Freshwater and Marine Environments in California Are a Reservoir of Carbapenem-Resistant Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Water Samples and Isolation of Carbapenem-Resistant Bacteria (CRB)
2.2. Selection, Identification, and Characterization of CRB Isolates
2.3. Identification of Carbapenemase-Producing CRB Isolates
2.4. Identification of Carbapenemase Genes
3. Results and Discussion
3.1. Distribution, Abundance, and Isolation of Carbapenem-Resistant Bacteria (CRB) in Diverse Aquatic Environments
3.2. Characterization of the Antibiotic Susceptibility Profile of Carbapenem-Resistant Bacteria (CRB) from Diverse Aquatic Environments
3.3. Detection of Carbapenemase Production and Carbapenemase Genes in CRB Isolates
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Papp-Wallace, K.M.; Endimiani, A.; Taracila, M.A.; Bonomo, R.A. Carbapenems: Past, present, and future. Antimicrob. Agents Chemother. 2011, 55, 4943–4960. [Google Scholar] [CrossRef] [PubMed]
- Bradley, J.S.; Garau, J.; Lode, H.; Rolston, K.V.; Wilson, S.E.; Quinn, J.P. Carbapenems in clinical practice: A guide to their use in serious infection. Int. J. Antimicrob. Agents 1999, 11, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Vardakas, K.Z.; Tansarli, G.S.; Rafailidis, P.I.; Falagas, M.E. Carbapenems versus alternative antibiotics for the treatment of bacteraemia due to Enterobacteriaceae producing extended-spectrum β-lactamases: A systematic review and meta-analysis. J. Antimicrob. Chemother. 2012, 67, 2793–2803. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.C.M.; Jones, M.; Snelling, T.L.; Duguid, R.; Moore, N.; Dickson, B.; Wu, Y.; Saunders, J.; Wijeratne, P.; Douangnouvong, A.; et al. Coverage gaps in empiric antibiotic regimens used to treat serious bacterial infections in neonates and children in Southeast Asia and the Pacific. Lancet Reg. Health Southeast Asia 2024, 22, 100291. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.I.; Kaye, K.M. Beta-lactam antibiotics: Newer formulations and newer agents. Infect. Dis. Clin. N. Am. 2004, 18, 603–619. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Dortet, L.; Poirel, L. Carbapenem resistance in Enterobacteriaceae: Here is the storm! Trends Mol. Med. 2012, 18, 263–272. [Google Scholar] [CrossRef]
- Jean, S.S.; Harnod, D.; Hsueh, P.R. Global Threat of Carbapenem-Resistant Gram-Negative Bacteria. Front. Cell Infect. Microbiol. 2022, 12, 823684. [Google Scholar] [CrossRef]
- Martirosov, D.M.; Lodise, T.P. Emerging trends in epidemiology and management of infections caused by carbapenem-resistant Enterobacteriaceae. Diagn. Microbiol. Infect. Dis. 2016, 85, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Livorsi, D.J.; Chorazy, M.L.; Schweizer, M.L.; Balkenende, E.C.; Blevins, A.E.; Nair, R.; Samore, M.H.; Nelson, R.E.; Khader, K.; Perencevich, E.N. A systematic review of the epidemiology of carbapenem-resistant Enterobacteriaceae in the United States. Antimicrob. Resist. Infect. Control 2018, 7, 55. [Google Scholar] [CrossRef]
- Queenan, A.M.; Bush, K. Carbapenemases: The versatile beta-lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef]
- Fisher, J.F.; Meroueh, S.O.; Mobashery, S. Bacterial resistance to beta-lactam antibiotics: Compelling opportunism, compelling opportunity. Chem. Rev. 2005, 105, 395–424. [Google Scholar] [CrossRef] [PubMed]
- Walther-Rasmussen, J.; Hoiby, N. Class A carbapenemases. J. Antimicrob. Chemother. 2007, 60, 470–482. [Google Scholar] [CrossRef]
- Prabaker, K.; Weinstein, R.A. Trends in antimicrobial resistance in intensive care units in the United States. Curr. Opin. Crit. Care 2011, 17, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Rhomberg, P.R.; Jones, R.N. Summary trends for the Meropenem Yearly Susceptibility Test Information Collection Program: A 10-year experience in the United States (1999–2008). Diagn. Microbiol. Infect. Dis. 2009, 65, 414–426. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Limbago, B.M.; Patel, J.B.; Kallen, A.J. Carbapenem-resistant Enterobacteriaceae: Epidemiology and prevention. Clin. Infect. Dis. 2011, 53, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Guh, A.Y.; Bulens, S.N.; Mu, Y.; Jacob, J.T.; Reno, J.; Scott, J.; Wilson, L.E.; Vaeth, E.; Lynfield, R.; Shaw, K.M.; et al. Epidemiology of Carbapenem-Resistant Enterobacteriaceae in 7 US Communities, 2012–2013. JAMA 2015, 314, 1479–1487. [Google Scholar] [CrossRef] [PubMed]
- van Duijn, P.J.; Dautzenberg, M.J.; Oostdijk, E.A. Recent trends in antibiotic resistance in European ICUs. Curr. Opin. Crit. Care 2011, 17, 658–665. [Google Scholar] [CrossRef]
- Rodríguez-Martínez, J.M.; Poirel, L.; Nordmann, P. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2009, 53, 4783–4788. [Google Scholar] [CrossRef]
- Correa, A.; Montealegre, M.C.; Mojica, M.F.; Maya, J.J.; Rojas, L.J.; De La Cadena, E.P.; Ruiz, S.J.; Recalde, M.; Rosso, F.; Quinn, J.P.; et al. First report of a Pseudomonas aeruginosa isolate coharboring KPC and VIM carbapenemases. Antimicrob. Agents Chemother. 2012, 56, 5422–5423. [Google Scholar] [CrossRef]
- Cuzon, G.; Naas, T.; Villegas, M.V.; Correa, A.; Quinn, J.P.; Nordmann, P. Wide dissemination of Pseudomonas aeruginosa producing beta-lactamase blaKPC-2 gene in Colombia. Antimicrob. Agents Chemother. 2011, 55, 5350–5353. [Google Scholar] [CrossRef]
- Rizek, C.; Fu, L.; Dos Santos, L.C.; Leite, G.; Ramos, J.; Rossi, F.; Guimaraes, T.; Levin, A.S.; Costa, S.F. Characterization of carbapenem-resistant Pseudomonas aeruginosa clinical isolates, carrying multiple genes coding for this antibiotic resistance. Ann. Clin. Microbiol. Antimicrob. 2014, 13, 43. [Google Scholar] [CrossRef] [PubMed]
- Khuntayaporn, P.; Montakantikul, P.; Mootsikapun, P.; Thamlikitkul, V.; Chomnawang, M.T. Prevalence and genotypic relatedness of carbapenem resistance among multidrug-resistant P. aeruginosa in tertiary hospitals across Thailand. Ann. Clin. Microbiol. Antimicrob. 2012, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Kallen, A.J.; Hidron, A.I.; Patel, J.; Srinivasan, A. Multidrug resistance among gram-negative pathogens that caused healthcare-associated infections reported to the National Healthcare Safety Network, 2006–2008. Infect. Control Hosp. Epidemiol. 2010, 31, 528–531. [Google Scholar] [CrossRef] [PubMed]
- Queenan, A.M.; Pillar, C.M.; Deane, J.; Sahm, D.F.; Lynch, A.S.; Flamm, R.K.; Peterson, J.; Davies, T.A. Multidrug resistance among Acinetobacter spp. in the USA and activity profile of key agents: Results from CAPITAL Surveillance 2010. Diagn. Microbiol. Infect. Dis. 2012, 73, 267–270. [Google Scholar] [CrossRef] [PubMed]
- Viehman, J.A.; Nguyen, M.H.; Doi, Y. Treatment options for carbapenem-resistant and extensively drug-resistant Acinetobacter baumannii infections. Drugs 2014, 74, 1315–1333. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Carbapenem-Resistant Enterobacteriaceae in Healthcare Settings. Available online: https://www.cdc.gov/hai/organisms/cre/index.html (accessed on 10 April 2024).
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2019; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2019. [CrossRef]
- Chagas, T.P.; Seki, L.M.; da Silva, D.M.; Asensi, M.D. Occurrence of KPC-2-producing Klebsiella pneumoniae strains in hospital wastewater. J. Hosp. Infect. 2011, 77, 281. [Google Scholar] [CrossRef] [PubMed]
- White, L.; Hopkins, K.L.; Meunier, D.; Perry, C.L.; Pike, R.; Wilkinson, P.; Pickup, R.W.; Cheesbrough, J.; Woodford, N. Carbapenemase-producing Enterobacteriaceae in hospital wastewater: A reservoir that may be unrelated to clinical isolates. J. Hosp. Infect. 2016, 93, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Nasri, E.; Subirats, J.; Sanchez-Melsio, A.; Mansour, H.B.; Borrego, C.M.; Balcazar, J.L. Abundance of carbapenemase genes (blaKPC, blaNDM and blaOXA-48) in wastewater effluents from Tunisian hospitals. Environ. Pollut. 2017, 229, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-negative Bacteria. Clin. Infect. Dis. 2019, 69, S521–S528. [Google Scholar] [CrossRef]
- Kehl, K.; Schallenberg, A.; Szekat, C.; Albert, C.; Sib, E.; Exner, M.; Zacharias, N.; Schreiber, C.; Parcina, M.; Bierbaum, G. Dissemination of carbapenem resistant bacteria from hospital wastewater into the environment. Sci. Total Environ. 2022, 806, 151339. [Google Scholar] [CrossRef]
- Tacão, M.; Correia, A.; Henriques, I.S. Low Prevalence of Carbapenem-Resistant Bacteria in River Water: Resistance Is Mostly Related to Intrinsic Mechanisms. Microb. Drug Resist. 2015, 21, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Girlich, D.; Poirel, L.; Nordmann, P. Novel ambler class A carbapenem-hydrolyzing beta-lactamase from a Pseudomonas fluorescens isolate from the Seine River, Paris, France. Antimicrob. Agents Chemother. 2010, 54, 328–332. [Google Scholar] [CrossRef]
- Poirel, L.; Barbosa-Vasconcelos, A.; Simões, R.R.; Da Costa, P.M.; Liu, W.; Nordmann, P. Environmental KPC-producing Escherichia coli isolates in Portugal. Antimicrob. Agents Chemother. 2012, 56, 1662–1663. [Google Scholar] [CrossRef] [PubMed]
- Potron, A.; Poirel, L.; Bussy, F.; Nordmann, P. Occurrence of the carbapenem-hydrolyzing beta-lactamase gene blaOXA-48 in the environment in Morocco. Antimicrob. Agents Chemother. 2011, 55, 5413–5414. [Google Scholar] [CrossRef]
- Isozumi, R.; Yoshimatsu, K.; Yamashiro, T.; Hasebe, F.; Nguyen, B.M.; Ngo, T.C.; Yasuda, S.P.; Koma, T.; Shimizu, K.; Arikawa, J. bla(NDM-1)-positive Klebsiella pneumoniae from environment, Vietnam. Emerg. Infect. Dis. 2012, 18, 1383–1385. [Google Scholar] [CrossRef]
- Zurfluh, K.; Hachler, H.; Nuesch-Inderbinen, M.; Stephan, R. Characteristics of extended-spectrum beta-lactamase- and carbapenemase-producing Enterobacteriaceae Isolates from rivers and lakes in Switzerland. Appl. Environ. Microbiol. 2013, 79, 3021–3026. [Google Scholar] [CrossRef]
- Di, D.Y.; Jang, J.; Unno, T.; Hur, H.G. Emergence of Klebsiella variicola positive for NDM-9, a variant of New Delhi metallo-beta-lactamase, in an urban river in South Korea. J. Antimicrob. Chemother. 2017, 72, 1063–1067. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Henriques, I.S.; Araujo, S.; Azevedo, J.S.; Alves, M.S.; Chouchani, C.; Pereira, A.; Correia, A. Prevalence and diversity of carbapenem-resistant bacteria in untreated drinking water in Portugal. Microb. Drug Resist. 2012, 18, 531–537. [Google Scholar] [CrossRef]
- Ash, R.J.; Mauck, B.; Morgan, M. Antibiotic resistance of gram-negative bacteria in rivers, United States. Emerg. Infect. Dis. 2002, 8, 713–716. [Google Scholar] [CrossRef]
- Aubron, C.; Poirel, L.; Ash, R.J.; Nordmann, P. Carbapenemase-producing Enterobacteriaceae, U.S. rivers. Emerg. Infect. Dis. 2005, 11, 260–264. [Google Scholar] [CrossRef]
- Harmon, D.E.; Miranda, O.A.; McCarley, A.; Eshaghian, M.; Carlson, N.; Ruiz, C. Prevalence and characterization of carbapenem-resistant bacteria in water bodies in the Los Angeles-Southern California area. Microbiologyopen 2019, 8, e00692. [Google Scholar] [CrossRef] [PubMed]
- Lopez, N.V.; Farsar, C.J.; Harmon, D.E.; Ruiz, C. Urban and agricultural soils in Southern California are a reservoir of carbapenem-resistant bacteria. Microbiologyopen 2020, 9, 1247–1263. [Google Scholar] [CrossRef]
- Harbor Beaches of Ventura County (Kiddie Beach and Hobie Beach) Bacteria Total Maximum Daily Load; California State Water Resources Control Board: Sacramento, CA, USA, 2007.
- Geosyntec. Bacteria Total Maximum Daily Load Compliance Report; GeoSyntec Consultants: Portland, OR, USA, 2016. [Google Scholar]
- Staff. Bacterial Levels Up at Six Ventura County Beaches; Public Warned to Avoid Ocean Water. VC Star. 2018. Available online: https://www.vcstar.com/story/news/2018/03/28/bacteria-levels-up-six-ventura-county-beaches-public-warned-avoid-ocean-water/467287002/ (accessed on 13 April 2024).
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; CLSI Supplement M100: Wayne, PA, USA, 2018. [Google Scholar]
- Denton, M.; Kerr, K.G. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 1998, 11, 57–80. [Google Scholar] [CrossRef]
- Holt, J.G. Bergey’s Manual of Determinative Bacteriology, 9th ed.; LWW: Baltimore, MD, USA, 1994. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Hudzicki, J. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol. Am. Soc. Microbiol. 2009, 15, 1–23. [Google Scholar]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters; The European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2018. [Google Scholar]
- Dortet, L.; Poirel, L.; Nordmann, P. Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob. Agents Chemother. 2012, 56, 6437–6440. [Google Scholar] [CrossRef]
- Dortet, L.; Poirel, L.; Nordmann, P. Rapid detection of carbapenemase-producing Pseudomonas spp. J. Clin. Microbiol. 2012, 50, 3773–3776. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L.; Dortet, L. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2012, 18, 1503–1507. [Google Scholar] [CrossRef]
- Pierce, V.M.; Simner, P.J.; Lonsway, D.R.; Roe-Carpenter, D.E.; Johnson, J.K.; Brasso, W.B.; Bobenchik, A.M.; Lockett, Z.C.; Charnot-Katsikas, A.; Ferraro, M.J.; et al. Modified Carbapenem Inactivation Method for Phenotypic Detection of Carbapenemase Production among Enterobacteriaceae. J. Clin. Microbiol. 2017, 55, 2321–2333. [Google Scholar] [CrossRef] [PubMed]
- Bottoni, C.; Marcoccia, F.; Compagnoni, C.; Colapietro, M.; Sabatini, A.; Celenza, G.; Segatore, B.; Maturo, M.G.; Amicosante, G.; Perilli, M. Identification of New Natural CphA Metallo-beta-Lactamases CphA4 and CphA5 in Aeromonas veronii and Aeromonas hydrophila Isolates from Municipal Sewage in Central Italy. Antimicrob. Agents Chemother. 2015, 59, 4990–4993. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, P.L.; Ko, W.C.; Wu, C.J. Complexity of β-lactamases among clinical Aeromonas isolates and its clinical implications. J. Microbiol. Immunol. Infect. 2012, 45, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Drk, S.; Puljko, A.; Dzelalija, M.; Udikovic-Kolic, N. Characterization of Third Generation Cephalosporin- and Carbapenem-Resistant Aeromonas Isolates from Municipal and Hospital Wastewater. Antibiotics 2023, 12, 513. [Google Scholar] [CrossRef] [PubMed]
- Libisch, B.; Giske, C.G.; Kovacs, B.; Toth, T.G.; Fuzi, M. Identification of the first VIM metallo-beta-lactamase-producing multiresistant Aeromonas hydrophila strain. J. Clin. Microbiol. 2008, 46, 1878–1880. [Google Scholar] [CrossRef] [PubMed]
- Massidda, O.; Rossolini, G.M.; Satta, G. The Aeromonas hydrophila cphA gene: Molecular heterogeneity among class B metallo-beta-lactamases. J. Bacteriol. 1991, 173, 4611–4617. [Google Scholar] [CrossRef] [PubMed]
- Rossolini, G.M.; Walsh, T.; Amicosante, G. The Aeromonas metallo-beta-lactamases: Genetics, enzymology, and contribution to drug resistance. Microb. Drug Resist. 1996, 2, 245–252. [Google Scholar] [CrossRef]
- Walsh, T.R.; Neville, W.A.; Haran, M.H.; Tolson, D.; Payne, D.J.; Bateson, J.H.; MacGowan, A.P.; Bennett, P.M. Nucleotide and amino acid sequences of the metallo-beta-lactamase, ImiS, from Aeromonas veronii bv. sobria. Antimicrob. Agents Chemother. 1998, 42, 436–439. [Google Scholar] [CrossRef]
- Belotti, P.T.; Thabet, L.; Laffargue, A.; Andre, C.; Coulange-Mayonnove, L.; Arpin, C.; Messadi, A.; M’Zali, F.; Quentin, C.; Dubois, V. Description of an original integron encompassing blaVIM-2, qnrVC1 and genes encoding bacterial group II intron proteins in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2015, 70, 2237–2240. [Google Scholar] [CrossRef]
- Romling, U.; Fiedler, B.; Bosshammer, J.; Grothues, D.; Greipel, J.; von der Hardt, H.; Tummler, B. Epidemiology of chronic Pseudomonas aeruginosa infections in cystic fibrosis. J. Infect. Dis. 1994, 170, 1616–1621. [Google Scholar] [CrossRef]
- Burgos-Diaz, C.; Pons, R.; Espuny, M.J.; Aranda, F.J.; Teruel, J.A.; Manresa, A.; Ortiz, A.; Marques, A.M. Isolation and partial characterization of a biosurfactant mixture produced by Sphingobacterium sp. isolated from soil. J. Colloid. Interface Sci. 2011, 361, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Lambiase, A.; Rossano, F.; Del Pezzo, M.; Raia, V.; Sepe, A.; de Gregorio, F.; Catania, M.R. Sphingobacterium respiratory tract infection in patients with cystic fibrosis. BMC Res. Notes 2009, 2, 262. [Google Scholar] [CrossRef] [PubMed]
- Byappanahalli, M.N.; Nevers, M.B.; Korajkic, A.; Staley, Z.R.; Harwood, V.J. Enterococci in the environment. Microbiol. Mol. Biol. Rev. 2012, 76, 685–706. [Google Scholar] [CrossRef] [PubMed]
- Moellering, R.C. Emergence of Enterococcus as a significant pathogen. Clin. Infect. Dis. 1992, 14, 1173–1176. [Google Scholar] [CrossRef] [PubMed]
- Solioz, M.; Stoyanov, J.V. Copper homeostasis in Enterococcus hirae. FEMS Microbiol. Rev. 2003, 27, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Morandi, S.; Silvetti, T.; Brasca, M. Biotechnological and safety characterization of Enterococcus lactis, a recently described species of dairy origin. Antonie Van. Leeuwenhoek 2013, 103, 239–249. [Google Scholar] [CrossRef]
- Loong, S.K.; Ishak, S.N.; Lim, F.S.; Khoo, J.J.; Tan, S.N.; Freddy-Jalin, E.-J.; Mohd-Taib, F.S.; Abubakar, S. Paenibacillus lautus, an opportunistic bacterial pathogen, isolated from Ixodes granulatus Supino (Acari: Ixodidae) collected from a Müller’s giant Sunda rat (Sundamys muelleri). Syst. Appl. Acarol. 2018, 23, 597–602. [Google Scholar] [CrossRef]
- Saez-Nieto, J.A.; Medina-Pascual, M.J.; Carrasco, G.; Garrido, N.; Fernandez-Torres, M.A.; Villalon, P.; Valdezate, S. Paenibacillus spp. isolated from human and environmental samples in Spain: Detection of 11 new species. New Microbes New Infect. 2017, 19, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Mushtaq, S.; Ge, Y.; Livermore, D.M. Doripenem versus Pseudomonas aeruginosa in vitro: Activity against characterized isolates, mutants, and transconjugants and resistance selection potential. Antimicrob. Agents Chemother. 2004, 48, 3086–3092. [Google Scholar] [CrossRef]
- Bassetti, M.; Nicolini, L.; Esposito, S.; Righi, E.; Viscoli, C. Current status of newer carbapenems. Curr. Med. Chem. 2009, 16, 564–575. [Google Scholar] [CrossRef]
- Avison, M.B.; Higgins, C.S.; von Heldreich, C.J.; Bennett, P.M.; Walsh, T.R. Plasmid location and molecular heterogeneity of the L1 and L2 beta-lactamase genes of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2001, 45, 413–419. [Google Scholar] [CrossRef] [PubMed]
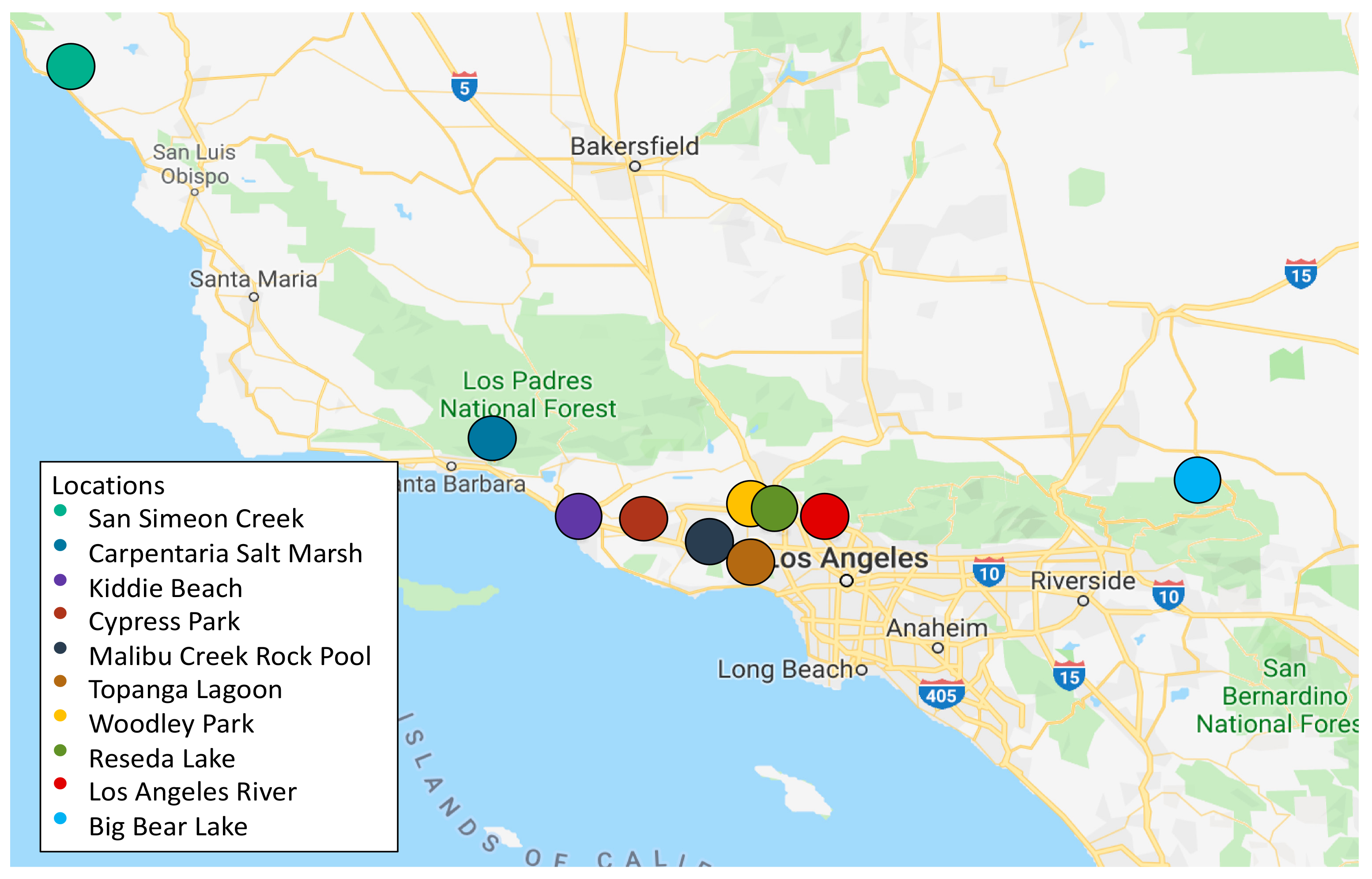



| Sample | Location | Collection Date (D/M/Y) | Location Type | Coordinates | Total Bacteria (CFU/mL) | CRB (CFU/mL) |
|---|---|---|---|---|---|---|
| C | Carpinteria Salt Marsh a | 24/4/18 | Marine estuary | 34.401244, 119.5401337 | 5.1 × 103 | 5.2 × 103 |
| K | Kiddie Beach b | 7/5/18 | Beach harbor | 34.1601144, 119.2235778 | ≤10 | ≤10 |
| LAR | LA River c | 4/6/18 | Urban river | 34.156572, 118.291143 | 2.2 × 103 | ≤10 |
| CP | Cypress Park d | 18/6/18 | Natural creek | 34.167243, 118.962229 | 5.2 × 102 | ≤10 |
| SS | San Simeon Creek e | 24/6/18 | Coastal wetlands | 35.5971832, 121.1218039 | 1 × 102 | ≤10 |
| TL | Topanga Lagoon f | 23/7/18 | Natural pool | 34.038536, 118.583085 | 7 × 102 | ≤10 |
| BBL | Big Bear Lake g | 21/8/18 | Natural freshwater lake | 34.245278, 116.917086 | 8 × 101 | ≤10 |
| WL | Woodley Lake h | 18/3/19 | Reclaimed water | 34.175326, 118.472829 | 3.1 × 103 | 10 |
| RL | Reseda Lake i | 18/3/19 | Artificial lake | 34.188714, 118.534383 | 2.4 × 102 | 20 |
| MRP | Malibu Rock Pool j | 28/3/19 | Natural pool | 34.096555, 118.729879 | 1.1 × 102 | ≤10 |
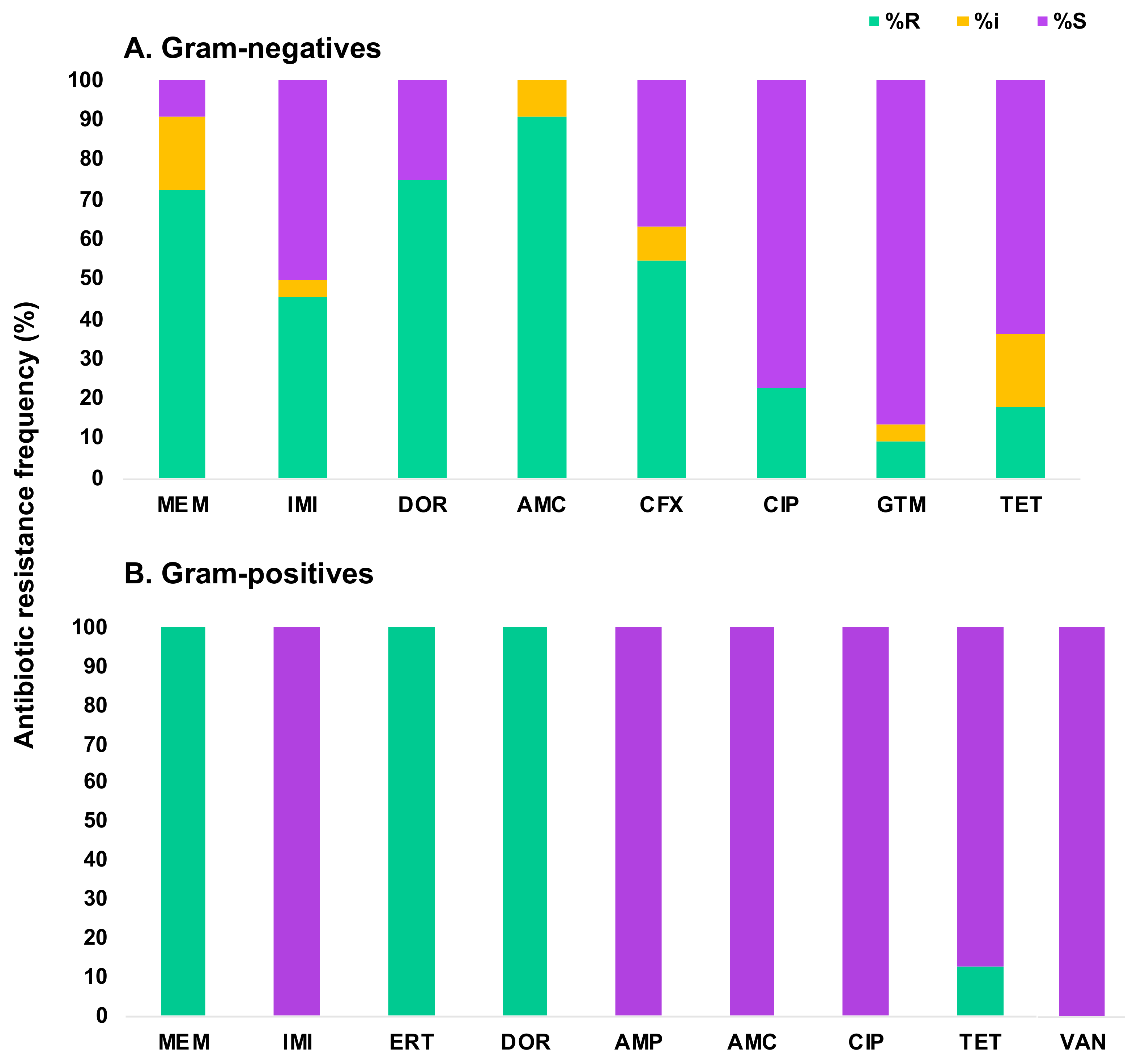
| Isolate a | Closest Species Identified by BLAST of 16S rRNA Gene | CarbaNP c | CP d | Inhibition Zone (Diameter in mm) b | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MEM | IMI | DOR | AMC | CFX | CIP | GTM | TET | ||||
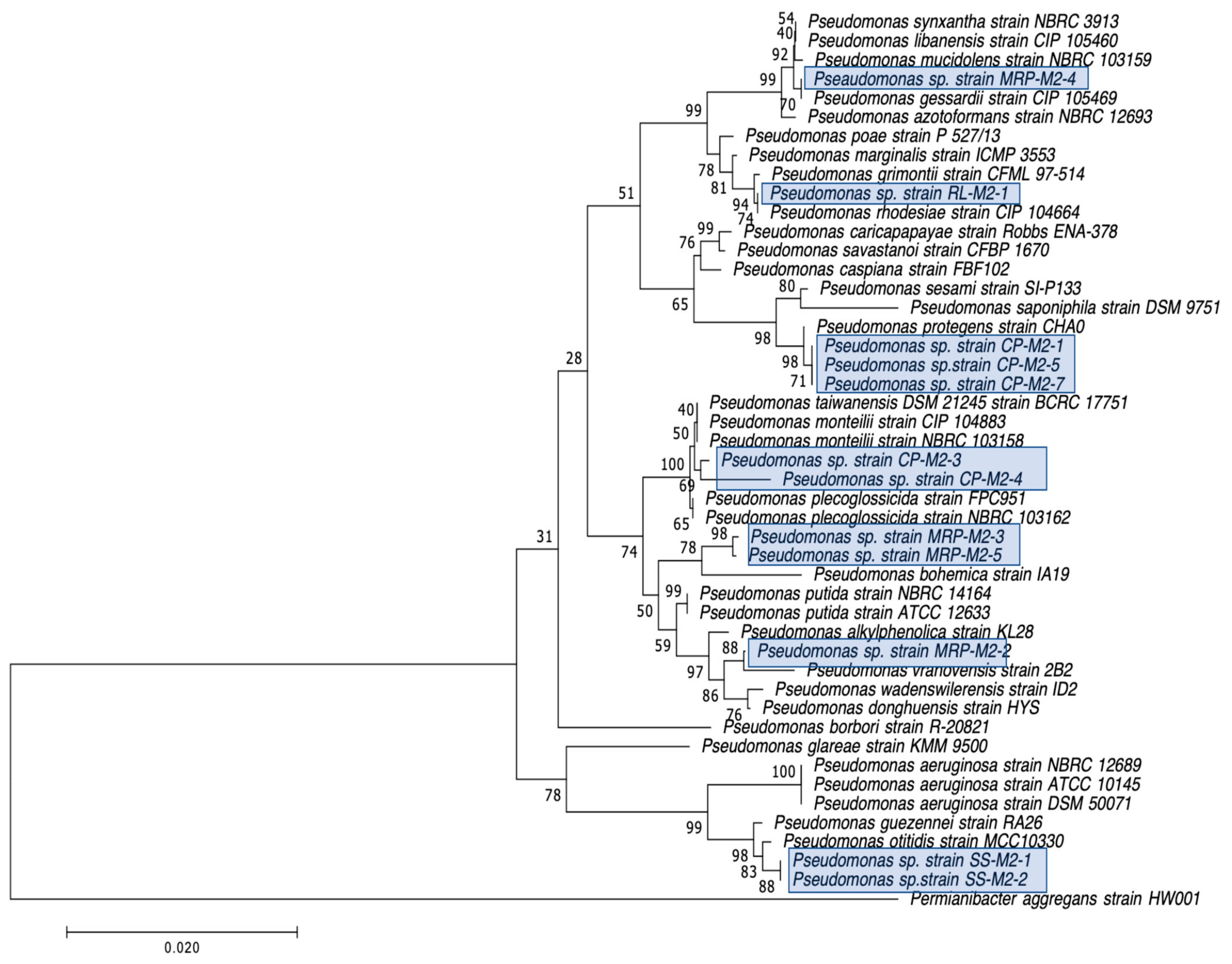
| CP-M2-1 | Pseudomonas sp. 18 | − | N/D | 0 | 16.7 | 16.7 | 0 | 5 | 22.6 | 18.3 | 6 |
| CP-M2-3 | Pseudomonas plecoglossicida | − | N/D | 5.3 | 27 | 19 | 0 | 10 | 25.3 | 18.3 | 14 |
| CP-M2-4 | Pseudomonas putida | − | N/D | 0 | 26 | 14.5 | 0 | 4.3 | 22.7 | 21.3 | 12 |
| CP-M2-5 | Pseudomonas spp. GC04 | − | N/D | 0 | 28.3 | 20.8 | 0 | 3.7 | 26 | 26 | 12.3 |
| CP-M2-7 | Pseudomonas sp. CHZYR63 | − | N/D | 0 | 19 | 16.3 | 0 | 0 | 23.3 | 17 | 7 |
| SS-M2-1 | Pseudomonas sp. J4AJ | − | N/D | 18.7 | 24.7 | 20.6 | 10 | 25.3 | 38.8 | 24.7 | 25.3 |
| SS-M2-2 | Pseudomonas sp. P7 | − | N/D | 9.7 | 27.3 | 18.4 | 5.7 | 25.3 | 33 | 23.7 | 16 |
| SS-M2-3 | Aeromonas veronii | + | UNK | 7 | 8 | N/D | 8 | 35 | 39 | 25 | 25.3 |
| BBL-M2-1 | Aeromonas veronii | + | UNK | 24.7 | 20.7 | N/D | 15 | 36.7 | 36.7 | 20.3 | 27.3 |
| BBL-M2-2 | Stenotrophomonas maltophilia | + | blaL1 | 0 | 0 | N/D | 0 | 0 | 26.7 | 0 | 11.7 |
| BBL-M2-3 | Aeromonas veronii | + | UNK | 23.5 | 24.7 | N/D | 9 | 34 | 31.8 | 17.8 | 27 |
| MRP-M2-1 | Stenotrophomonas maltophilia | + | blaL1 | 0 | 0 | N/D | 0 | 0 | 26 | 7.3 | 10.7 |
| MRP-M2-2 | Pseudomonas sp. W15Feb40A | − | N/D | 19.8 | 30.7 | 28.5 | 3.3 | 15.3 | 24.7 | 22.8 | 16 |
| MRP-M2-3 | Pseudomonas putida | − | N/D | 17 | 29.7 | 27.3 | 0 | 15 | 31.3 | 23.3 | 19.3 |
| MRP-M2-4 | Pseudomonas fluorescens | − | N/D | 19.7 | 21 | 21 | 0 | 4.7 | 35.7 | 32 | 34 |
| MRP-M2-5 | Pseudomonas putida | − | N/D | 12 | 25.3 | 22 | 0 | 13 | 28.7 | 23.7 | 17.3 |
| RL-M2-1 | Pseudomonas rhodesiae | − | N/D | 0 | 13.5 | 13 | 0 | 3 | 36 | 30 | 21.7 |
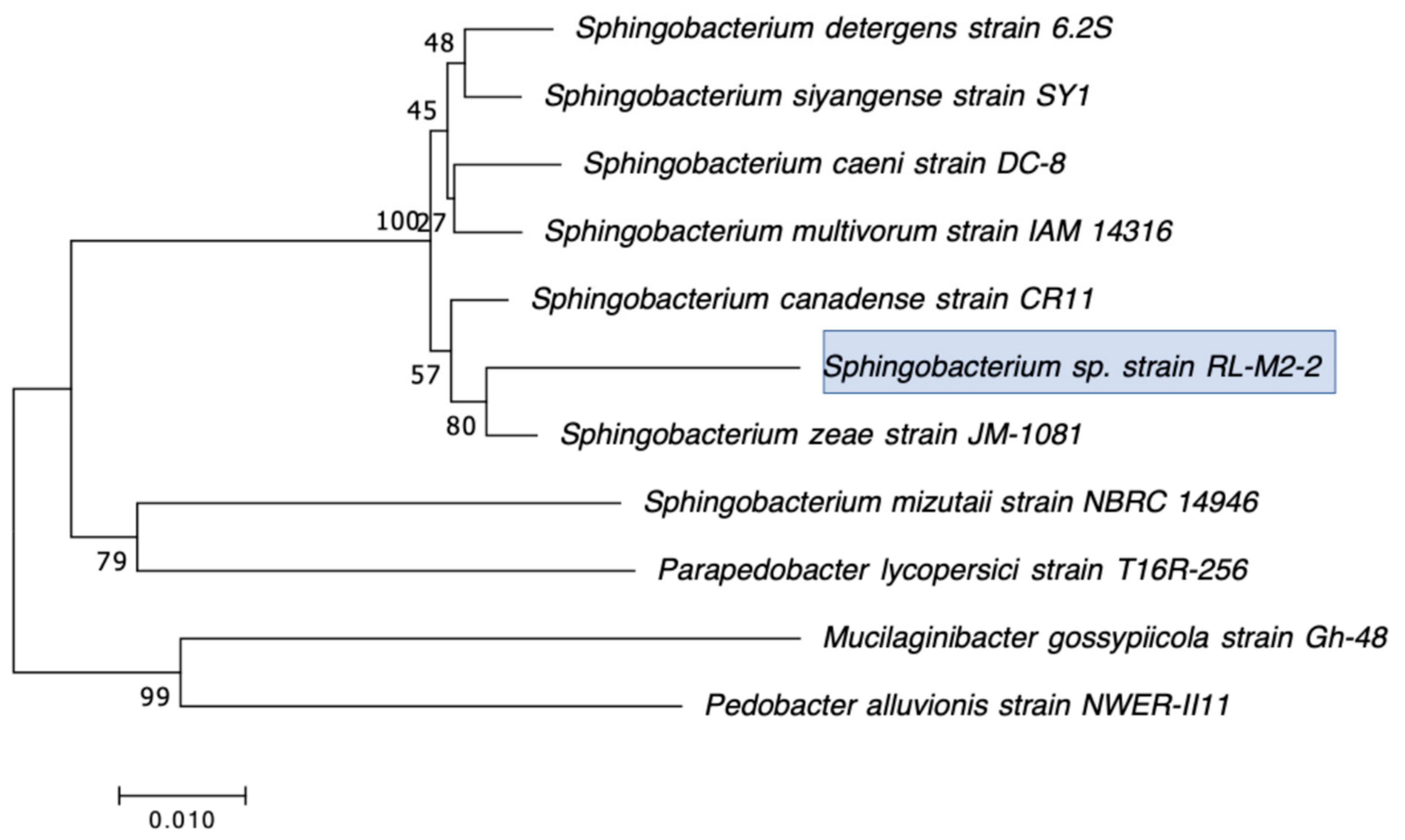
| RL-M2-2 | Sphingobacterium siyangensis | − | N/D | 21.2 | 15.8 | N/D | 13.7 | 26 | 29.7 | 15 | 24.5 |
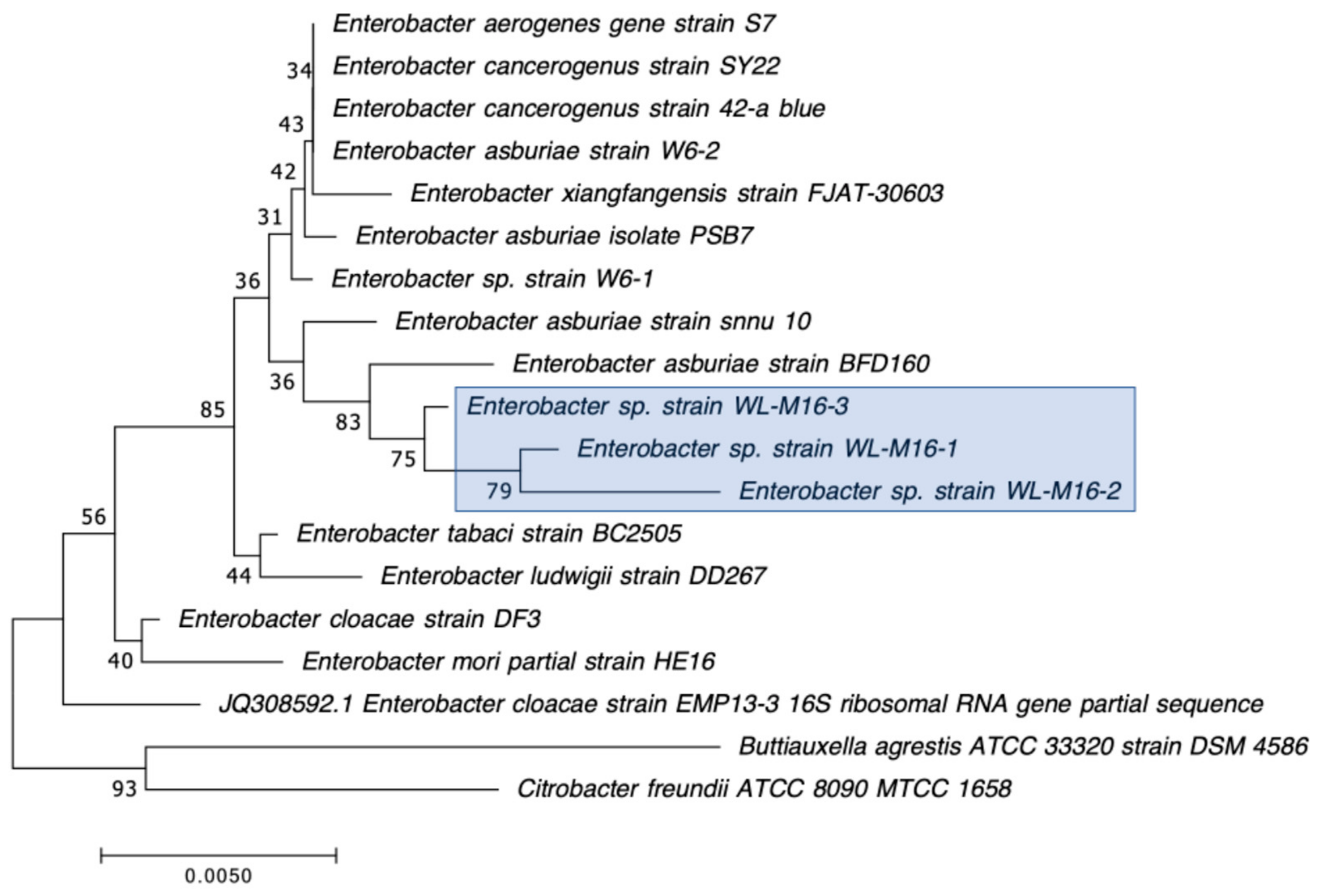
| WL-M2-1 | Enterobacter asburiae | + | blaIMI-2 | 2.2 | 0 | N/D | 0 | 32 | 35.7 | 21.3 | 23.3 |
| WL-M2-2 | Enterobacter asburiae | + | blaIMI-2 | 2.2 | 0 | N/D | 0 | 32.3 | 35.7 | 21.3 | 23.3 |
| WL-M2-3 | Enterobacter asburiae | + | blaIMI-2 | 2.2 | 0 | N/D | 0 | 31.7 | 36 | 21.7 | 24 |
| WL-M2-4 | Stenotrophomonas maltophilia | + | blaL1 | 0 | 0 | N/D | 0 | 0 | 26.7 | 13.3 | 10.8 |
| Isolate a | Closest Species Identified by BLAST of 16S rRNA Gene | CarbaNP c | Inhibition Zone (Diameter in mm) b | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MEM | IMI | ERT | DOR | AMP | AMC | CIP | TET | VAN | |||
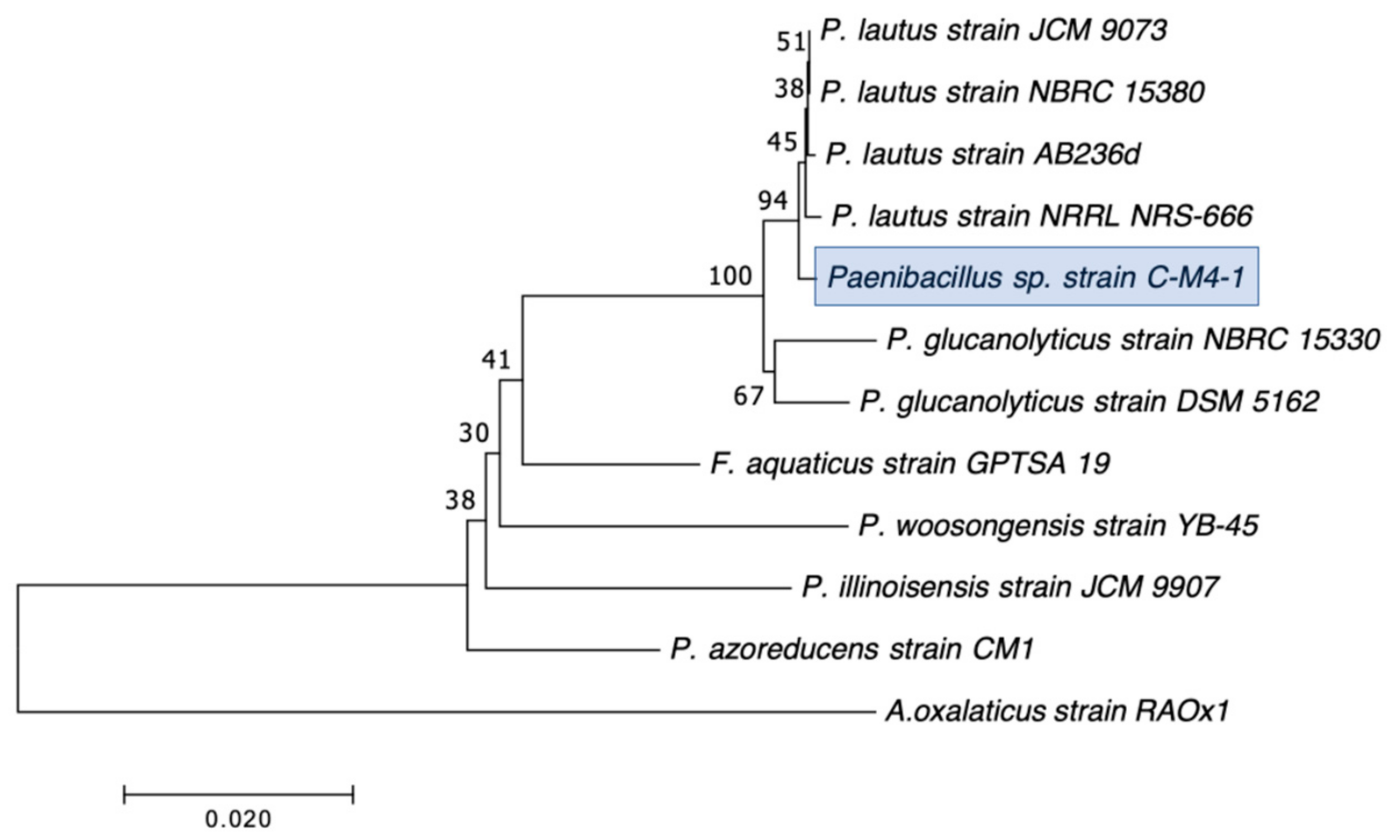
| C-M4-1 | Paenibacillus lautus | − | 5 | 25 | 0 | 12.3 | 23.3 | 26.7 | 22.3 | 34.3 | 26.3 |
| K-M2-1 | Enterococcus lactis | − | 2.7 | 23.7 | 2.3 | 11.7 | 25.7 | 31.3 | 23.6 | 30 | 24.3 |
| K-M2-2 | Enterococcus faecium | − | 2 | 24.3 | 0 | 9.6 | 21 | 29 | 24 | 29.3 | 24.8 |
| K-M2-3 | Enterococcus faecium | − | 4.7 | 22.7 | 4.7 | 9.7 | 21.7 | 27.3 | 23.3 | 27.7 | 24 |
| K-M2-4 | Enterococcus faecium | − | 0 | 20.3 | 0 | 6.7 | 19.3 | 27.3 | 23.3 | 30.7 | 25.3 |
| LAR-M2-1 | Enterococcus hirae | − | 5.5 | 24.7 | 8.3 | 10.7 | 22 | 27 | 24 | 26 | 22.5 |
| CP-M2-6 | Enterococcus hirae | − | 11 | 28 | 13 | 16.2 | 29.3 | 32 | 23 | 2.8 | 22.3 |
| RL-M2-3 | Enterococcus lactis | − | 14 | 27 | 11.7 | 11.3 | 25.7 | 27.3 | 25 | 30.3 | 25.3 |
| Isolate | Closest Species Identified by 16S rRNA Gene | CarbaNP | mCIM | eCIM | Carbapenemase Gene (% DNA Identity) | Carbapenemase (% Amino Acid Similarity) |
|---|---|---|---|---|---|---|
| SS-M2-3 | Aeromonas veronii | + | + | + | Unknown | Unknown |
| BBL-M2-1 | Aeromonas veronii | + | + | + | Unknown | Unknown |
| BBL-M2-2 | Stenotrophomonas maltophilia | + | N/D | N/D | blaL1 (88.6%) | L1 (96.9%) |
| BBL-M2-3 | Aeromonas veronii | + | + | + | Unknown | Unknown |
| MRP-M2-1 | Stenotrophomonas maltophilia | + | N/D | N/D | blaL1 (87.7%) | L1 (96.0%) |
| WL-M16-1 | Enterobacter asburiae | + | N/D | N/D | blaIMI-2 (99.2%) | IMI-2 (98.6%) |
| WL-M16-2 | Enterobacter asburiae | + | N/D | N/D | blaIMI-2 (98.3%) | IMI-2 (97.7%) |
| WL-M16-3 | Enterobacter asburiae | + | N/D | N/D | blaIMI-2 (99.8%) | IMI-2 (100%) |
| WL-M16-4 | Stenotrophomonas maltophilia | + | N/D | N/D | blaL1 (88.2%) | L1 (97.0%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McCarley, A.; Espejo, M.L.; Harmon, D.E.; Ruiz, C. Freshwater and Marine Environments in California Are a Reservoir of Carbapenem-Resistant Bacteria. Microorganisms 2024, 12, 802. https://doi.org/10.3390/microorganisms12040802
McCarley A, Espejo ML, Harmon DE, Ruiz C. Freshwater and Marine Environments in California Are a Reservoir of Carbapenem-Resistant Bacteria. Microorganisms. 2024; 12(4):802. https://doi.org/10.3390/microorganisms12040802
Chicago/Turabian StyleMcCarley, Ashley, Manuel Luis Espejo, Dana E. Harmon, and Cristian Ruiz. 2024. "Freshwater and Marine Environments in California Are a Reservoir of Carbapenem-Resistant Bacteria" Microorganisms 12, no. 4: 802. https://doi.org/10.3390/microorganisms12040802
APA StyleMcCarley, A., Espejo, M. L., Harmon, D. E., & Ruiz, C. (2024). Freshwater and Marine Environments in California Are a Reservoir of Carbapenem-Resistant Bacteria. Microorganisms, 12(4), 802. https://doi.org/10.3390/microorganisms12040802






