequencing, Resistance Phenotype Prediction, and Downstream Molecular Applications. J. Clin. Microbiol. 2022, 60, e0101222. [Google Scholar] [CrossRef]
- Sophonsri, A.; Lou, M.; Ny, P.; Minejima, E.; Nieberg, P.; Wong-Beringer, A. Machine learning to identify risk factors associated with the development of ventilated hospital-acquired pneumonia and mortality: Implications for antibiotic therapy selection. Front. Med. 2023, 10, 1268488. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.G.; Aung, A.H.; Lian, W.; Lye, D.C.; Ooi, C.K.; Chow, A. Risk prediction models to guide antibiotic prescribing: A study on adult patients with uncomplicated upper respiratory tract infections in an emergency department. Antimicrob. Resist. Infect. Control 2020, 9, 171. [Google Scholar] [CrossRef] [PubMed]
- Rich, S.N.; Jun, I.; Bian, J.; Boucher, C.; Cherabuddi, K.; Morris, J.G., Jr.; Prosperi, M. Development of a Prediction Model for Antibiotic-Resistant Urinary Tract Infections Using Integrated Electronic Health Records from Multiple Clinics in North-Central Florida. Infect. Dis. Ther. 2022, 11, 1869–1882. [Google Scholar] [CrossRef] [PubMed]
- Yelin, I.; Snitser, O.; Novich, G.; Katz, R.; Tal, O.; Parizade, M.; Chodick, G.; Koren, G.; Shalev, V.; Kishony, R. Personal clinical history predicts antibiotic resistance of urinary tract infections. Nat. Med. 2019, 25, 1143–1152. [Google Scholar] [CrossRef] [PubMed]
- Mancini, A.; Vito, L.; Marcelli, E.; Piangerelli, M.; De Leone, R.; Pucciarelli, S.; Merelli, E. Machine learning models predicting multidrug resistant urinary tract infections using “DsaaS”. BMC Bioinform. 2020, 21, 347. [Google Scholar] [CrossRef] [PubMed]
- Gadalla, A.A.H.; Friberg, I.M.; Kift-Morgan, A.; Zhang, J.; Eberl, M.; Topley, N.; Weeks, I.; Cuff, S.; Wootton, M.; Gal, M.; et al. Identification of clinical and urine biomarkers for uncomplicated urinary tract infection using machine learning algorithms. Sci. Rep. 2019, 9, 19694. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Borkum, T.; Shprits, S.; Boen, J.; Arshavsky-Graham, S.; Rofman, B.; Strauss, M.; Colodner, R.; Sulam, J.; Halachmi, S.; et al. Accurate Prediction of Antimicrobial Susceptibility for Point-of-Care Testing of Urine in Less than 90 Minutes via iPRISM Cassettes. Adv. Sci. 2023, 10, e2303285. [Google Scholar] [CrossRef] [PubMed]
- Stracy, M.; Snitser, O.; Yelin, I.; Amer, Y.; Parizade, M.; Katz, R.; Rimler, G.; Wolf, T.; Herzel, E.; Koren, G.; et al. Minimizing treatment-induced emergence of antibiotic resistance in bacterial infections. Science 2022, 375, 889–894. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Seo, Y.; Kim, J.H.; Han, S.B.; Im, J.H.; Jung, C.Y.; Durey, A. Machine learning model for predicting ciprofloxacin resistance and presence of ESBL in patients with UTI in the ED. Sci. Rep. 2023, 13, 3282. [Google Scholar] [CrossRef]
- Choi, M.H.; Kim, D.; Park, Y.; Jeong, S.H. Development and validation of artificial intelligence models to predict urinary tract infections and secondary bloodstream infections in adult patients. J. Infect. Public Health 2024, 17, 10–17. [Google Scholar] [CrossRef]
- Rawson, T.M.; Hernandez, B.; Moore, L.S.P.; Blandy, O.; Herrero, P.; Gilchrist, M.; Gordon, A.; Toumazou, C.; Sriskandan, S.; Georgiou, P.; et al. Supervised machine learning for the prediction of infection on admission to hospital: A prospective observational cohort study. J. Antimicrob. Chemother. 2019, 74, 1108–1115. [Google Scholar] [CrossRef]
- Jin, C.; Jia, C.; Hu, W.; Xu, H.; Shen, Y.; Yue, M. Predicting antimicrobial resistance in E. coli with discriminative position fused deep learning classifier. Comput. Struct. Biotechnol. J. 2024, 23, 559–565. [Google Scholar] [CrossRef]
- Luterbach, C.L.; Qiu, H.; Hanafin, P.O.; Sharma, R.; Piscitelli, J.; Lin, F.C.; Ilomaki, J.; Cober, E.; Salata, R.A.; Kalayjian, R.C.; et al. A Systems-Based Analysis of Mono- and Combination Therapy for Carbapenem-Resistant Klebsiella pneumoniae Bloodstream Infections. Antimicrob. Agents Chemother. 2022, 66, e0059122. [Google Scholar] [CrossRef]
- Choi, M.H.; Kim, D.; Kim, J.; Song, Y.G.; Jeong, S.H. Shift in risk factors for mortality by period of the bloodstream infection timeline. J. Microbiol. Immunol. Infect. 2024, 57, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Burgaya, J.; Marin, J.; Royer, G.; Condamine, B.; Gachet, B.; Clermont, O.; Jaureguy, F.; Burdet, C.; Lefort, A.; de Lastours, V.; et al. The bacterial genetic determinants of Escherichia coli capacity to cause bloodstream infections in humans. PLoS Genet. 2023, 19, e1010842. [Google Scholar] [CrossRef]
- McFadden, B.R.; Inglis, T.J.J.; Reynolds, M. Machine learning pipeline for blood culture outcome prediction using Sysmex XN-2000 blood sample results in Western Australia. BMC Infect. Dis. 2023, 23, 552. [Google Scholar] [CrossRef] [PubMed]
- Caglayan, C.; Barnes, S.L.; Pineles, L.L.; Harris, A.D.; Klein, E.Y. A Data-Driven Framework for Identifying Intensive Care Unit Admissions Colonized with Multidrug-Resistant Organisms. Front. Public Health 2022, 10, 853757. [Google Scholar] [CrossRef] [PubMed]
- You, S.; Chitwood, M.H.; Gunasekera, K.S.; Crudu, V.; Codreanu, A.; Ciobanu, N.; Furin, J.; Cohen, T.; Warren, J.L.; Yaesoubi, R. Predicting resistance to fluoroquinolones among patients with rifampicin-resistant tuberculosis using machine learning methods. PLoS Digit. Health 2022, 1, e0000059. [Google Scholar] [CrossRef]
- Soltan, A.A.S.; Kouchaki, S.; Zhu, T.; Kiyasseh, D.; Taylor, T.; Hussain, Z.B.; Peto, T.; Brent, A.J.; Eyre, D.W.; Clifton, D.A. Rapid triage for COVID-19 using routine clinical data for patients attending hospital: Development and prospective validation of an artificial intelligence screening test. Lancet Digit. Health 2021, 3, e78–e87. [Google Scholar] [CrossRef]
- Wilson, L.; Chang, J.W.; Meier, S.; Ganief, T.; Ganief, N.; Oelofse, S.; Baillie, V.; Nunes, M.C.; Madhi, S.A.; Blackburn, J.; et al. Proteomic Profiling of Urine from Hospitalized Patients with Severe Pneumonia due to SARS-CoV-2 vs Other Causes: A Preliminary Report. Open Forum Infect. Dis. 2023, 10, ofad451. [Google Scholar] [CrossRef]
- Rawson, T.M.; Hernandez, B.; Wilson, R.C.; Ming, D.; Herrero, P.; Ranganathan, N.; Skolimowska, K.; Gilchrist, M.; Satta, G.; Georgiou, P.; et al. Supervised machine learning to support the diagnosis of bacterial infection in the context of COVID-19. JAC Antimicrob. Resist. 2021, 3, dlab002. [Google Scholar] [CrossRef] [PubMed]
- Feretzakis, G.; Sakagianni, A.; Loupelis, E.; Kalles, D.; Skarmoutsou, N.; Martsoukou, M.; Christopoulos, C.; Lada, M.; Petropoulou, S.; Velentza, A.; et al. Machine Learning for Antibiotic Resistance Prediction: A Prototype Using Off-the-Shelf Techniques and Entry-Level Data to Guide Empiric Antimicrobial Therapy. Healthc. Inform. Res. 2021, 27, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Oonsivilai, M.; Mo, Y.; Luangasanatip, N.; Lubell, Y.; Miliya, T.; Tan, P.; Loeuk, L.; Turner, P.; Cooper, B.S. Using machine learning to guide targeted and locally-tailored empiric antibiotic prescribing in a children’s hospital in Cambodia. Wellcome Open Res. 2018, 3, 131. [Google Scholar] [CrossRef] [PubMed]
- Pascucci, M.; Royer, G.; Adamek, J.; Asmar, M.A.; Aristizabal, D.; Blanche, L.; Bezzarga, A.; Boniface-Chang, G.; Brunner, A.; Curel, C.; et al. AI-based mobile application to fight antibiotic resistance. Nat. Commun. 2021, 12, 1173. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Han, L.; Cai, H.X.; Cui, Z.H.; Yang, R.S.; Zhang, R.M.; Bai, S.C.; Liu, X.W.; Wei, R.; Chen, L.; et al. AI-Blue-Carba: A Rapid and Improved Carbapenemase Producer Detection Assay Using Blue-Carba with Deep Learning. Front. Microbiol. 2020, 11, 585417. [Google Scholar] [CrossRef] [PubMed]
- McFadden, B.R.; Reynolds, M.; Inglis, T.J.J. Developing machine learning systems worthy of trust for infection science: A requirement for future implementation into clinical practice. Front. Digit. Health 2023, 5, 1260602. [Google Scholar] [CrossRef] [PubMed]
- Theodosiou, A.A.; Read, R.C. Artificial intelligence, machine learning and deep learning: Potential resources for the infection clinician. J. Infect. 2023, 87, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, U.; Pappalardo, M.; Chine, V.; Gismondi, P.; Neglia, C.; Argentiero, A.; Calderaro, A.; Prati, A.; Esposito, S. Role of Artificial Intelligence in Fighting Antimicrobial Resistance in Pediatrics. Antibiotics 2020, 9, 767. [Google Scholar] [CrossRef] [PubMed]
- Geirhos, R.; Jacobsen, J.H.; Michaelis, C.; Zemel, R.; Brendel, W.; Bethge, M.; Wichmann, F.A. Shortcut learning in deep neural networks. Nat. Mach. Intell. 2020, 2, 665–673. [Google Scholar] [CrossRef]
- Coxe, T.; Azad, R.K. Silicon versus Superbug: Assessing Machine Learning’s Role in the Fight against Antimicrobial Resistance. Antibiotics 2023, 12, 1604. [Google Scholar] [CrossRef]
- Ali, T.; Ahmed, S.; Aslam, M. Artificial Intelligence for Antimicrobial Resistance Prediction: Challenges and Opportunities towards Practical Implementation. Antibiotics 2023, 12, 523. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Q.; Wang, T.; Xu, N.; Lu, T.; Hong, W.; Penuelas, J.; Gillings, M.; Wang, M.; Gao, W.; et al. Assessment of global health risk of antibiotic resistance genes. Nat. Commun. 2022, 13, 1553. [Google Scholar] [CrossRef] [PubMed]
- Cravo Oliveira Hashiguchi, T.; Ait Ouakrim, D.; Padget, M.; Cassini, A.; Cecchini, M. Resistance proportions for eight priority antibiotic-bacterium combinations in OECD, EU/EEA and G20 countries 2000 to 2030: A modelling study. Eurosurveillance 2019, 24, 1800445. [Google Scholar] [CrossRef]
- Kim, J.; Rupasinghe, R.; Halev, A.; Huang, C.; Rezaei, S.; Clavijo, M.J.; Robbins, R.C.; Martinez-Lopez, B.; Liu, X. Predicting antimicrobial resistance of bacterial pathogens using time series analysis. Front. Microbiol. 2023, 14, 1160224. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, R.; Rakholia, V.; Agrawal, S.; Dhingra, L.S.; Nagori, A.; Kaur, H.; Sethi, T. Estimating the impact of health systems factors on antimicrobial resistance in priority pathogens. J. Glob. Antimicrob. Resist. 2022, 30, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Oliver, A.; Xue, Z.; Villanueva, Y.T.; Durbin-Johnson, B.; Alkan, Z.; Taft, D.H.; Liu, J.; Korf, I.; Laugero, K.D.; Stephensen, C.B.; et al. Association of Diet and Antimicrobial Resistance in Healthy U.S. Adults. mBio 2022, 13, e0010122. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Stockdale, J.E.; Naidu, V.; Hatherell, H.; Stimson, J.; Stagg, H.R.; Abubakar, I.; Colijn, C. Transmission analysis of a large tuberculosis outbreak in London: A mathematical modelling study using genomic data. Microb. Genom. 2020, 6, mgen000450. [Google Scholar] [CrossRef]
- Vilne, B.; Meistere, I.; Grantina-Ievina, L.; Kibilds, J. Machine Learning Approaches for Epidemiological Investigations of Food-Borne Disease Outbreaks. Front. Microbiol. 2019, 10, 1722. [Google Scholar] [CrossRef] [PubMed]
- Bayliss, S.C.; Locke, R.K.; Jenkins, C.; Chattaway, M.A.; Dallman, T.J.; Cowley, L.A. Rapid geographical source attribution of Salmonella enterica serovar Enteritidis genomes using hierarchical machine learning. eLife 2023, 12, e84167. [Google Scholar] [CrossRef]
- Atkinson, A.; Ellenberger, B.; Piezzi, V.; Kaspar, T.; Salazar-Vizcaya, L.; Endrich, O.; Leichtle, A.B.; Marschall, J. Extending outbreak investigation with machine learning and graph theory: Benefits of new tools with application to a nosocomial outbreak of a multidrug-resistant organism. Infect. Control Hosp. Epidemiol. 2023, 44, 246–252. [Google Scholar] [CrossRef]
- Contreas, L.; Hook, A.L.; Winkler, D.A.; Figueredo, G.; Williams, P.; Laughton, C.A.; Alexander, M.R.; Williams, P.M. Linear Binary Classifier to Predict Bacterial Biofilm Formation on Polyacrylates. ACS Appl. Mater. Interfaces 2023, 15, 14155–14163. [Google Scholar] [CrossRef] [PubMed]
- Mikulskis, P.; Hook, A.; Dundas, A.A.; Irvine, D.; Sanni, O.; Anderson, D.; Langer, R.; Alexander, M.R.; Williams, P.; Winkler, D.A. Prediction of Broad-Spectrum Pathogen Attachment to Coating Materials for Biomedical Devices. ACS Appl. Mater. Interfaces 2018, 10, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Al-Hindawi, A.; Abdulaal, A.; Rawson, T.M.; Alqahtani, S.A.; Mughal, N.; Moore, L.S.P. COVID-19 Prognostic Models: A Pro-con Debate for Machine Learning vs. Traditional Statistics. Front. Digit. Health 2021, 3, 637944. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).