Who Is the Intermediate Host of RNA Viruses? A Study Focusing on SARS-CoV-2 and Poliovirus
Abstract
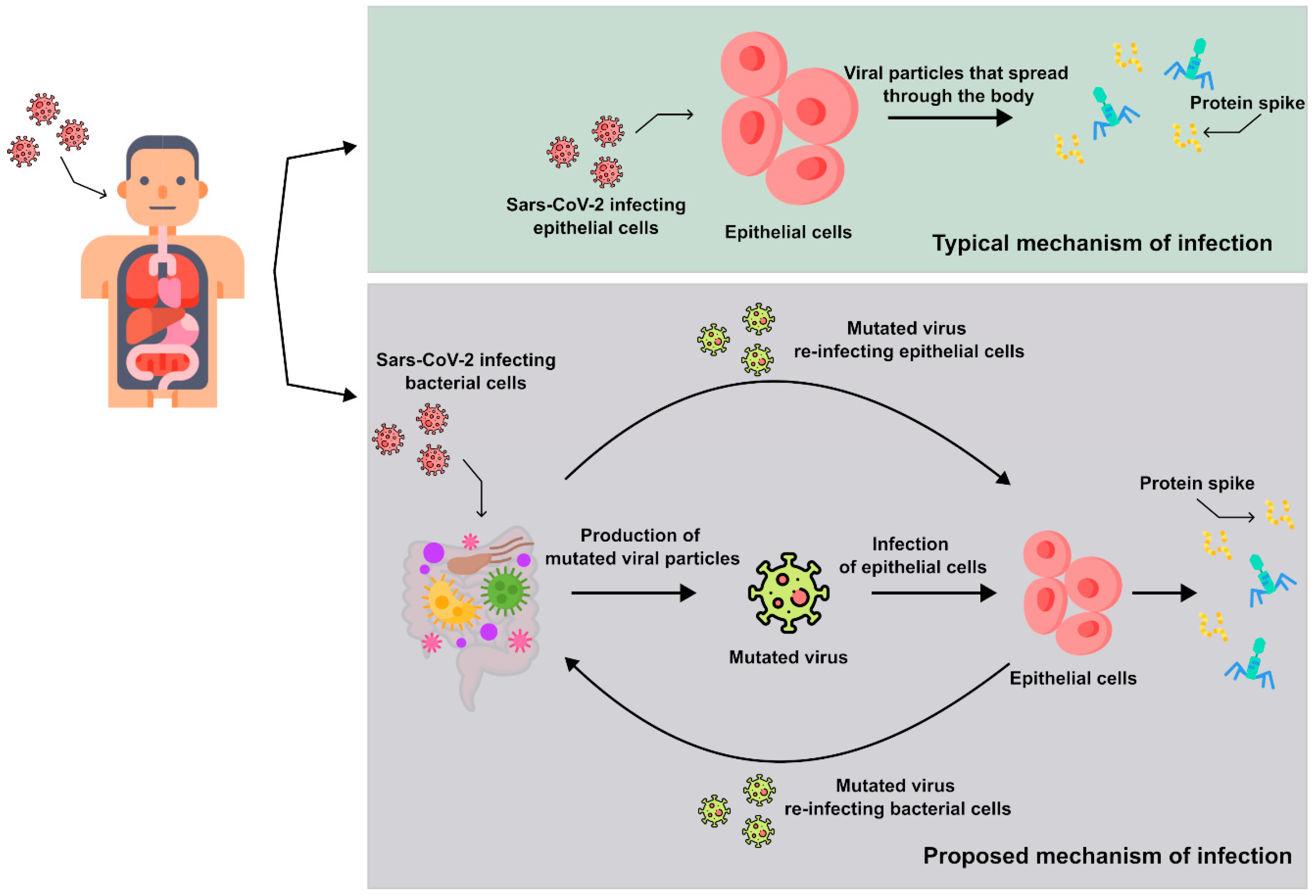
1. Introduction
2. Overview of the Results on the Bacteriophage Behavior of SARS-CoV-2
2.1. Initial Molecular Investigation of Viral RNA in Bacterial Cultures
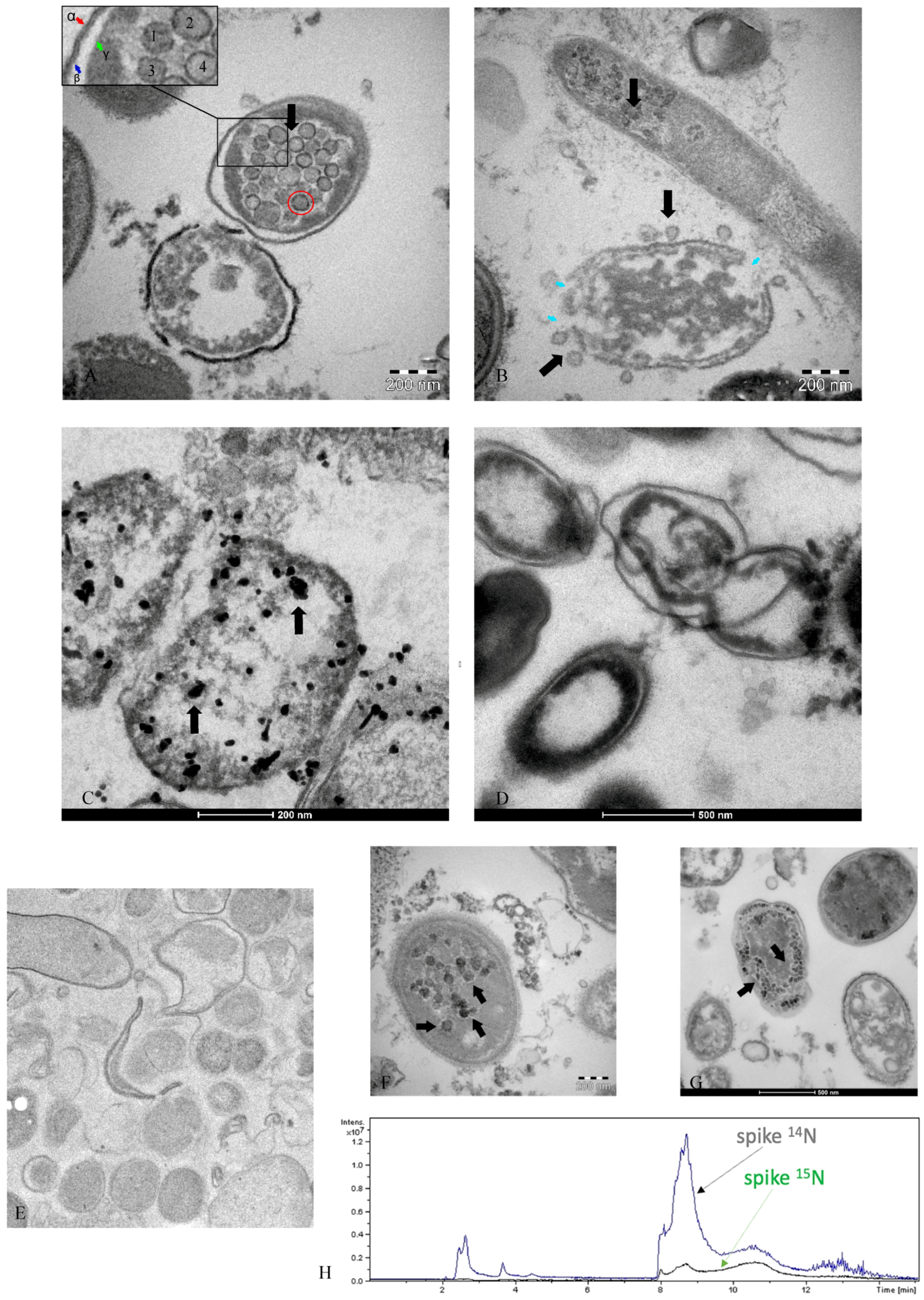
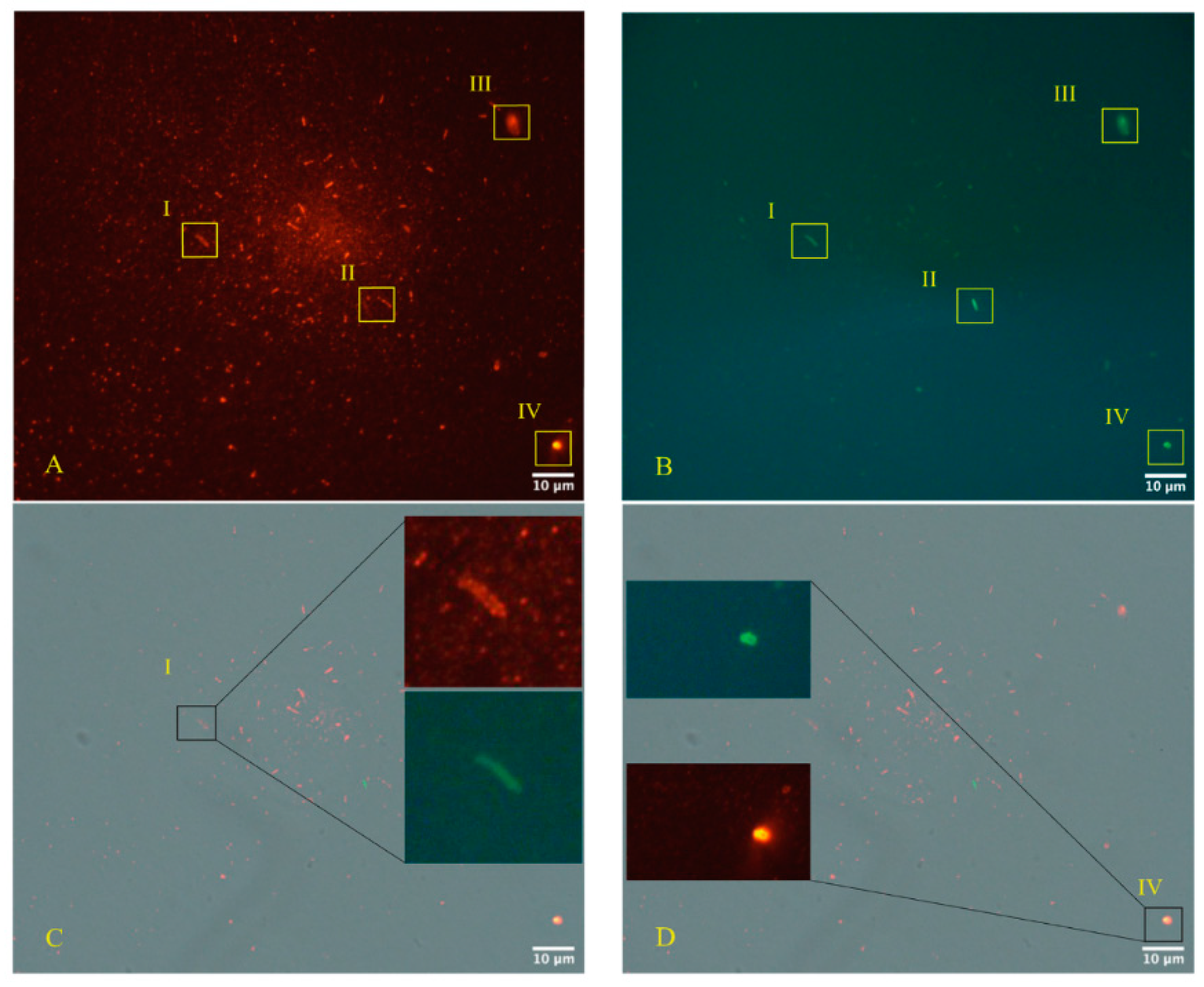
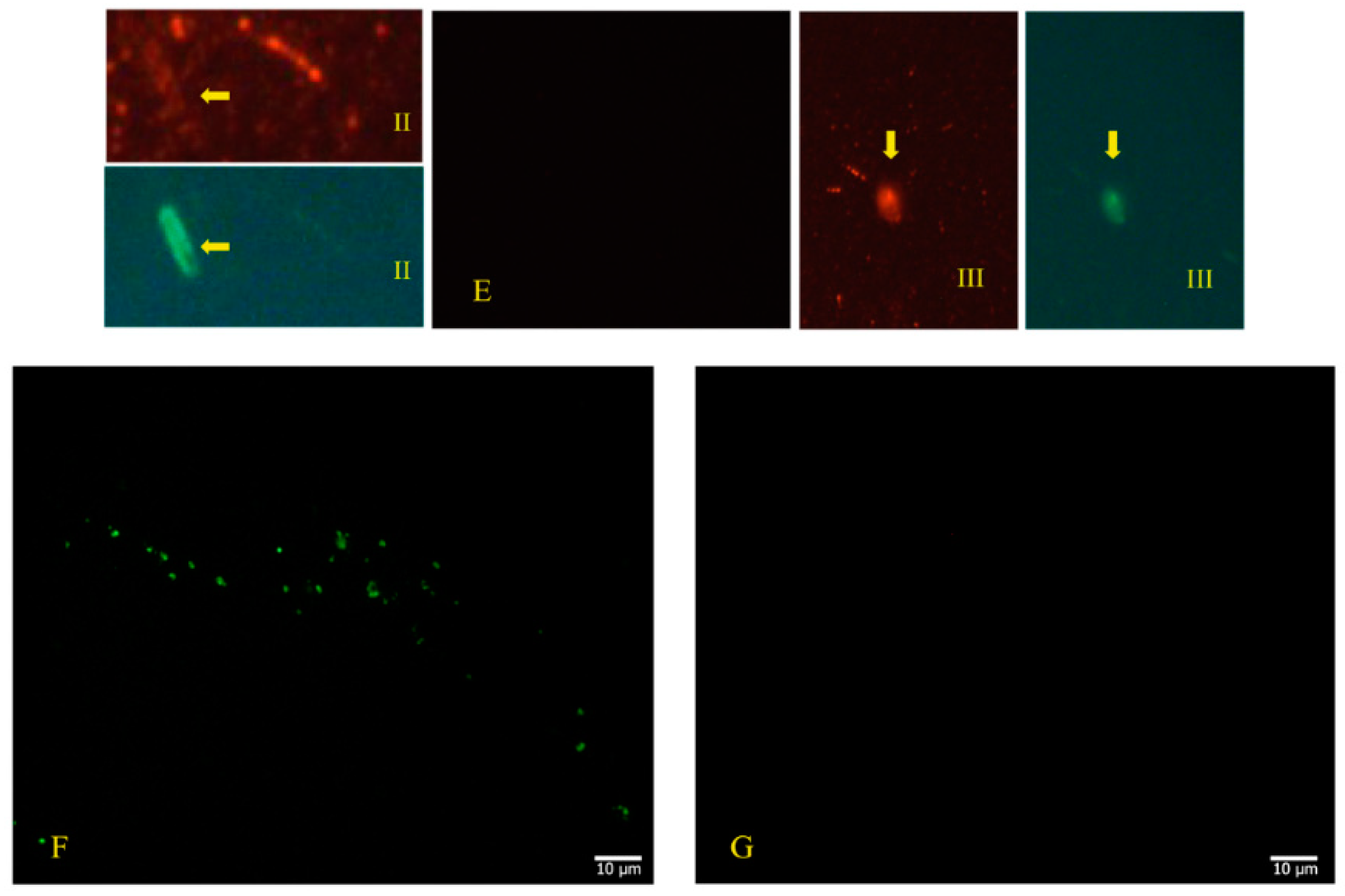
2.2. Use of Electron Microscopy and Immunofluorescence Microscopy as Confirmation of the Bacteriophage Aspect of SARS-CoV-2
2.3. Analysis and Exclusion of Other Bacteriophages Present in Bacterial Cultures
2.4. Evidence of Bacterial Lysis on a Petri Dish
2.5. The Use of Radioisotopes to Ultimately Confirm the Reclassification of RNA Viruses as Bacteriophages
3. Discussion
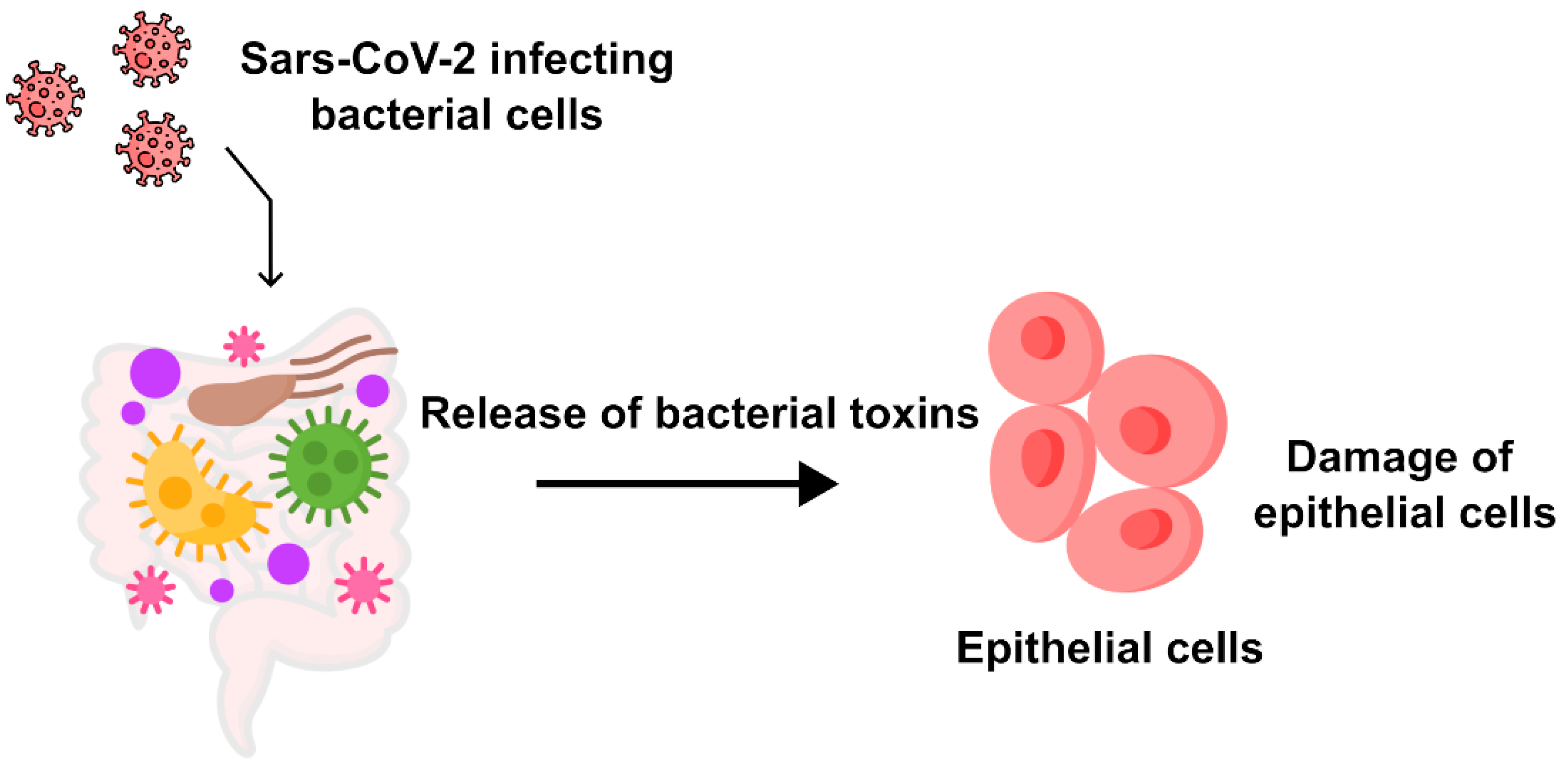
3.1. The Evaluation of the Integration of Microscopic, Genetic, and Proteomic Data on SARS-CoV-2 to Determine the Major Cells Infected
3.2. Considerations on the Importance of Radioisotope Testing as the Most Immediate Investigation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Number of COVID-19 Cases Reported to WHO. Available online: https://covid19.who.int/ (accessed on 5 May 2023).
- Wang, C.C.; Prather, K.A.; Sznitman, J.; Jimenez, J.L.; Lakdawala, S.S.; Tufekci, Z.; Marr, L.C. Airborne transmission of respiratory viruses. Science 2021, 373, eabd9149. [Google Scholar] [CrossRef] [PubMed]
- Leung, N.H.L. Transmissibility and transmission of respiratory viruses. Nat. Rev. Microbiol. 2021, 19, 528–545. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Tao, W.; Flavell, R.A.; Zhu, S. Potential intestinal infection and faecal-oral transmission of SARS-CoV-2. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 269–283. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Tao, W.; Flavell, R.A.; Zhu, S. Reply to: Rectally shed SARS-CoV-2 lacks infectivity: Time to rethink faecal-oral transmission? Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 669–670. [Google Scholar] [CrossRef]
- Cuicchi, D.; Lazzarotto, T.; Poggioli, G. Fecal-oral transmission of SARS-CoV-2: Review of laboratory-confirmed virus in gastrointestinal system. Int. J. Colorectal. Dis. 2021, 36, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.-H.; Ni, W.; Wu, Q.; Li, W.-J.; Li, G.-J.; Wang, W.-D.; Tong, J.-N.; Song, X.-F.; Wong, G.W.-K.; Xing, Q.-S. Prolonged viral shedding in feces of pediatric patients with coronavirus disease 2019. J. Microbiol. Immunol. Infect. 2020, 53, 473–480. [Google Scholar] [CrossRef]
- Pedersen, R.M.; Tornby, D.S.; Bang, L.L.; Madsen, L.W.; Skov, M.N.; Jensen, T.G.; Johansen, I.S.; Andersen, T.E. Rectally shed SARS-CoV-2 lacks infectivity: Time to rethink faecal–oral transmission? Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 669. [Google Scholar] [CrossRef]
- Li, H.; Li, B.; Liang, Q.; Jin, X.; Tang, L.; Ding, Q.; Wang, Z.; Wei, Z. Porcine deltacoronavirus infection alters bacterial communities in the colon and feces of neonatal piglets. Microbiologyopen 2020, 9, e1036. [Google Scholar] [CrossRef]
- Felten, S.; Klein-Richers, U.; Unterer, S.; Bergmann, M.; Leutenegger, C.M.; Pantchev, N.; Balzer, J.; Zablotski, Y.; Hofmann-Lehmann, R.; Hartmann, K. Role of Feline Coronavirus as Contributor to Diarrhea in Cats from Breeding Catteries. Viruses 2022, 14, 858. [Google Scholar] [CrossRef]
- Meazzi, S.; Stranieri, A.; Lauzi, S.; Bonsembiante, F.; Ferro, S.; Paltrinieri, S.; Giordano, A. Feline gut microbiota composition in association with feline coronavirus infection: A pilot study. Res. Vet. Sci. 2019, 125, 272–278. [Google Scholar] [CrossRef]
- Storz, J.; Lin, X.; Purdy, C.W.; Chouljenko, V.N.; Kousoulas, K.G.; Enright, F.M.; Gilmore, W.C.; Briggs, R.E.; Loan, R.W. Coronavirus and Pasteurella infections in bovine shipping fever pneumonia and Evans’ criteria for causation. J. Clin. Microbiol. 2000, 38, 3291–3298. [Google Scholar] [CrossRef]
- Saif, L.J. Bovine respiratory coronavirus. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 349–364. [Google Scholar] [CrossRef]
- Bhatnagar, R.; Johnson, G.R.; Christian, R.G. Electron microscopy of rapid identification of animal viruses in hematoxylin-eosin sections. Can. J. Comp. Med. 1977, 41, 416–419. [Google Scholar] [PubMed]
- Sabin, A.B. Problems in the natural history of poliomyelitis. Ann. Intern. Med. 1949, 30, 40–54. [Google Scholar] [CrossRef] [PubMed]
- Sabin, A.B. Present status of attenuated live virus poliomyelitis vaccine. Bull. N. Y. Acad. Med. 1957, 33, 17–39. [Google Scholar] [CrossRef] [PubMed]
- Howitt, B.F.; Barnett, V.H. Use of penicillin and streptomycin in the isolation of poliomyelitis virus from fecal specimens. J. Lab. Clin. Med. 1948, 33, 1402–1409. [Google Scholar] [PubMed]
- Tsunoda, A. Chemoprophylaxis of poliomyelitis in mice with quinomycin. (Studies on the antibiotic substances from Actinomycetes. XLVI). J. Antibiot. 1962, 15, 60–66. [Google Scholar]
- Acevedo, M.A.W.; Pfeiffer, J.K. Microbiota-independent antiviral effects of antibiotics on Poliovirus and coxsackievirus. Virology 2020, 546, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Tracking COVID-19 through Wastewater. Available online: https://covid19.nih.gov/news-and-stories/tracking-covid-19-through-wastewater-testing (accessed on 20 February 2024).
- Papajová, I.; Šmigová, J.; Gregová, G.; Šoltys, J.; Venglovský, J.; Papaj, J.; Szabóová, T.; Dančová, N.; Ihnacik, L.; Schusterová, I.; et al. Effect of Wastewater Treatment on Bacterial Community, Antibiotic-Resistant Bacteria and Endoparasites. Int. J. Environ. Res. Public Health 2022, 19, 2750. [Google Scholar] [CrossRef]
- Zhang, D.; Zhou, Y.; Ma, Y.; Chen, P.; Tang, J.; Yang, B.; Li, H.; Liang, M.; Xue, Y.; Liu, Y.; et al. Gut Microbiota Dysbiosis Correlates with Long COVID-19 at One-Year After Discharge. J. Korean Med. Sci. 2023, 38, e120. [Google Scholar] [CrossRef]
- Zollner, A.; Koch, R.; Jukic, A.; Pfister, A.; Meyer, M.; Rössler, A.; Kimpel, J.; Adolph, T.E.; Tilg, H. Postacute COVID-19 is Characterized by Gut Viral Antigen Persistence in Inflammatory Bowel Diseases. Gastroenterology 2022, 163, 495–506.e8. [Google Scholar] [CrossRef] [PubMed]
- Bernard-Raichon, L.; Venzon, M.; Klein, J.; Axelrad, J.E.; Zhang, C.; Sullivan, A.P.; Hussey, G.A.; Casanovas-Massana, A.; Noval, M.G.; Valero-Jimenez, A.M.; et al. Gut microbiome dysbiosis in antibiotic-treated COVID-19 patients is associated with microbial translocation and bacteremia. Nat. Commun. 2022, 13, 5926. [Google Scholar] [CrossRef]
- Alexandersen, S.; Chamings, A.; Bhatta, T. SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. Nat. Commun. 2020, 11, 6059. [Google Scholar] [CrossRef] [PubMed]
- Seah, I.; Agrawal, R. Can the Coronavirus Disease 2019 (COVID-19) Affect the Eyes? A Review of Coronaviruses and Ocular Implications in Humans and Animals. Ocul. Immunol. Inflamm. 2020, 28, 391–395. [Google Scholar] [CrossRef] [PubMed]
- Douglas, K.A.; Douglas, V.P.; Moschos, M.M. Ocular Manifestations of COVID-19 (SARS-CoV-2): A Critical Review of Current Literature. In Vivo 2020, 34, 1619–1628. [Google Scholar] [CrossRef]
- Bostanci, C.B.; Ozates, S. Ocular manifestations of coronavirus disease 2019. Graefes Arch. Clin. Exp. Ophthalmol. 2020, 258, 1959–1963. [Google Scholar] [CrossRef]
- Vabret, A.; Mourez, T.; Dina, J.; van der Hoek, L.; Gouarin, S.; Petitjean, J.; Brouard, J.; Freymuth, F. Human coronavirus NL63, France. Emerg. Infect. Dis. 2005, 11, 1225–1229. [Google Scholar] [CrossRef]
- Petrillo, M.; Brogna, C.; Cristoni, S.; Querci, M.; Piazza, O.; Eede, G.V.D. Increase of SARS-CoV-2 RNA load in faecal samples prompts for rethinking of SARS-CoV-2 biology and COVID-19 epidemiology. F1000Research 2021, 10, 370. [Google Scholar] [CrossRef]
- Dunbar, S.A. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin. Chim. Acta 2006, 363, 71–82. [Google Scholar] [CrossRef]
- Floridia, M.; Cristoni, S. A powerful platform for instrument calibration and quantification. Rapid. Commun. Mass Spectrom. 2014, 28, 536–544. [Google Scholar] [CrossRef]
- Lehman, G.J.; McGill, S.M. The importance of normalization in interpreting surface electromyography: A proof of principle. J. Manip. Physiol. Ther. 1999, 22, 444–446. [Google Scholar] [CrossRef] [PubMed]
- Bandiera, S.; Lebas, A.; Canizares-Martinello, L.; Guinchard, F.; Lyonnais, C.; Perrin, S.; Nicolas, M.; Uhlrich, S.; Chabaud-Riou, M. A single immunogenicity assay for testing potency of combination DTaP vaccines: Simultaneous quantitation of anti-DT, anti-TT, anti-PTxd and anti-FHA antibodies in Guinea-pig serum with a Luminex®-xMAP® bead-based serological assay. Biologicals 2019, 61, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Brogna, C.; Cristoni, S.; Petrillo, M.; Querci, M.; Piazza, O.; Eede, G.V.D. Toxin-like peptides in plasma, urine and faecal samples from COVID-19 patients. F1000Research 2021, 10, 550. [Google Scholar] [CrossRef]
- Brogna, C.; Cristoni, S.; Brogna, B.; Bisaccia, D.R.; Marino, G.; Viduto, V.; Montano, L.; Piscopo, M. Toxin-like Peptides from the Bacterial Cultures Derived from Gut Microbiome Infected by SARS-CoV-2-New Data for a Possible Role in the Long COVID Pattern. Biomedicines 2022, 11, 87. [Google Scholar] [CrossRef] [PubMed]
- Pistollato, F.; Petrillo, M.; Clerbaux, L.-A.; Leoni, G.; Ponti, J.; Bogni, A.; Brogna, C.; Cristoni, S.; Sanges, R.; Gyves, E.M.-D.; et al. Effects of spike protein and toxin-like peptides found in COVID-19 patients on human 3D neuronal/glial model undergoing differentiation: Possible implications for SARS-CoV-2 impact on brain development. Reprod. Toxicol. 2022, 111, 34–48. [Google Scholar] [CrossRef]
- Brogna, C.; Brogna, B.; Bisaccia, D.R.; Lauritano, F.; Marino, G.; Montano, L.; Cristoni, S.; Prisco, M.; Piscopo, M. Could SARS-CoV-2 Have Bacteriophage Behavior or Induce the Activity of Other Bacteriophages? Vaccines 2022, 10, 708. [Google Scholar] [CrossRef]
- Brogna, C.; Costanzo, V.; Brogna, B.; Bisaccia, D.R.; Brogna, G.; Giuliano, M.; Montano, L.; Viduto, V.; Cristoni, S.; Fabrowski, M.; et al. Analysis of Bacteriophage Behavior of a Human RNA Virus, SARS-CoV-2, through the Integrated Approach of Immunofluorescence Microscopy, Proteomics and D-Amino Acid Quantification. Int. J. Mol. Sci. 2023, 24, 3929. [Google Scholar] [CrossRef]
- Petrillo, M.; Querci, M.; Brogna, C.; Ponti, J.; Cristoni, S.; Markov, P.V.; Valsesia, A.; Leoni, G.; Benedetti, A.; Wiss, T.; et al. Evidence of SARS-CoV-2 bacteriophage potential in human gut microbiota [version 1; peer review: 1 approved with reservations]. F1000Research 2022, 11, 292. [Google Scholar] [CrossRef]
- Brogna, C.; Cristoni, S. A new absolute quantitative method for peptide and metabolite detection. J. Mass Spectrom. 2023, 59, e4991. [Google Scholar] [CrossRef]
- Kohda, K.; Li, X.; Soga, N.; Nagura, R.; Duerna, T.; Nakajima, S.; Nakagawa, I.; Ito, M.; Ikeuchi, A. An In Vitro Mixed Infection Model with Commensal and Pathogenic Staphylococci for the Exploration of Interspecific Interactions and Their Impacts on Skin Physiology. Front. Cell Infect. Microbiol. 2021, 11, 712360. [Google Scholar] [CrossRef]
- Kameli, N.; Borman, R.; Lpez-Iglesias, C.; Savelkoul, P.; Stassen, F.R.M. Characterization of Feces-Derived Bacterial Membrane Vesicles and the Impact of Their Origin on the Inflammatory Response. Front. Cell Infect. Microbiol. 2021, 11, 667987. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Ni, C.; Gao, R.; Wang, Y.; Yang, L.; Wei, J.; Lv, T.; Liang, J.; Zhang, Q.; Xu, W. Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell 2020, 11, 771–775. [Google Scholar] [CrossRef] [PubMed]
- Cristoni, S.; Rubini, S.; Bernardi, L.R. Development and applications of surface-activated chemical ionization. Mass Spectrom. Rev. 2007, 26, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yu, L.; Steill, J.D.; Oomens, J.; Polfer, N.C. Effect of peptide fragment size on the propensity of cyclization in collision-induced dissociation: Oligoglycine b(2)-b(8). J. Am. Chem. Soc. 2009, 131, 18272–18282. [Google Scholar] [CrossRef] [PubMed]
- Madama, S.; Falletta, E.; Malvandi, A.M.; Arzoni, K.; Brogna, C.; Varelli, M.; Bertelli, M.; Conti, M.; Larini, M.; Guidugli, F.; et al. q value and parent energy optimization using a low-voltage ionization approach increases resolution in linear ion trap mass spectrometry. J. Mass Spectrom. 2022, 57, e4876. [Google Scholar] [CrossRef] [PubMed]
- Alluwaimi, A.M.; Alshubaith, I.H.; Al-Ali, A.M.; Abohelaika, S. The Coronaviruses of Animals and Birds: Their Zoonosis, Vaccines, and Models for SARS-CoV and SARS-CoV2. Front. Vet. Sci. 2020, 7, 582287. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, M.J.; Kory, R.C.; Siegesmund, K.A.; Pedersen, H.J.; Sullivan, E.J.; Peckinpaugh, R.O. Electron microscope methods for the identification of adenoviruses isolated in micro tissue cultures. Appl. Microbiol. 1972, 23, 141–144. [Google Scholar] [CrossRef]
- Chen, Q.; Li, G.; Stasko, J.; Thomas, J.T.; Stensland, W.R.; Pillatzki, A.E.; Gauger, P.C.; Schwartz, K.J.; Madson, D.; Yoon, K.-J.; et al. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 2014, 52, 234–243. [Google Scholar] [CrossRef]
- Schweer, W.P.; Schwartz, K.; Burrough, E.R.; Yoon, K.J.; Sparks, J.C.; Gabler, N.K. The effect of porcine reproductive and respiratory syndrome virus and porcine epidemic diarrhea virus challenge on growing pigs I: Growth performance and digestibility. J. Anim. Sci. 2016, 94, 514–522. [Google Scholar] [CrossRef]
- Pizuorno, A.; Brim, H.; Ashktorab, H. Gastrointestinal manifestations and SARS-CoV-2 infection. Curr. Opin. Pharmacol. 2021, 61, 114–119. [Google Scholar] [CrossRef]
- Is Conjunctivitis a ‘New’ COVID Symptom? Here’s What Experts and Studies Say|5 Points. Available online: https://www.cnbctv18.com/healthcare/covid-new-symptom-variant-arcturus-omicron-conjunctivitis-itchy-eyes-children-16399051.htm (accessed on 20 February 2024).
- Doughri, A.M.; Storz, J.; Hajer, I.; Fernando, H.S. Morphology and morphogenesis of a coronavirus infecting intestinal epithelial cells of newborn calves. Exp. Mol. Pathol. 1976, 25, 355–370. [Google Scholar] [CrossRef] [PubMed]
- Young, K.D. Bacterial morphology: Why have different shapes? Curr. Opin. Microbiol. 2007, 10, 596–600. [Google Scholar] [CrossRef] [PubMed]
- Calomeni, E.; Satoskar, A.; Ayoub, I.; Brodsky, S.; Rovin, B.H.; Nadasdy, T. Multivesicular bodies mimicking SARS-CoV-2 in patients without COVID-19. Kidney Int. 2020, 98, 233–234. [Google Scholar] [CrossRef] [PubMed]
- Mihelc, E.M.; Baker, S.C.; Lanman, J.K. Coronavirus infection induces progressive restructuring of the endoplasmic reticulum involving the formation and degradation of double membrane vesicles. Virology 2021, 556, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Ryter, A. Contribution of electron microscopy in the study of the interactions between pathogenic bacteria and their host cells. J. Struct. Biol. 1990, 104, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Brieger, E.M.; Crowe, G.R.; Cosslett, V.E. Electron microscopy of bacteria. Nature 1947, 160, 864. [Google Scholar] [CrossRef] [PubMed]
- McFarlane, A.S. Electron microscopy of bacteria and viruses. Br. Med. J. 1949, 2, 1247–1250. [Google Scholar] [CrossRef]
- Baker, S.J.; Mathan, M.; Mathan, V.I.; Jesudoss, S.; Swaminathan, S.P. Chronic enterocyte infection with coronavirus. One possible cause of the syndrome of tropical sprue? Dig. Dis. Sci. 1982, 27, 1039–1043. [Google Scholar] [CrossRef]
- Mathan, M.; Swaminathan, S.; Mathan, V.; Yesudoss, S.; Baker, S. Pleomorphic virus-like particles in human faeces. Lancet 1975, 305, 1068–1069. [Google Scholar] [CrossRef]
- Orenstein, J.M.; Banach, B.S.; Baker, S.C. Morphogenesis of Coronavirus HCoV-NL63 in Cell Culture: A Transmission Electron Microscopic Study. Open Infect. Dis. J. 2009, 2, 52–58. [Google Scholar] [CrossRef]
- Bullock, H.A.; Goldsmith, C.S.; Miller, S.E. Best practices for correctly identifying coronavirus by transmission electron microscopy. Kidney Int. 2021, 99, 824–827. [Google Scholar] [CrossRef]
- Li, M.; Ren, Y.; Aw, Z.Q.; Chen, B.; Yang, Z.; Lei, Y.; Cheng, L.; Liang, Q.; Hong, J.; Yang, Y.; et al. Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses. Nat. Commun. 2022, 13, 7957. [Google Scholar] [CrossRef]
- Shang, C.; Liu, Z.; Zhu, Y.; Lu, J.; Ge, C.; Zhang, C.; Li, N.; Jin, N.; Li, Y.; Tian, M.; et al. SARS-CoV-2 Causes Mitochondrial Dysfunction and Mitophagy Impairment. Front. Microbiol. 2022, 12, 780768. [Google Scholar] [CrossRef]
- Goldsmith, C.S.; Tatti, K.M.; Ksiazek, T.G.; Rollin, P.E.; Comer, J.A.; Lee, W.W.; Rota, P.A.; Bankamp, B.; Bellini, W.J.; Zaki, S.R. Ultrastructural Characterization of SARS Coronavirus. Emerg. Infect. Dis. 2004, 10, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Sars Nucleocapsid Protein Antibody. Available online: https://www.rockland.com/categories/primary-antibodies/sars-nucleocapsid-protein-antibody-200-401-A50/#productReferenceSectionWrapper (accessed on 20 February 2024).
- Available online: https://www.genetex.com/Product/Detail/Gram-Positive-Bacteria-antibody-BDI380/GTX42630 (accessed on 20 February 2024).
- Du, R. Mutual exclusion between related phages. J. Bacteriol. 1952, 63, 209–217. [Google Scholar] [CrossRef]
- Szeberényi, J. The Meselson-Stahl experiment. Biochem. Mol. Biol. Educ. 2012, 40, 209–211. [Google Scholar] [CrossRef] [PubMed]


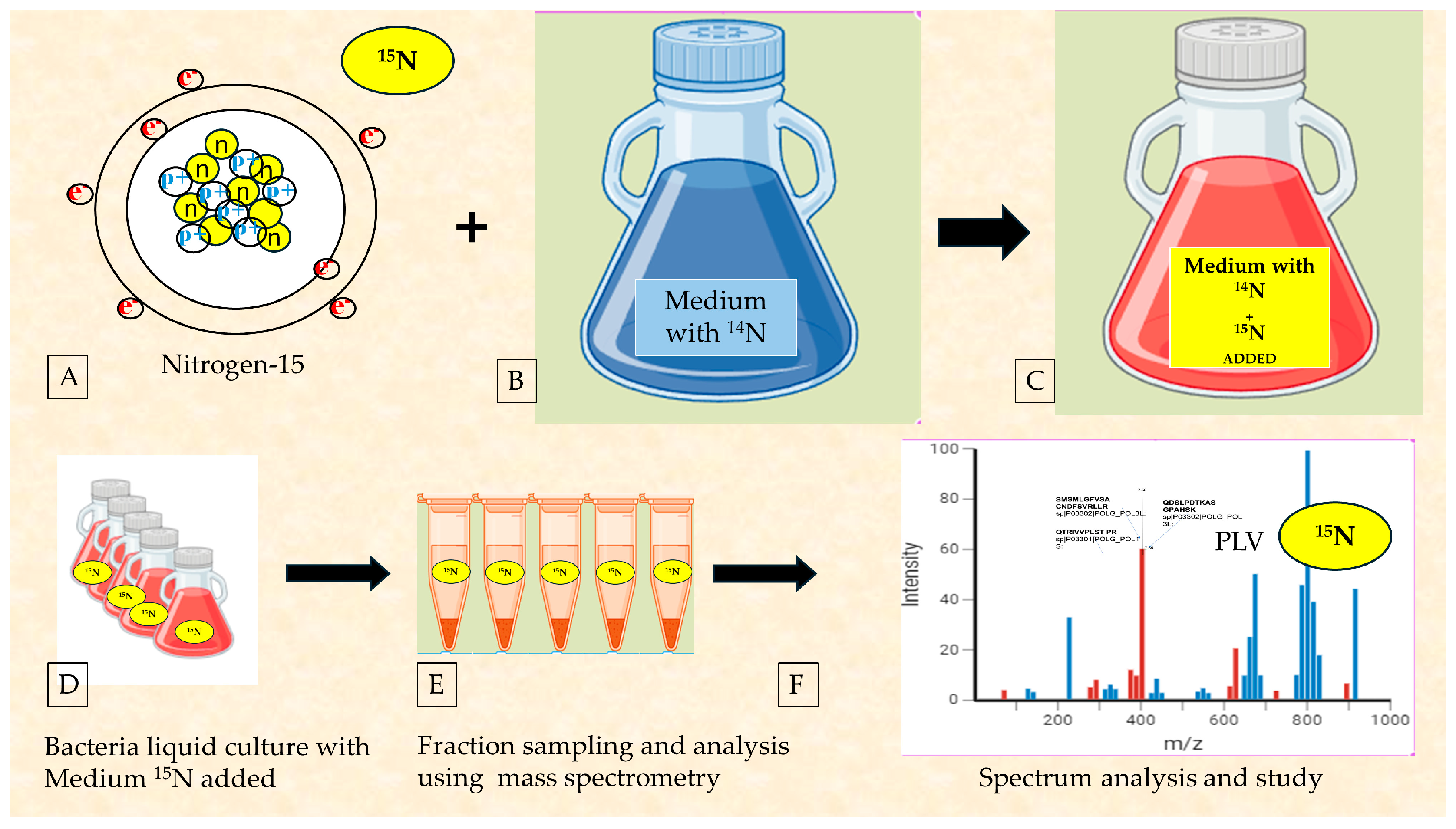
| Group | N° Volunteers | N° Excreted Virus | N° of Days Each Excreted Virus | Peak Virus Titers in Stool Log10, TCD50, per Gram |
|---|---|---|---|---|
| No antibody | 11 | 11 | 10, 10, 10, 21+ *, | 3.7, 3.7, 3.7, 3.7, 3.7, 3.7 |
| 25+, 26+, 26+, 41, 77 | 4.2, 4.2, 4.2, 4.2, 4.2 | |||
| Antibody after Salk vaccine | 9, 10, 10, 13, 14, | 2.7, 3.2, 3.4, 3.7 | ||
| 8 | 8 | 21+, 26+, 42 | 4.2, 4.2, 4.7, 4.7 | |
| Naturally acquired antibody | ||||
| 8 | 1 | 10 | 4.2 |
| Putative Bacteriophage Sequences | Size and Form | Tail Yes/No |
|---|---|---|
| BK010646.1 TPA_exp: Bacteroides phage p00 | 50 to 70 nm in diameter and most tail lengths ranged from 120 to 200 nm | Yes |
| LR596903.1 Roseburia phage Shimadzu | Similar to a Syphoviridae | Yes |
| MT121960.1 Phage DP SC_2_H4_2017 DNA polymerase 1 and recombinase A genes, complete cds | Diameter. 75 nm, while the tail length is 125 nm | Yes |
| NC_024711.1 Uncultured crAssphage | Icosahedral capsids with short tails (type I, with head diameters of 76.5 nm and short tails; and type II, with a similar head size but head-tail collar structures and slightly longer tails) | Yes |
| NC_027980.1 Enterobacter phage phiKDA1, | Family T1 phage | Yes |
| NC_047910.1 Faecalibacterium phage FP_Epona | Phage with contractile tails | Yes |
| NC_047916.1 Faecalibacterium phage FP_oengus | Phage with long non- contractile tails | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brogna, C.; Bisaccia, D.R.; Costanzo, V.; Lettieri, G.; Montano, L.; Viduto, V.; Fabrowski, M.; Cristoni, S.; Prisco, M.; Piscopo, M. Who Is the Intermediate Host of RNA Viruses? A Study Focusing on SARS-CoV-2 and Poliovirus. Microorganisms 2024, 12, 643. https://doi.org/10.3390/microorganisms12040643
Brogna C, Bisaccia DR, Costanzo V, Lettieri G, Montano L, Viduto V, Fabrowski M, Cristoni S, Prisco M, Piscopo M. Who Is the Intermediate Host of RNA Viruses? A Study Focusing on SARS-CoV-2 and Poliovirus. Microorganisms. 2024; 12(4):643. https://doi.org/10.3390/microorganisms12040643
Chicago/Turabian StyleBrogna, Carlo, Domenico Rocco Bisaccia, Vincenzo Costanzo, Gennaro Lettieri, Luigi Montano, Valentina Viduto, Mark Fabrowski, Simone Cristoni, Marina Prisco, and Marina Piscopo. 2024. "Who Is the Intermediate Host of RNA Viruses? A Study Focusing on SARS-CoV-2 and Poliovirus" Microorganisms 12, no. 4: 643. https://doi.org/10.3390/microorganisms12040643
APA StyleBrogna, C., Bisaccia, D. R., Costanzo, V., Lettieri, G., Montano, L., Viduto, V., Fabrowski, M., Cristoni, S., Prisco, M., & Piscopo, M. (2024). Who Is the Intermediate Host of RNA Viruses? A Study Focusing on SARS-CoV-2 and Poliovirus. Microorganisms, 12(4), 643. https://doi.org/10.3390/microorganisms12040643










