Comparison of Copper-Tolerance Genes between Different Groups of Acidovorax citrulli
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids, and Antibiotics
2.2. Selection of Copper-Tolerant Representative Strains
2.3. Construction and Verification of Deletion Mutants and Complementary Strains for Putative Copper-Tolerance Genes
2.4. Determination of Copper MIC
2.5. Analysis of Gene Expression Related to Copper Tolerance
2.6. Data Analysis
3. Results
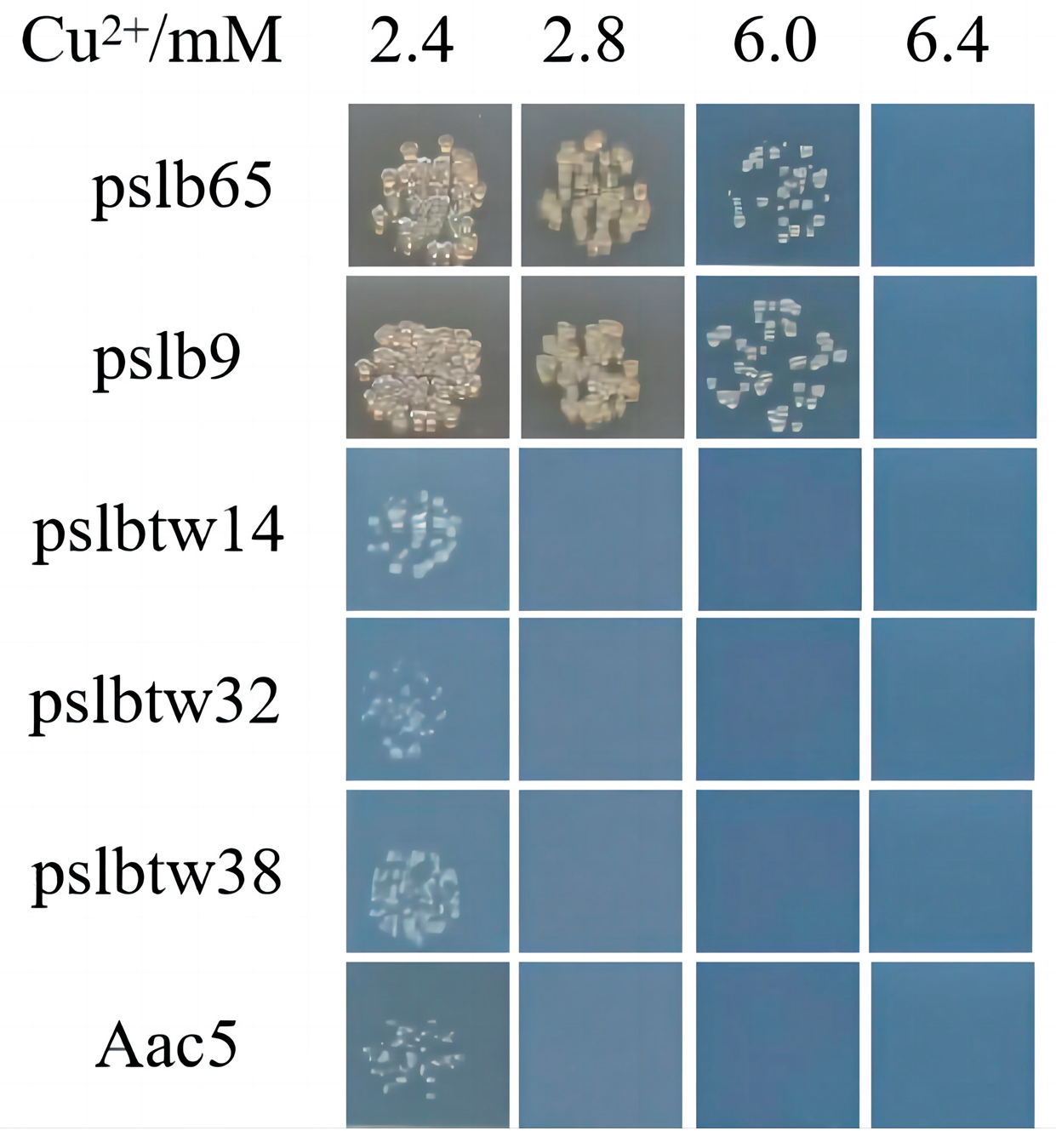
3.1. Identification of Copper-Tolerant A. citrulli Strains
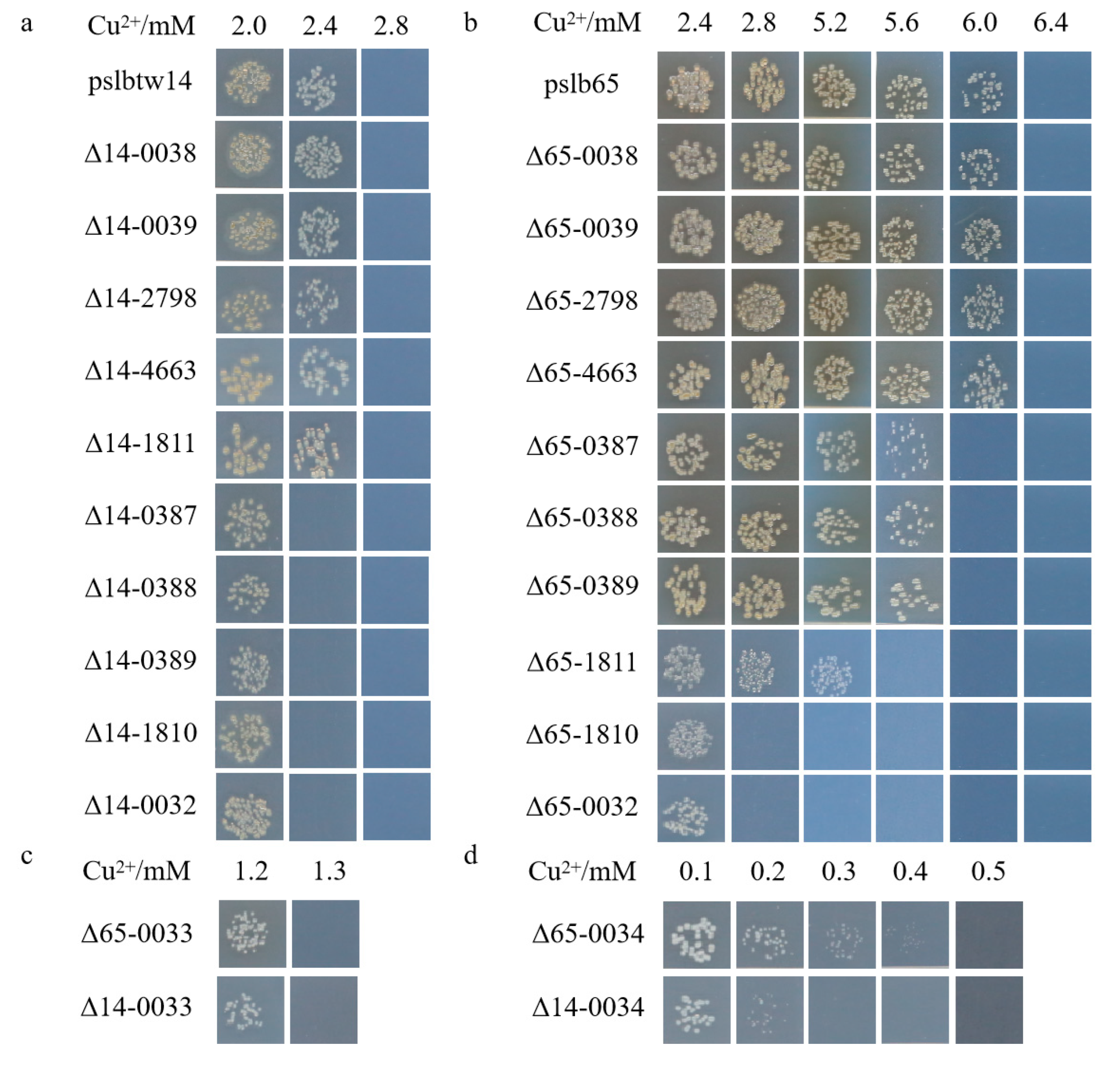
3.2. Verification of Deletion Mutants and Complementary Strains for Putative Copper-Tolerance Genes
3.3. Copper-Tolerance Phenotypes of Representative Strains and Mutants
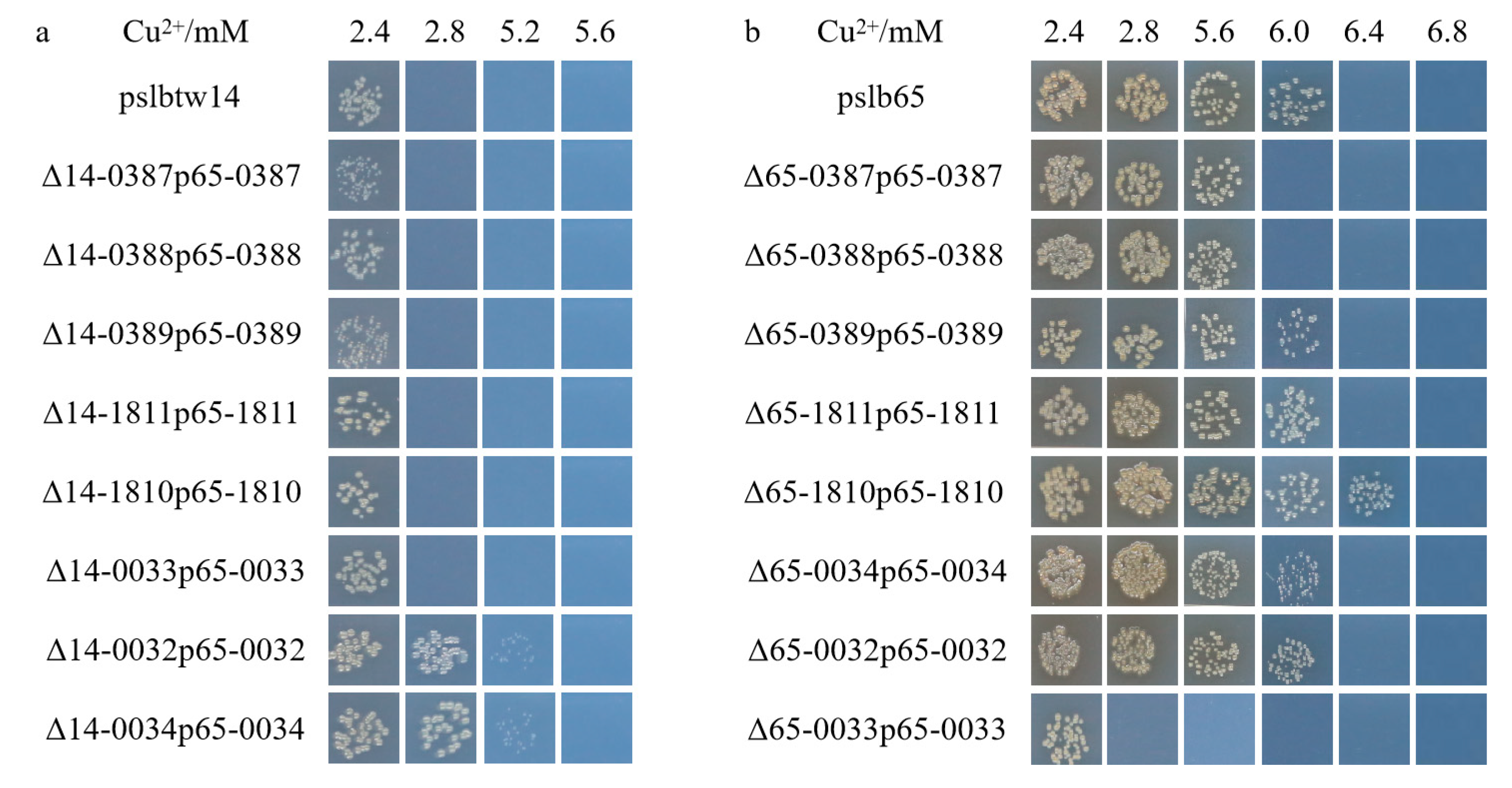
3.4. Copper-Tolerant Phenotype of the Complementary Strains
3.5. Analysis of Copper-Tolerance-Related Genes Expression in Wildtype Strains
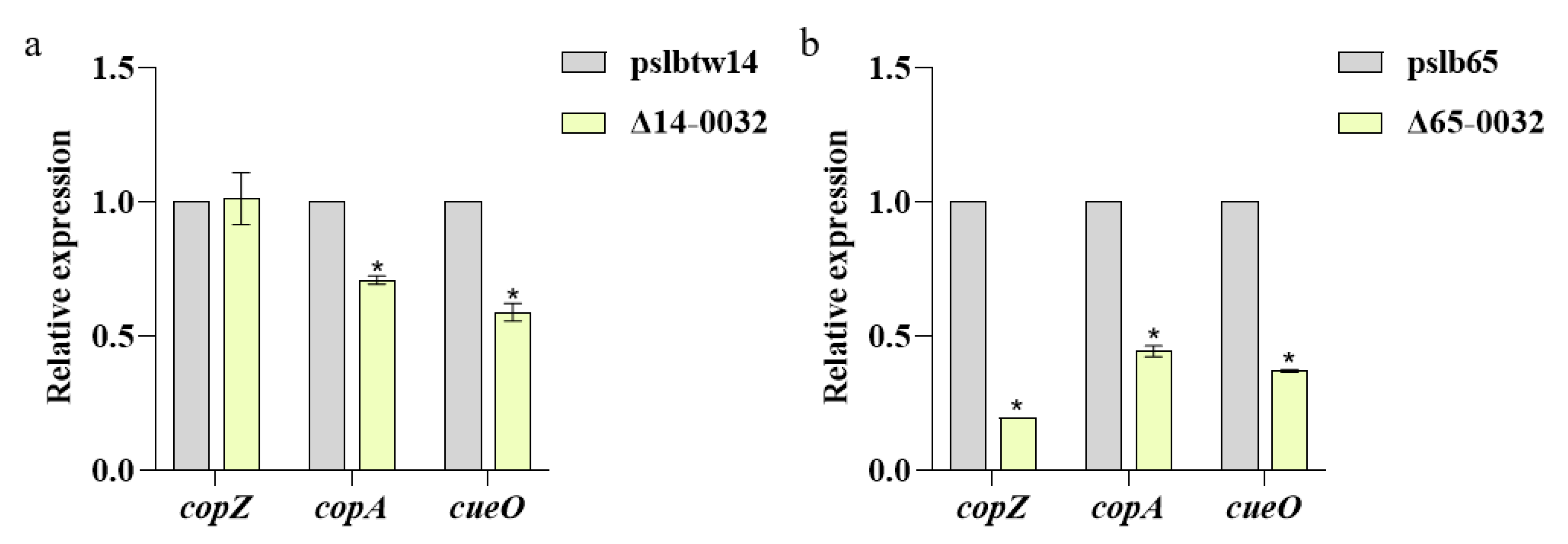
3.6. Results of Analysis on the Expression of Copper-Tolerance-Related Genes in Mutant Strains
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schaad, N.W.; Postnikova, E.; Sechler, A.; Claflin, L.E.; Vidaver, A.K.; Jones, J.B.; Agarkova, I.; Ignatov, A.; Dickstein, E.; Ramundo, B.A. Reclassification of subspecies of Acidovorax avenae as A. Avenae (Manns, 1905) emend., A. cattleyae (Pavarino, 1911) comb nov., A citrulli (Schaad, 1978) comb. nov., and proposal of A. oryzae sp. nov. Syst. Appl. Microbiol. 2008, 31, 434–446. [Google Scholar] [CrossRef]
- Schaad, N.W.; Sowell, G.; Goth, R.W.; Colwell, R.R.; Webb, R.E. Pseudomonas pseudoalcaligenes subsp. citrulli subsp. nov. Int. J. Syst. Bacteriol. 1978, 28, 117–125. [Google Scholar] [CrossRef]
- Willems, A.; Goor, M.; Thielemans, S.; Gillis, M.; Kersters, K.; De Ley, J. Transfer of several phytopathogenic Pseudomonas species to Acidovorax as Acidovorax avenae subsp. avenae subsp. nov. comb. nov., Acidovorax avenae subsp. citrulli, Acidovorax avenae subsp. cattleyae, and Acidovorax konjaci. Int. J. Syst. Bacteriol. 1992, 42, 107. [Google Scholar] [CrossRef]
- Xie, H.T.; Li, Z.B.; Qin, B.X.; Yang, S.A.; Cui, L.X.; Deng, T.J.; Cai, J.H. Identification of pathogen causing melon bacterial fruit blotch in Guangxi and analysis of its 16S rDNA sequence. J. South. Agric. 2016, 47, 1698–1703. (In Chinese) [Google Scholar]
- Chalupowicz, L.; Dror, O.; Reuven, M.; Burdman, S.; Manulis-Sasson, S. Cotyledons are the main source of secondary spread of Acidovorax citrulli in melon nurseries. Plant Pathol. 2014, 64, 528–536. [Google Scholar] [CrossRef]
- Yang, Y.W.; Zhao, M.; Zhang, L.Q.; Qiao, P.; Bai, X.; Zhang, X.X.; Walcott, R.R.; Guan, W.; Zhao, T.C. Development of a multiplex PCR assay based on the pilA gene sequences to detect different types of Acidovorax citrulli. J. Microbiol. Method 2019, 158, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Qiao, P.; Wang, T.; Ji, W.; Fei, N.; Zhang, L.; Guan, W.; Zhao, T.C. Further characterization of host preference of Acidovorax citrulli based on growth competition between group I and group II Strains. Horticulturae 2022, 8, 1173. [Google Scholar] [CrossRef]
- Hopkins, D.L. Copper-containing fungicides reduce the spread of bacterial fruit blotch of watermelon in the greenhouse. Phytopathology 1995, 85, 510–516. [Google Scholar]
- Hopkins, D.L. Control of bacterial fruit blotch of watermelon with cupric hydroxide. Phytopathology 1991, 81, 1228–1234. [Google Scholar]
- Zhao, W.L.; Yang, Y.W.; Wang, T.L.; Jiang, J.; Liu, H.Q.; Zhao, T.C. Sensitivity test and analysis of Acidovorax citrulli to copper sulfate. Plant Prot. 2013, 39, 100–105. (In Chinese) [Google Scholar]
- Puig, S.; Thiele, D.J. Molecular mechanisms of copper uptake and distribution. Curr. Opin. Chem. Biol. 2002, 6, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Osman, D.; Cavet, J.S. Chapter 8 copper homeostasis in bacteria. Adv. Appl. Microbiol. 2008, 65, 217–247. [Google Scholar] [PubMed]
- Rensing, C.; Grass, G. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 2003, 27, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Li, Q. Functional Analysis of cueO Gene in the Bacterial Fruit Blotch of Watermelon; Inner Mongolia Agricultural University: Hohhot, China, 2014. (In Chinese) [Google Scholar]
- Liu, Y.F.; Guan, W.; Wang, T.L.; Li, Z.H.; Yang, Y.W.; Liu, H.Q.; Zhao, T.C. Functional analysis of copZ involved in copper metabolism of Acidovorax citrulli. Acta Phytopathol. Sin. 2022, 52, 377–387. (In Chinese) [Google Scholar]
- Song, J.D. Identification and Functional Analysis of Copper Resistance Genes cusABC and zneB in Acidovorax citrulli; Xinjiang Agricultural University: Urumqi, China, 2021. (In Chinese) [Google Scholar]
- Liu, X.; Wang, X.D.; Liu, J. Functional analysis of a RND family efflux transporter component-cusB gene associated with copper resistance in Acidovorax citrulli. Microbiol. China Tongbao 2016, 43, 97–106. (In Chinese) [Google Scholar]
- Huang, C.W. Identification and Functional Analysis of Copper Resistance Genes tolC and gntR in Acidovorax citrulli; Xinjiang Agricultural University: Urumqi, China, 2018. (In Chinese) [Google Scholar]
- Xie, B.B. Identification and Functional Analysis of Copper Resistance Genes cueR and copZ in Acidovorax citrulli; Xinjiang Agricultural University: Urumqi, China, 2020. (In Chinese) [Google Scholar]
- Fei, N.Y.; Ji, W.Q.; Yang, L.L.; Yu, C.; Qiao, P.; Yan, J.; Guan, W.; Yang, Y.; Zhao, T. Hcp of the Type VI Secretion System (T6SS) in Acidovorax citrulli group II strain Aac5 has a dual role as a core structural protein and an effector protein in colonization, growth ability, competition, biofilm formation, and ferric iron absorption. Int. J. Mol. Sci. 2022, 23, 9632. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Zhao, M.; Yan, J.P.; Yang, L.L.; Yang, Y.W.; Guan, W.; Walcott, R.R.; Zhao, T.C. Involvement of hrpX and hrpG in the virulence of Acidovorax citrulli strain Aac5, causal agent of bacterial fruit blotch in cucurbits. Front. Microbiol. 2018, 9, 507. [Google Scholar] [CrossRef]
- Schafer, A.; Tauch, A.; Jager, W.; Kalinowski, J.; Thierbach, G.; Pühler, A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Yang, Y.; Fei, N.; Ji, W.; Qiao, P.; Yang, L.; Liu, D.; Guan, W.; Zhao, T. pilA gene contributes to virulence, motility, biofilm formation, and interspecific competition of bacteria in Acidovorax citrulli. Microorganisms 2023, 11, 1806. [Google Scholar] [CrossRef]
- Zhang, A.P.; Zhang, X.X.; Wu, L.N.; Yu, C.; Qiao, P.; Yang, Y.W.; Guan, W.; Zhao, T.C. Construction of chemotaxis and flagella gene double mutant ΔcheAΔfliC and its functional analysis in Acidovorax citrulli. J. Agric. Biotechnol. 2017, 25, 1838–1850. (In Chinese) [Google Scholar]
- Ren, Z.G.; Hou, L.; Song, Z.G.; Zhang, L.Q. Screening of the pathogenicity mutants of Acidovorax avenae subsp. citrulli and cloning of the hrcR gene. Acta Phytopathol. Sin. 2009, 39, 53–58. (In Chinese) [Google Scholar]
- Walcott, R.R.; Gitaitis, R.D. Detection of Acidovorax avenae subsp. citrulli in watermelon seed using immunomagnetic separation and the polymerase chain reaction. Plant Dis. 2000, 84, 470–474. [Google Scholar] [CrossRef]
- Hu, X.M. Research on Function of Putative Copper Resistance Gene Cluster copSRABCD of Ralstonia solanacearum; Chinese Academy of Agricultural Sciences: Beijing, China, 2016. (In Chinese) [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the (2−ΔΔCT) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Liu, D.; Zhao, M.; Qiao, P.; Li, Z.; Chen, G.; Guan, W.; Bai, Q.; Walcott, R.; Yang, Y.; Zhao, T. NtrC Contributes to Nitrogen Utilization, Stress Tolerance, and Virulence in Acidovorax citrulli. Microorganisms 2023, 11, 767. [Google Scholar] [CrossRef]
- Tanabe, M.; Szakonyi, G.; Brown, K.A.; Henderson, P.J.F.; Nield, J.; Byrne, B. The multidrug resistance efflux complex, EmrAB from Escherichia coli forms a dimer in vitro. Biochem. Biophys. Res. Commun. 2009, 380, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Turlin, E.; Heuck, G.; Brando, M.I.S.; Szili, N.; Mellin, J.; Lange, N.; Wandersman, C. Protoporphyrin (PPIX) efflux by the MacAB-TolC pump in Escherichia coli. Microbiol. Open 2014, 3, 849–859. [Google Scholar] [CrossRef]
- Rensing, C.; Fan, B.; Sharma, R. CopA: An Escherichia coli Cu(I)-translocating P-type ATPase. Proc. Natl. Acad. Sci. USA 2000, 97, 652. [Google Scholar] [CrossRef]
- Wen, Q. Construction of an Efficient Markerless Gene Disruption/Integration System in Acidithiobacillus thiooxidans and Study on Its Copper Tolerance Mechanisms; Shandong University: Jinan, China, 2021. (In Chinese) [Google Scholar]
- Xie, B.B.; Liu, J.; Youlituzi, N.B.; Zhang, C.B. Bioinformatics analysis and functional verification of copper resistance gene cueR in Acidovorax citrulli. Microbiol. Bull. 2020, 47, 1534–1543. (In Chinese) [Google Scholar]



| Primers | Primer Sequence (5′-3′) | Length/bp | Source |
|---|---|---|---|
| rpoB-F | GCGACAGCGTGCTCAAAGTG | 134 | [30] |
| rpoB-R | GCCTTCGTTGGTGCGTTTCT | ||
| cueR-QF | CGCATGGTCCGCCACTAC | 173 | [19] |
| cueR-QR | TCCTGCCAGAGCCCGAG | ||
| copZ-QF | TGACCTGCGGCCATTGC | 118 | [19] |
| copZ-QR | CGAGGGCTGTCGCTTTCC | ||
| copA-QR | TGTCGCTGTGGCTGTGGTTC | 144 | [19] |
| copA-QF | CTTCCGTGGTCTGCCGCTTG | ||
| cusA-QR | AGGGCTTCAACCTGTCGCT | 143 | This study |
| cusA-QF | GTTGAGTTGCCCCTTGACG | ||
| cusB-QR | TTCACGGAAGGCAGCGAC | 200 | This study |
| cusB-QF | ACCGCGTTGTCGTACTCCTG | ||
| cusC-QR | GCTGCCAACGCCAACATC | 111 | This study |
| cusC-QF | GCCGCCCTTGAACAGACC | ||
| cueO-QR | GACCAACCACCCCATCCAC | 187 | This study |
| cueO-QF | GTGGTGGCTCTTGTGGCAGT | ||
| tolC-QR | GGCCATTCCGAAATCAAGC | 101 | This study |
| tolC-QF | TCATTCACAAGCCCCTACGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Zhao, M.; Qiao, P.; Liu, D.; Bai, Q.; Guan, W.; Yang, Y.; Zhao, T. Comparison of Copper-Tolerance Genes between Different Groups of Acidovorax citrulli. Microorganisms 2024, 12, 682. https://doi.org/10.3390/microorganisms12040682
Zhang M, Zhao M, Qiao P, Liu D, Bai Q, Guan W, Yang Y, Zhao T. Comparison of Copper-Tolerance Genes between Different Groups of Acidovorax citrulli. Microorganisms. 2024; 12(4):682. https://doi.org/10.3390/microorganisms12040682
Chicago/Turabian StyleZhang, Min, Mei Zhao, Pei Qiao, Dehua Liu, Qingrong Bai, Wei Guan, Yuwen Yang, and Tingchang Zhao. 2024. "Comparison of Copper-Tolerance Genes between Different Groups of Acidovorax citrulli" Microorganisms 12, no. 4: 682. https://doi.org/10.3390/microorganisms12040682
APA StyleZhang, M., Zhao, M., Qiao, P., Liu, D., Bai, Q., Guan, W., Yang, Y., & Zhao, T. (2024). Comparison of Copper-Tolerance Genes between Different Groups of Acidovorax citrulli. Microorganisms, 12(4), 682. https://doi.org/10.3390/microorganisms12040682






