Ruminal Microbiome Differences in Angus Steers with Differing Feed Efficiencies during the Feedlot Finishing Phase
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Experimental Design, and Management
2.2. Sample Collection, DNA Extraction and Sequencing
2.3. Volatile Fatty Acid Determination
2.4. Sequencing Data
2.5. Statistical Analyses
3. Results
3.1. Steer Growth and Efficiency Performance
3.2. Volatile Fatty Acid Concentrations
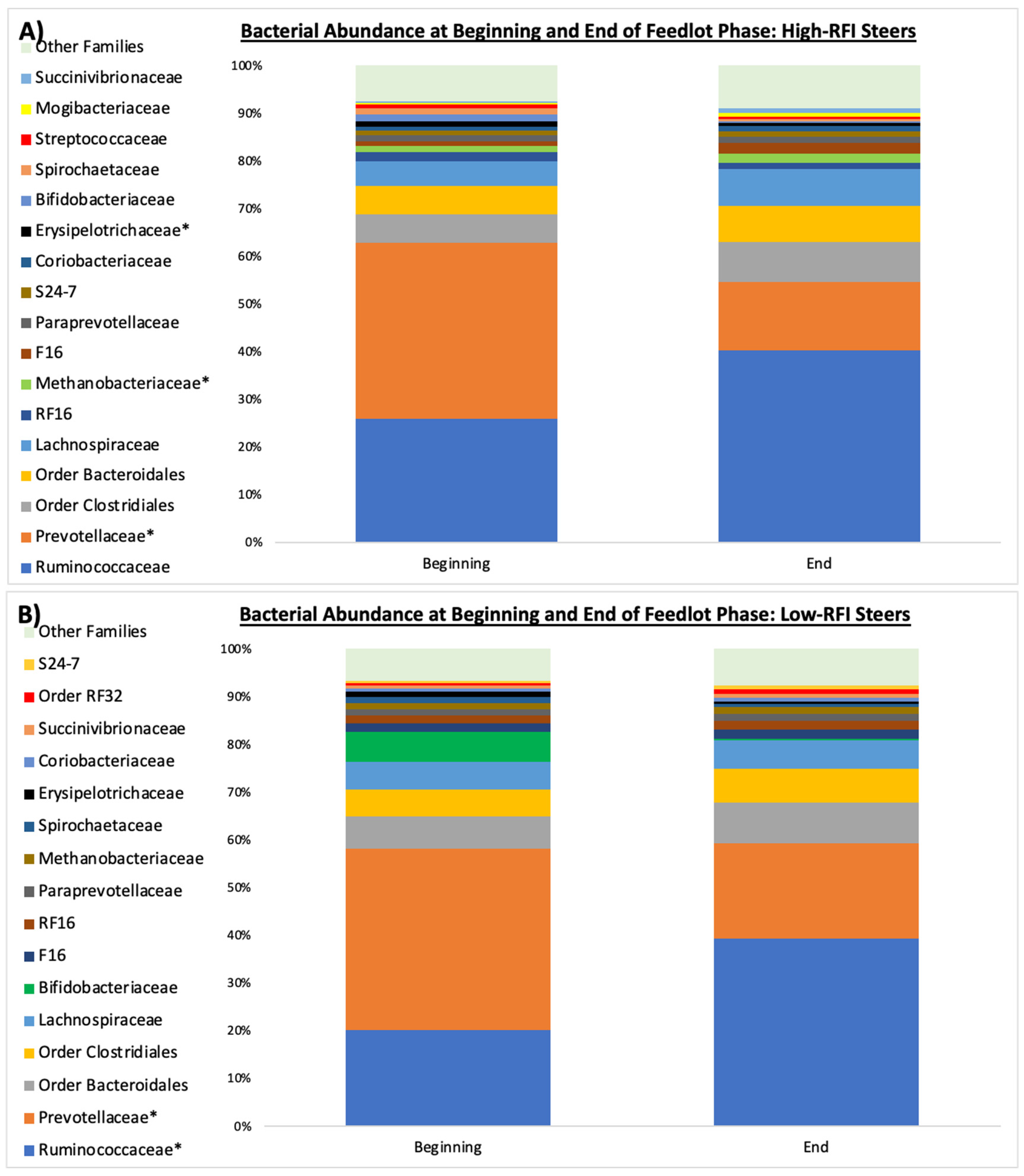
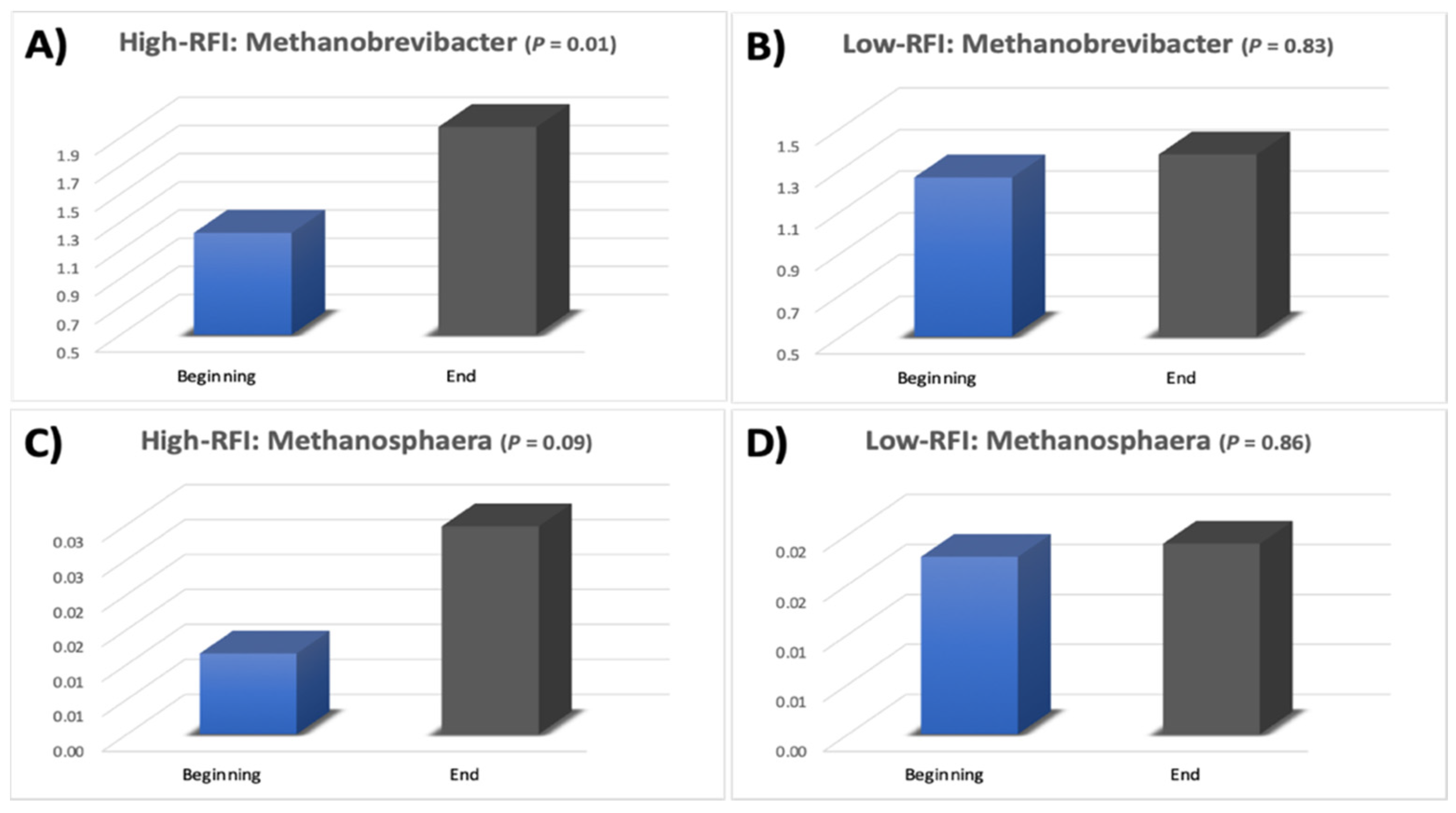
3.3. Microbial Community Structure
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hungate, R.E. CHAPTER V—The Rumen as a Continuous Fermentation System. In The Rumen and Its Microbes; Hungate, R.E., Ed.; Academic Press: Cambridge, MA, USA, 1966; pp. 206–244. [Google Scholar]
- Hungate, R.E. Studies on cellulose fermentation. I. The culture and physiology of an anaerobic cellulose-digesting bacterium. J. Bact. 1944, 48, 499–512. [Google Scholar] [CrossRef]
- Hungate, R.E. The anaerobic mesophilic cellulolytic bacteria. Bact. Rev. 1950, 14, 1–49. [Google Scholar] [CrossRef]
- Russell, J.B.; Rychlik, J.L. Factors that alter rumen microbial ecology. Science 2001, 292, 1119–1122. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.A.; Johnson, D.E. Methane emissions from cattle. J. Anim. Sci. 1995, 73, 2483–2492. [Google Scholar] [CrossRef]
- Ellis, J.L.; Dijkstra, J.; Kebreab, E.; Bannink, A.; Odongo, N.E.; McBride, B.W.; France, J. Aspects of rumen microbiology central to mechanistic modelling of methane production in cattle. J. Agric. Sci. 2008, 146, 213–233. [Google Scholar] [CrossRef]
- Bowen, J.M.; Cormican, P.; Lister, S.J.; McCabe, M.S.; Duthie, C.-A.; Roehe, R.; Dewhurst, R.J. Links between the rumen microbiota, methane emissions and feed efficiency of finishing steers offered dietary lipid and nitrate supplementation. PLoS ONE 2020, 15, e0231759. [Google Scholar] [CrossRef]
- Lancaster, P.A.; Carstens, G.E.; Crews, D.H.; Welsh, T.H.; Forbes, T.D.A.; Forrest, D.W.; Tedeschi, L.O.; Randel, R.D.; Rouquette, F.M. Phenotypic and genetic relationships of residual feed intake with performance and ultrasound carcass traits in Brangus heifers1. J. Anim. Sci. 2009, 87, 3887–3896. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.A. Feed Efficiency in the Beef Industry; John Wiley & Sons: Hoboken, NJ, USA, 2012. [Google Scholar]
- USDA. Cattle and Beef: Sector at a Glance. 2019. Available online: https://www.ers.usda.gov/topics/animal-products/cattle-beef/ (accessed on 22 February 2024).
- Huntington, G.B. Starch utilization by ruminants: From basics to the bunk. J. Anim. Sci. 1997, 75, 852. [Google Scholar] [CrossRef]
- Owens, F.N.; Secrist, D.S.; Hill, W.J.; Gill, D.R. Acidosis in cattle: A review. J. Anim. Sci. 1998, 76, 275–286. [Google Scholar] [CrossRef]
- Nocek, J.E.; Tamminga, S. Site of Digestion of Starch in the Gastrointestinal Tract of Dairy Cows and Its Effect on Milk Yield and Composition. J. Dariy Sci. 1991, 74, 3598–3629. [Google Scholar] [CrossRef]
- Nkrumah, J.D.; Okine, E.K.; Mathison, G.W.; Schmid, K.; Li, C.; Basarab, J.A.; Price, M.A.; Wang, Z.; Moore, S.S. Relationships of feedlot feed efficiency, performance, and feeding behavior with metabolic rate, methane production, and energy partitioning in beef cattle1. J. Anim. Sci. 2006, 84, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Sanabria, E.; Goonewardene, L.A.; Wang, Z.; Durunna, O.N.; Moore, S.S.; Guan, L.L. Impact of Feed Efficiency and Diet on Adaptive Variations in the Bacterial Community in the Rumen Fluid of Cattle. Appl. Environ. Microbiol. 2012, 78, 1203–1214. [Google Scholar] [CrossRef] [PubMed]
- Shabat, S.K.B.; Sasson, G.; Doron-Faigenboim, A.; Durman, T.; Yaacoby, S.; Miller, M.E.B.; A White, B.; Shterzer, N.; Mizrahi, I. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 2016, 10, 2958–2972. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra-Hijar, G.; Abo-Ismail, M.; Carstens, G.E.; Guan, L.L.; Hegarty, R.; Kenny, D.A.; McGee, M.; Plastow, G.; Relling, A.; Ortigues-Marty, I. Review: Biological determinants of between-animal variation in feed efficiency of growing beef cattle. Animal 2018, 12, s321–s335. [Google Scholar] [CrossRef] [PubMed]
- North, S.; Bowman, B.; American Angus Association. By the Numbers. 2010. Available online: http://www.angus.org/nce/documents/bythenumbersradg.pdf (accessed on 22 February 2024).
- Association, A.A. General Minimum Guidelines for Recording Individual Feed Intake in Growing Bulls, Steer and Heifer Progeny. 2015. Available online: http://www.angus.org/Performance/Documents/FeedIntakeGuidelines.pdf (accessed on 22 February 2024).
- Detweiler, R.A.; Pringle, T.D.; Rekaya, R.; Wells, J.B.; Segers, J.R. The impact of selection using residual average daily gain and marbling EPDs on growth, performance, and carcass traits in Angus steers1. J. Anim. Sci. 2019, 97, 2450–2459. [Google Scholar] [CrossRef] [PubMed]
- National Research Council. Nutrient Requirements of Beef Cattle, 7th ed.; Update 2000; The National Academies Press: Washington, DC, USA, 2000. [Google Scholar]
- Wang, Z.; Nkrumah, J.D.; Li, C.; Basarab, J.A.; Goonewardene, L.A.; Okine, E.K.; Crews, D.H.; Moore, S.S. Test duration for growth, feed intake, and feed efficiency in beef cattle using the GrowSafe System. J. Anim. Sci. 2006, 84, 2289–2298. [Google Scholar] [CrossRef]
- Mendes, E.D.M.; Carstens, G.E.; Tedeschi, L.O.; Pinchak, W.E.; Friend, T.H. Validation of a system for monitoring feeding behavior in beef cattle. J. Anim. Sci. 2011, 89, 2904–2910. [Google Scholar] [CrossRef]
- DeVries, T.J.; Von Keyserlingk, M.A.G.; Weary, D.M.; Beauchemin, K.A. Validation of a system for monitoring feeding behavior of dairy cows. J. Dairy. Sci. 2003, 86, 3571–3574. [Google Scholar] [CrossRef]
- Lourenco, J.M.; Callaway, T.R.; Kieran, T.J.; Glenn, T.C.; McCann, J.C.; Stewart, R.L. Analysis of the Rumen Microbiota of Beef Calves Supplemented During the Suckling Phase. Front. Microbiol. 2019, 10, 1131. [Google Scholar] [CrossRef]
- Rothrock, M.J., Jr.; Hiett, K.L.; Gamble, J.; Caudill, A.C.; Cicconi-Hogan, K.M.; Caporaso, J.G. A hybrid DNA extraction method for the qualitative and quantitative assessment of bacterial communities from poultry production samples. J. Vis. Exp. 2014, 94, e52161. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Lourenco, J.M.; Kieran, T.J.; Seidel, D.S.; Glenn, T.C.; Silveira, M.F.D.; Callaway, T.R.; Stewart, R.L., Jr. Comparison of the ruminal and fecal microbiotas in beef calves supplemented or not with concentrate. PLoS ONE 2020, 15, e0231533. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Benchaar, C.; Pomar, C.; Chiquette, J. Evaluation of dietary strategies to reduce methane production in ruminants: A modelling approach. Can. J. Anim. Sci. 2001, 81, 563–574. [Google Scholar] [CrossRef]
- Ferraretto, L.F.; Crump, P.; Shaver, R. Effect of cereal grain type and corn grain harvesting and processing methods on intake, digestion, and milk production by dairy cows through a meta-analysis. J. Dairy. Sci. 2013, 96, 533–550. [Google Scholar] [CrossRef] [PubMed]
- Elolimy, A.A.; Abdelmegeid, M.K.; McCann, J.C.; Shike, D.W.; Loor, J.J. Residual feed intake in beef cattle and its association with carcass traits, ruminal solid-fraction bacteria, and epithelium gene expression. J. Anim. Sci. Biotechnol. 2018, 9, 67. [Google Scholar] [CrossRef] [PubMed]
- Lourenco, J.M.; Welch, C.B. Using microbiome information to understand and improve animal performance. Ital. J. Anim. Sci. 2022, 21, 899–913. [Google Scholar] [CrossRef]
- Holmes, D.E.; Smith, J.A. Biologically Produced Methane as a Renewable Energy Source; Elsevier: Amsterdam, The Netherlands, 2016; pp. 1–61. [Google Scholar]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027. [Google Scholar] [CrossRef]
- McCann, J.C. New Perspectives on Adapting Cattle to Finishing Diets Without Compromising Rumen Health. In Proceedings of the 30th Annual Florida Ruminant Nutrition Symposium, Gainesville, FL, USA, 5–7 February 2018; pp. 61–66. [Google Scholar]
- Mackie, R.I.; Gilchrist, F.M. Changes in Lactate-Producing and Lactate-Utilizing Bacteria in Relation to pH in the Rumen of Sheep During Stepwise Adaptation to a High-Concentrate Diet. Appl. Environ. Microbiol. 1979, 38, 422–430. [Google Scholar] [CrossRef]
- Slyter, L.L. Ability of pH-Selected Mixed Ruminal Microbial Populations to Digest Fiber at Various pHs. Appl. Environ. Microbiol. 1986, 52, 390–391. [Google Scholar] [CrossRef]
- Goad, D.W.; Goad, C.L.; Nagaraja, T.G. Ruminal microbial and fermentative changes associated with experimentally induced subacute acidosis in steers. J. Anim. Sci. 1998, 76, 234–241. [Google Scholar] [CrossRef]
- Leschine, S.B. Cellulose degradation in anaerobic environments. Annu. Rev. Microbiol. 1995, 49, 399–426. [Google Scholar] [CrossRef] [PubMed]
- Rainey, F. Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Cotta, M.A. Interaction of ruminal bacteria in the production and utilization of maltooligosaccharides from starch. Appl. Environ. Microbiol. 1992, 58, 48–54. [Google Scholar] [CrossRef]
- Matsui, H.; Ogata, K.; Tajima, K.; Nakamura, M.; Nagamine, T.; Aminov, R.I.; Benno, Y. Phenotypic characterization of polysaccharidases produced by four Prevotella type strains. Curr. Microbiol 2000, 41, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Kittelmann, S.; Pinares-Patiño, C.S.; Seedorf, H.; Kirk, M.R.; Ganesh, S.; McEwan, J.C.; Janssen, P.H. Two different bacterial community types are linked with the low-methane emission trait in sheep. PLoS ONE 2014, 9, e103171. [Google Scholar] [CrossRef]
- Morita, H.; Shiratori, C.; Murakami, M.; Takami, H.; Toh, H.; Kato, Y.; Nakajima, F.; Takagi, M.; Akita, H.; Masaoka, T.; et al. Sharpea azabuensis gen. nov., sp. nov., a Gram-positive, strictly anaerobic bacterium isolated from the faeces of thoroughbred horses. Int. J. Syst. Evol. Microbiol. 2008, 58, 2682–2686. [Google Scholar] [CrossRef] [PubMed]
- Kamke, J.; Kittelmann, S.; Soni, P.; Li, Y.; Tavendale, M.; Ganesh, S.; Janssen, P.H.; Shi, W.; Froula, J.; Rubin, E.M.; et al. Rumen metagenome and metatranscriptome analyses of low methane yield sheep reveals a Sharpea-enriched microbiome characterised by lactic acid formation and utilisation. Microbiome 2016, 4, 56. [Google Scholar] [CrossRef]
- van Houtert, M.F.J. The production and metabolism of volatile fatty acids by ruminants fed roughages: A review. Anim. Feed. Sci. Technol. 1993, 43, 189–225. [Google Scholar] [CrossRef]
- Russell, J.B. Rumen Microbiology and Its Role in Ruminant Nutrition; Cornell University: Ithaca, NY, USA, 2002. [Google Scholar]
- Arndt, C.; Powell, J.; Aguerre, M.; Crump, P.; Wattiaux, M. Feed conversion efficiency in dairy cows: Repeatability, variation in digestion and metabolism of energy and nitrogen, and ruminal methanogens. J. Dairy. Sci. 2015, 98, 3938–3950. [Google Scholar] [CrossRef]
- Lourenço, J.M.; Froetschel, M.A.; Segers, J.R.; Tucker, J.J.; Stewart, R.L., Jr. Utilization of canola and sunflower meals as replacements for soybean meal in a corn silage-based stocker system. Transl. Anim. Sci. 2017, 1, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.G.; Coleman, G.S. The rumen protozoa. In The Rumen Microbial Ecosystem; Hobson, P.N., Stewart, C.S., Eds.; Springer: Berlin/Heidelberg, Germany, 1997; pp. 73–139. [Google Scholar]
- Hino, T.; Russell, J.B. Effect of reducing-equivalent disposal and NADH/NAD on deamination of amino acids by intact rumen microorganisms and their cell extracts. Appl. Environ. Microbiol. 1985, 50, 1368–1374. [Google Scholar] [CrossRef]
- Rothschild, L.J.; Mancinelli, R.L. Life in extreme environments. Nature 2001, 409, 1092–1101. [Google Scholar] [CrossRef]
- Bryant, M.P.; Wolin, E.A.; Wolin, M.J.; Wolfe, R.S. Methanobacillus omelianskii, a symbiotic association of two species of bacteria. Arch. Mikrobiol. 1967, 59, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Iannotti, E.L.; Kafkewitz, D.; Wolin, M.J.; Bryant, M.P. Glucose fermentation products of Ruminococcus albus grown in continuous culture with Vibrio succinogenes: Changes caused by interspecies transfer of H2. J. Bacteriol. 1973, 114, 1231–1240. [Google Scholar] [CrossRef] [PubMed]
- Thiele, J.H.; Zeikus, J.G. Control of interspecies electron flow during anaerobic digestion: Significance of formate transfer versus hydrogen transfer during syntrophic methanogenesis in flocs. Appl. Environ. Microbiol. 1988, 54, 20–29. [Google Scholar] [CrossRef]
- Poulsen, M.; Schwab, C.; Jensen, B.B.; Engberg, R.M.; Spang, A.; Canibe, N.; Højberg, O.; Milinovich, G.; Fragner, L.; Schleper, C. Methylotrophic methanogenic Thermoplasmata implicated in reduced methane emissions from bovine rumen. Nat. Commun. 2013, 4, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Seedorf, H.; Kittelmann, S.; Henderson, G.; Janssen, P.H. RIM-DB: A taxonomic framework for community structure analysis of methanogenic archaea from the rumen and other intestinal environments. PeerJ 2014, 2, e494. [Google Scholar] [CrossRef] [PubMed]
- Jami, E.; White, B.A.; Mizrahi, I. Potential Role of the Bovine Rumen Microbiome in Modulating Milk Composition and Feed Efficiency. PLoS ONE 2014, 9, e85423. [Google Scholar] [CrossRef]
- Myer, P.R.; Smith, T.P.; Wells, J.E.; Kuehn, L.A.; Freetly, H.C. Rumen microbiome from steers differing in feed efficiency. PLoS ONE 2015, 10, e0129174. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Guan, L.L. Metatranscriptomic profiling reveals linkages between the active rumen microbiome and feed efficiency in beef cattle. AEM 2017, 83, e00061-17. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Hitch, T.C.A.; Chen, Y.; Creevey, C.J.; Guan, L.L. Comparative metagenomic and metatranscriptomic analyses reveal the breed effect on the rumen microbiome and its associations with feed efficiency in beef cattle. Microbiome 2019, 7, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Artegoitia, V.M.; Foote, A.P.; Lewis, R.M.; Freetly, H.C. Rumen Fluid Metabolomics Analysis Associated with Feed Efficiency on Crossbred Steers. Sci. Rep. 2017, 7, 2864. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, R.T.R.; Prado, R.M.D.; Porto, C.; dos Santos, G.T.; Huws, S.A.; Pilau, E.J. Exploring the rumen fluid metabolome using liquid chromatography-high-resolution mass spectrometry and Molecular Networking. Sci. Rep. 2018, 8, 17971. [Google Scholar] [CrossRef]
- Berry, D.P.; Meade, K.G.; Mullen, M.P.; Butler, S.; Diskin, M.G.; Morris, D.; Creevey, C.J. The integration of ‘omic’ disciplines and systems biology in cattle breeding. Animal 2011, 5, 493–505. [Google Scholar] [CrossRef] [PubMed]
| Transition Diet | Finishing Diet | |
|---|---|---|
| Ingredient, % DM | ||
| Corn | 41.12 | 56.20 |
| Dried distillers grains | 22.18 | 19.54 |
| Corn gluten feed | - | 7.08 |
| Soybean hulls | 15.80 | - |
| Barley straw | 6.15 | 4.36 |
| Vitamin/Mineral Premix | 4.47 | 4.76 |
| Corn silage | 10.27 | 8.05 |
| Total | 100.00 | 100.00 |
| Nutrient, % DM | ||
| Dry Matter, % | 62.00 | 62.00 |
| Feedlot NEm, Mcal/cwt | 91.84 | 95.17 |
| Feedlot NEg, Mcal/cwt | 62.10 | 65.00 |
| Crude Protein, % | 14.63 | 14.51 |
| Roughage, % | 16.43 | 12.40 |
| Rough NDF, % | 9.28 | 6.92 |
| Fat, % | 5.11 | 5.28 |
| Calcium, % | 0.75 | 0.70 |
| Phosphorus, % | 0.39 | 0.45 |
| Potassium, % | 0.90 | 0.71 |
| Magnesium, % | 0.22 | 0.21 |
| Sulfur, % | 0.25 | 0.26 |
| Trace Mineral Salt, % | 0.21 | 0.22 |
| RFI Classification | ||||
|---|---|---|---|---|
| Item | High | Low | SEM | p-Value 1 |
| Average body weight, kg 2 | 545.7 | 564.0 | 14.61 | 0.62 |
| Dry matter intake (DMI), kg/day | 13.02 | 10.89 | 0.52 | 0.03 |
| Residual feed Intake (RFI), kg | 0.76 | −1.09 | 0.37 | 0.003 |
| Feed/gain ratio, kg | 12.43 | 11.27 | 0.61 | 0.37 |
| Average daily gain (ADG), kg/day | 1.05 | 1.02 | 0.07 | 0.82 |
| VFA Concentration (mM) | ||||
|---|---|---|---|---|
| Item | Beginning | End | SEM | p-Value 1 |
| Less-efficient steers (high RFI) | ||||
| Acetate | 50.9 | 69.5 | 5.10 | 0.06 |
| Propionate | 16.5 | 23.7 | 1.76 | 0.03 |
| Butyrate | 11.1 | 16.8 | 1.27 | 0.01 |
| Isobutyrate | 0.8 | 1.1 | 0.09 | 0.06 |
| Valerate | 1.0 | 1.5 | 0.12 | 0.03 |
| Isovalerate | 2.1 | 2.9 | 0.28 | 0.19 |
| Caproate | 0.3 | 0.4 | 0.09 | 0.79 |
| Total VFA | 82.8 | 115.8 | 8.24 | 0.03 |
| More-efficient steers (low RFI) | ||||
| Acetate | 56.2 | 64.8 | 3.38 | 0.22 |
| Propionate | 21.0 | 22.4 | 1.66 | 0.70 |
| Butyrate | 14.1 | 15.9 | 1.13 | 0.47 |
| Isobutyrate | 0.7 | 1.1 | 0.07 | 0.01 |
| Valerate | 1.1 | 1.5 | 0.13 | 0.11 |
| Isovalerate | 1.9 | 3.0 | 0.22 | 0.01 |
| Caproate | 0.2 | 0.2 | 0.04 | 0.38 |
| Total VFA | 95.1 | 108.8 | 5.78 | 0.26 |
| Feedlot Finishing Period | ||||
|---|---|---|---|---|
| Item | Beginning (d 0) | End (d 82) | SEM | p-Value 1 |
| High-RFI Steers (n = 5) | ||||
| Number of OTUs | 1,587 | 1,732 | 54.20 | 0.14 |
| Chao1 | 2,366 | 2,575 | 75.27 | 0.07 |
| Faith’s Phylogenetic Diversity | 92.1 | 97.6 | 2.65 | 0.21 |
| Shannon Index | 7.19 | 7.89 | 0.18 | 0.10 |
| Low-RFI Steers (n = 5) | ||||
| Number of OTUs | 1,498 | 1,787 | 114.0 | 0.21 |
| Chao1 | 2,284 | 2,727 | 177.59 | 0.22 |
| Faith’s Phylogenetic Diversity | 89.0 | 100.8 | 4.73 | 0.20 |
| Shannon Index | 7.14 | 7.74 | 0.32 | 0.41 |
| Feedlot Finishing Period | Average Abundance | |||
|---|---|---|---|---|
| Bacterial Phyla | Beginning | End | p-Value 1 | |
| Firmicutes | 41.34 | 60.37 | 50.86 | 0.08 |
| Bacteroidetes | 47.34 | 25.98 | 36.66 | 0.09 |
| Actinobacteria | 3.01 | 1.56 | 2.28 | 0.19 |
| Proteobacteria | 0.97 | 2.27 | 1.62 | 0.12 |
| Euryarchaeota | 1.24 | 2.01 | 1.62 | 0.01 |
| TM7 | 1.05 | 2.12 | 1.59 | 0.09 |
| Spirochaetes | 1.40 | 0.53 | 0.97 | 0.06 |
| Tenericutes | 0.32 | 0.89 | 0.61 | 0.14 |
| Cyanobacteria | 0.49 | 0.44 | 0.46 | 0.81 |
| Planctomycetes | 0.35 | 0.39 | 0.37 | 0.82 |
| Other Phyla | 2.49 | 3.43 | 2.96 | 0.07 |
| Firmicutes/Bacteroidetes ratio | 1.14 | 2.80 | 1.97 | 0.05 |
| Feedlot Finishing Period | Average Abundance | |||
|---|---|---|---|---|
| Bacterial Phylum | Beginning | End | p-Value 1 | |
| Firmicutes | 34.95 | 55.14 | 45.04 | 0.02 |
| Bacteroidetes | 48.73 | 33.04 | 40.88 | 0.12 |
| Actinobacteria | 7.21 | 1.29 | 4.25 | 0.34 |
| TM7 | 1.81 | 2.06 | 1.94 | 0.85 |
| Proteobacteria | 1.48 | 2.31 | 1.90 | 0.43 |
| Euryarchaeota | 1.28 | 1.39 | 1.34 | 0.83 |
| Spirochaetes | 1.31 | 0.81 | 1.06 | 0.42 |
| Cyanobacteria | 0.39 | 0.33 | 0.36 | 0.74 |
| Tenericutes | 0.16 | 0.46 | 0.31 | 0.20 |
| Planctomycetes | 0.21 | 0.27 | 0.24 | 0.55 |
| Other Phyla | 2.46 | 2.91 | 2.69 | 0.55 |
| Firmicutes/Bacteroidetes ratio | 0.78 | 1.96 | 1.37 | 0.10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carmichael, M.N.; Dycus, M.M.; Lourenco, J.M.; Welch, C.B.; Davis, D.B.; Krause, T.R.; Rothrock, M.J.; Fluharty, F.L.; Pringle, T.D.; Callaway, T.R. Ruminal Microbiome Differences in Angus Steers with Differing Feed Efficiencies during the Feedlot Finishing Phase. Microorganisms 2024, 12, 536. https://doi.org/10.3390/microorganisms12030536
Carmichael MN, Dycus MM, Lourenco JM, Welch CB, Davis DB, Krause TR, Rothrock MJ, Fluharty FL, Pringle TD, Callaway TR. Ruminal Microbiome Differences in Angus Steers with Differing Feed Efficiencies during the Feedlot Finishing Phase. Microorganisms. 2024; 12(3):536. https://doi.org/10.3390/microorganisms12030536
Chicago/Turabian StyleCarmichael, Mia N., Madison M. Dycus, Jeferson M. Lourenco, Christina B. Welch, Dylan B. Davis, Taylor R. Krause, Michael J. Rothrock, Francis L. Fluharty, Timothy D. Pringle, and Todd R. Callaway. 2024. "Ruminal Microbiome Differences in Angus Steers with Differing Feed Efficiencies during the Feedlot Finishing Phase" Microorganisms 12, no. 3: 536. https://doi.org/10.3390/microorganisms12030536
APA StyleCarmichael, M. N., Dycus, M. M., Lourenco, J. M., Welch, C. B., Davis, D. B., Krause, T. R., Rothrock, M. J., Fluharty, F. L., Pringle, T. D., & Callaway, T. R. (2024). Ruminal Microbiome Differences in Angus Steers with Differing Feed Efficiencies during the Feedlot Finishing Phase. Microorganisms, 12(3), 536. https://doi.org/10.3390/microorganisms12030536







