Abstract
The Gram-negative marine bacterium GXY010T, which has been isolated from the surface seawater of the western Pacific Ocean, is aerobic, non-motile and non-flagellated. Strain GXY010T exhibits growth across a temperature range of 10–42 °C (optimal at 37 °C), pH tolerance from 7.0 to 11.0 (optimal at 7.5) and a NaCl concentration ranging from 1.0 to 15.0% (w/v, optimal at 5.0%). Ubiquinone-8 (Q-8) was the predominant isoprenoid quinone in strain GXY010T. The dominant fatty acids (>10%) of strain GXY010T were iso-C15:0 (14.65%), summed feature 9 (iso-C17:1 ω9c and/or 10-methyl C16:0) (12.41%), iso-C17:0 (10.85%) and summed feature 3 (C16:1 ω7c and/or C16:1 ω6c) (10.41%). Phosphatidylethanolamine (PE), phosphatidylglycerol (PG), diphosphatidylglycerol (DPG), unidentifiable glycolipid (GL) and four non-identifiable aminolipids (AL1-AL4) were the predominant polar lipids of strain GXY010T. The genomic DNA G+C content was identified as a result of 48.0% for strain GXY010T. The strain GXY010T genome consisted of 2,766,857 bp, with 2664 Open Reading Frames (ORFs), including 2586 Coding sequences (CDSs) and 78 RNAs. Strain GXY010T showed Average Nucleotide Identity (ANI) values of 73.4% and 70.6% and DNA–DNA hybridization (DDH) values of 19.2% and 14.5% with reference species Pseudidiomarina tainanensis MCCC 1A02633T (=PIN1T) and Pseudidiomarina taiwanensis MCCC 1A00163T (=PIT1T). From the results of the polyphasic analysis, a newly named species, Pseudidiomarina fusca sp. nov. within the genus Pseudidiomarina, was proposed. The type strain of Pseudidiomarina fusca is GXY010T (=JCM 35760T = MCCC M28199T = KCTC 92693T).
1. Introduction
The genus Pseudidiomarina was classified in the family Idiomarinaceae, belonging to the order Alteromonadales and the class Gammaproteobacteria. The description of the family was revised in 2006 by Jean et al. with the introduction of a newly introduced genus, Pseudidiomarina [1]. This classification persisted until 2009 when the genus was subsequently merged into the genus Idiomarina [2]. The genus Pseudidiomarina was subsequently recovered by Liu et al. based on a phylogenetic analysis [3]. The genus Pseudidiomarina has been found in diverse environments, including coastal and marine waters, coastal sands, coastal sediments and solar salt flats, all of which are saline areas with an extensive range of salinity [4,5,6,7]. The genus Pseudidiomarina currently includes 18 validly published species (https://lpsn.dsmz.de/genus/pseudidiomarina (accessed on 1 December 2023)) [8]. Members of this genus are characterized as chemo-heterotrophic, mesophilic, non-sporulating, Gram-negative rods and iso-branched fatty acids of both 15- and 17-carbon atoms are the predominant components of this genus. Predominant fatty acids are iso-branched and polar lipids include phosphatidylethanolamine (PE), diphosphatidylglycerol (DPG) and phosphatidylglycerol (PG) [9]. In the present study, we describe a bacterium isolated from a sample of surface seawater from the western Pacific Ocean and characterized using polyphasic taxonomic identification. The results indicate that the isolated bacterium is a novel species of the genus Pseudidiomarina.
2. Materials and Methods
2.1. Isolation and Cultivation Conditions
In September 2020, a bacterial strain designated as GXY010T was obtained from the surface seawater of the western Pacific (162°14′ E, 23°13′ N), where the temperature was 28.2 °C and the salinity was 3.45%. The bacterial isolation process involved adding 1 mL of seawater sample to 30 mL of 2216E liquid medium (Qingdao Hope Bio-Technology Co., Ltd., Qingdao, China). The mixture was then incubated at 28 °C for one week for enrichment. Subsequently, three concentration gradients of 10−2, 10−4 and 10−6 were prepared by dilution, and 200 μL of each gradient was spread onto 2216E agar plates (Qingdao Hope Bio-Technology Co., Ltd., Qingdao, China). After incubating the plates at 28 °C until a large number of colonies grew, the purification process was iteratively conducted to isolate a monoclonal strain. Single colonies were picked individually using an inoculation loop and inoculated on pre-prepared test tube slant 2216E agar; all strains, including GXY010T, were cultured until moss grew. Preservation solution was prepared with 85% saline (0.85%, w/v) and 15% glycerol, sterilized and added to the slant medium to rinse the moss and shake it to form a bacterial suspension, which was dispensed into freezing tubes and stored in an ultra-low temperature freezer at −80 °C.
2.2. Molecular Analysis
Moore’s method was employed for the extraction of genomic DNA from strain GXY010T [10]. The 16S rRNA gene of strain GXY010T was amplified using a PCR instrument (2720 Thermal Cycler, Applied Biosystems, Foster, CA, USA) and generic bacterial primers (B8F, as well as B1510R) [11]. Pairwise similarity values comparing strain GXY010T with its related reference strains were calculated using the EzBioCloud server (http://www.ezbiocloud.net (accessed on 25 August 2023)), following the method of Yoon et al. [12]. Pseudidiomarina tainanensis MCCC 1A02633T and Pseudidiomarina taiwanensis MCCC 1A00163T were selected as reference strains and were obtained from the Marine Culture Collection of China (MCCC). We used the EzBioCloud database to obtain the 16S rRNA gene sequences of the relevant strains, and they were analyzed using the Clustal X version 2.0 program [13]. Phylogenetic trees were built using neighbor joining (NJ), maximum likelihood (ML) and maximum parsimony (MP) methods with MEGA X (version 10.2.6) [14,15,16,17]. NJ and ML analyses were conducted using Kimura’s 2-parameter (K2-P) model [18], while MP analysis employed the method TBR (Tree Bisection Reconnection). Phylogenetic tree topologies were assessed using bootstrap resampling and 1000 replicates. Escherichia coli ATCC 11775T (GenBank accession: AB681728) was used as an outgroup in all phylogenetic analyses.
2.3. Genome Sequencing, Annotation and Analysis
The Illumina HiSeq platform (Illumina, San Diego, CA, USA) was used for whole genome sequencing (WGS). The reads were assembled into sequences using SOAPdenovo2 software (http://sourceforge.net/projects/soapdenovo2/ (accessed on 10 August 2023)) [19]. The genome data of the most similar strain were retrieved from the GenBank database for comparison of genome relatedness. Phylogenetic analyses of the core genomes involved aligning 39 species using MAFFT (version 7.505) software with default settings, followed by trimming to the length of the shortest sequence. Phylogenetic trees of the core genomes were generated using FastTree (version 2.1.10) with JTT+CAT parameters and 1000 bootstrap replicates [20]. E. coli ATCC 11775T was used as an outgroup in the phylogenetic analyses. The resulting trees were visualized, annotated and managed using the web-based tool Chiplot (https://www.chiplot.online/ (accessed on 15 October 2023)). Genomic similarity was evaluated using the Average Nucleotide Identity (ANI) value obtained through orthoANI (https://www.ezbiocloud.net/tools/orthoani (accessed on 10 August 2023)). At the same time, genome-to-genome distance calculator (GGDC) software version 2.1 and Formula 2 were employed to calculate DNA–DNA hybridization (DDH) value. Annotation was performed using BLAST+ 2.2.24 and Rapid Annotation using Subsystem Technology (RAST) [21,22]. With reference to Tatusov et al., the Cluster of Orthologous Groups (COG) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases were utilized for functional gene annotation of GXY010T [23].
2.4. Phenotypic Characterization
The shape, size, gloss, margins and color of the colonies on the plates were directly observed and recorded. Referring to the standard method of Beveridge et al. [24], Gram and flagellum staining of strain GXY010T was performed; the results were observed and recorded using a Nikon Eclipse E600 light microscope (Trusco Nakayama Corporation, Shinagawa, Japan). Following staining of strain GXY010T with 1% (w/v) phosphotungstic acid, cell morphology was observed using transmission electron microscopy (JEM-1200EX, JEOL Ltd., Akishima, Japan) (Figure S1). The experiments to determine the optimum temperature and pH for GXY010T were conducted using 2216E liquid medium, while the assay for optimal salinity was performed using a modified 2216E liquid medium [25], in which K+ was substituted for Na+ (formulated as 4.78 g K2SO4, 0.664 g KCl, 0.096 g KBr, 0.04 g SrCl2·6H2O, 0.0053 g KF, 4.981 g MgCl2·6H2O, 0.95 g CaCl2, 0.228 g KHCO3, 0.026 g H3BO3, 1 L distilled water). The chemical reagents used in the modified 2216E liquid medium were all sourced from Shanghai Sinopharm Chemical Reagent Ltd. in China Shanghai.
The absorbance of the bacterial suspension was measured at 590 nm (OD590) to assess the growth of the strain. Three parallel samples were established for each condition of temperature, salinity and pH, and OD590 was measured following the experimental procedure. The average value was calculated to identify the optimal conditions for strain growth. In temperature experiments (different temperature, 170 rpm, salinity 3.5%, pH 7.5), cultures were incubated at 0 and 4 °C for 4 weeks with measurements taken every 4 days and at 10, 16, 20, 24, 28, 32, 37, 42 and 45 °C for 1 week and measured daily. For the salinity assay (28 °C, 170 rpm, pH 7.5), the NaCl concentration was varied to 0, 0.5, 1, 2, 2.5, 3, 3.5 and 4 to 15% (w/v, with 1.0% intervals), and measurements were taken continuously for one week. In the pH range experiment (28 °C, 170 rpm, salinity 3.5%), growth was assessed using a pH buffer system with concentrations set at 20 mM for the following buffers: H3PO4/KH2PO4 (pH 2.0), sodium ethanoate/ethanoic acid (pH 3.0–5.0), MES (pH 5.5, 6.0 and 6.5), MOPS (pH 7.0 and 7.5), TRICINE (pH 8.0 and 8.5), TAPS (pH 9.0) and CAPSO (pH 9.5 and 10.0) buffers [26,27]. Measurements were taken continuously for a week.
2.5. Enzyme Activity Characterization
Agar media were prepared according to established procedures [28], encompassing media with starch, chitin, casein, gelatin, cellulose, sodium alginate and Tween 20, 40, 80. The only deviation from standard methods was the substitution of distilled water with filtered seawater. To assess the DNase activity of the strains, DNase agar plates from Qingdao Hope Biotechnology were utilized. The degradation efficacy was assessed based on the formation of transparent zones surrounding the colony or when the plates were immersed in the relevant solution. Bacterial cultures were spotted onto enzyme activity agar plates and incubated under optimal conditions. The ability of the strains to hydrolyze Tween 20, 40, 80, chitin and gelatin was assessed by direct observation of the transparent circle surrounding the colonies. Additional enzyme activities were examined using specific identification solutions: Lugol’s iodine solution for amylase activity, 1 M HCl for DNase activity, 3% trichloroacetic acid for caseinase activity, 1 mg/mL Congo red solution with 1 M NaCl for cellulase activity and 10% CaCl2 for sodium alginate enzyme activity. Oxidase activity was tested using 1% tetramethyl-p-phenylenediamine dihydrochloride (TMPD), and catalase activity was assessed using 3% hydrogen peroxide solution. Using API 20E, 20NE and ZYM test strips (bioMérieux Shanghai Ltd., Shanghai, China), as well as GEN III MicroPlates (Biolog Inc., Hayward, CA, USA), the constitutive enzyme activities, fermentation/oxidation profile, acid production and the utilization rate of substrates as the sole carbon and energy source were determined for GXY010T following the manufacturer’s instructions.
2.6. Chemotaxonomy
During the exponential phase, GXY010T and its reference strains were cultivated concurrently in 2216E liquid medium and subsequently harvested for the analysis of cellular fatty acid types and contents using the method reported by Komagata and Suzuki [29]. Fatty acid methyl esters were extracted and analyzed following the standard MIDI protocol, and the TSBA 6.0 database was employed for identification [30]. After incubation in 2216E liquid medium, GXY010T and the reference strains were harvested, lyophilized and subjected to analysis for respiratory quinones and polar lipids. Respiratory quinones were extracted from the strains using chloroform/methanol (2:1, v/v), separated via TLC and identified through LC-MS [31]. The polar lipids were separated using the two-dimensional TLC method on 60 F254 silica gel plates (Merck KGaA, Darmstadt, Germany), following Minnikin et al.’s method [32]. The first-dimensional solution consisted of chloroform/methanol/water in a volume ratio of 65:25:4, while the second-dimensional solution comprised chloroform/methanol/acetic acid/water at a volume ratio of 80:12:15:4 [33]. Additionally, the identified extracted lipids were characterized by spraying the plates with suitable detection reagents for amino groups (ninhydrin/n-butyl alcohol, Dragendoff’s reagent), glycolipids (alpha-naphthol/sulfuric acid) and total lipids (molybdatophosphoric acid).
2.7. Repositories
The GenBank 16S rRNA gene and genome sequence accession numbers for strain GXY010T are OP351365 and JANFPJ000000000, respectively.
3. Results
3.1. Physiological Characteristics
GXY010T thrived under aerobic conditions on 2216E agar plates, displaying colonies that were smooth, rounded and light brown in color. The cells were Gram-staining negative and non-motile with a lack of flagella. The strains were grown at temperatures ranging from 10 to 42 °C (optimum 37 °C). After 72 h of incubation on 2216E agar at 37 °C, the cell morphology was observed to be rod-shaped, with a length of about 2.0 to 3.0 μm and a width of about 0.4 to 0.6 μm. Strain GXY010T was able to grow at a salinity of 1.0% to 15.0% (w/v) NaCl (optimal salinity of 5.0%) and a pH of 7.0 to 11.0 (optimal pH of 7.5). The morphological, physiological and biochemical characteristics of GXY010T are summarized in Table 1. The strain demonstrated oxidase and catalase activities and showed the ability to hydrolyze Tween 20, Tween 40, gelatin and DNA. The following constitutive enzyme activities were positive in API ZYM tests: esterase (C4), esterase lipase (C8), lipase (C14), valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, acid phosphatase, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase and β-fucosidase. The results of the enzyme activity assay demonstrated that strains GXY010T, P. tainanensis MCCC 1A02633T, P. taiwanensis MCCC 1A00163T and Pseudidiomarina marina PIM1T were positive for oxidase, catalase, esterase (C4), esterase lipase (C8), lipase (C14), valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin and acid phosphatase. However, these strains tested negative for the hydrolysis of casein, chitin, starch, cellulose and sodium alginate.

Table 1.
Different characteristics between strain GXY010T and the reference strains.
The results of API 20E and 20NE strips were positive for a reduction in acetoin (Voges–Proskauer reaction) and activity of urease and arginine dihydrolase; they were negative for a reduction in nitrate, utilization of citric acid, production of H2S, indole, the activity of β-galactosidase, gelatinase, lysine decarboxylase, ornithine decarboxylase, tryptophan deaminase and β-glucosidase, assimilation of d-glucose, l-arabinose, mannose, mannitol, N-acetyl-glucosamine, maltose, potassium gluconate, capric acid, adipic acid, malic acid, trisodium citrate and phenylacetic acid, and the fermentation of glucose, mannitol, inositol, sorbitol, rhamnose, sucrose, melibiose, amygdalin and arabinose.
The results of Biolog GEN III microplates were positive for utilization of dextrin, d-glucose-6-PO4, d-fructose-6-PO4, p-hydroxy-phenylacetic acid, l-lactic acid, d-glucuronic acid, Tween 40, acetoacetic acid, d-malic acid, l-malic acid, propionic acid and acetic acid; they were negative for utilization of d-maltose, gentiobiose, d-turanose, d-fructose, d-galactose, mucic acid, 3-methyl glucose, d-fucose, l-fucose, glycyl-l-proline, l-arginine, α-keto-butyric acid, l-aspartic acid, l-glutamic acid, d-galacturonic acid, l-galactonic acid lactone, l-rhamnose, d-trehalose, d-melibiose, d-cellobiose, sucrose, stachyose, d-raffinose, α-d-lactose, β-methyl-d-glucoside, d-salicin, N-acetyl-d-glucosamine, N-acetyl-β-d-mannosamine, α-d-glucose, N-acetyl-d-galactosamine, N-acetyl neuraminic acid, d-mannose, inosine, d-sorbitol, d-mannitol, d-arabitol, myo-inositol, glycerol, d-aspartic acid, d-serine, gelatin, d-alanine, l-histidine, l-pyroglutamic acid, l-serine, pectin, d-gluconic acid, glucuronamide, quinic acid, d-saccharic acid, methyl pyruvate, d-lactic acid methyl ester, citric acid, α-keto-glutaric acid, bromo-succinic acid, γ-amino-butryric acid, α-hydroxy-butyric acid, formic acid and β-hydroxy-d,l-butyric acid.
3.2. Chemotaxonomic Characteristics
The dominant fatty acids (>10%) of strain GXY010T were iso-C15:0 (14.65%), summed feature 9 (iso-C17:1 ω9c and/or 10-methyl C16:0) (12.41%), iso-C17:0 (10.85%) and summed feature 3 (C16:1 ω7c and/or C16:1 ω6c) (10.41%). Moreover, compared to the two reference strains, strain GXY010T had higher amounts of C17:0, anteiso-C17:0, C17:1 ω8c and summed feature 3 (C16:1ω7c or/and C16:1ω6c) but less iso-C15:0 and iso-C15:1 F. Table 2 provides a detailed comparison of the cellular fatty acid type and content between strain GXY010T and the reference strains. Ubiquinone-8 (Q-8) was determined to be the abundant isoprenoid quinone in strain GXY010T, the same as other strains in the family Idiomarinaceae. Figure S5a illustrates that the primary polar lipids of strain GXY010T include phosphatidylethanolamine (PE), phosphatidylglycerol (PG), diphosphatidylglycerol (DPG), unidentified glycolipid (GL) and four non-identified aminolipids (AL1-AL4). In comparison, the major polar lipids of strain P. tainanensis MCCC 1A02633T were PE, PG, DPG and four non-identified aminolipids (AL1-AL4), as depicted in Figure S5b. Figure S5c displays that the primary polar lipids of strain P. taiwanensis MCCC 1A00163T include PE, PG, DPG and three non-identified aminolipids (AL1-AL3). A distinctive feature observed in the polar lipids of strain GXY010T is the presence of an unidentified glycolipid (GL), setting it apart from the other two reference strains.

Table 2.
Cellular fatty acid content (%) of strain GXY010T and reference strains.
3.3. 16S rRNA Gene Sequences Analysis
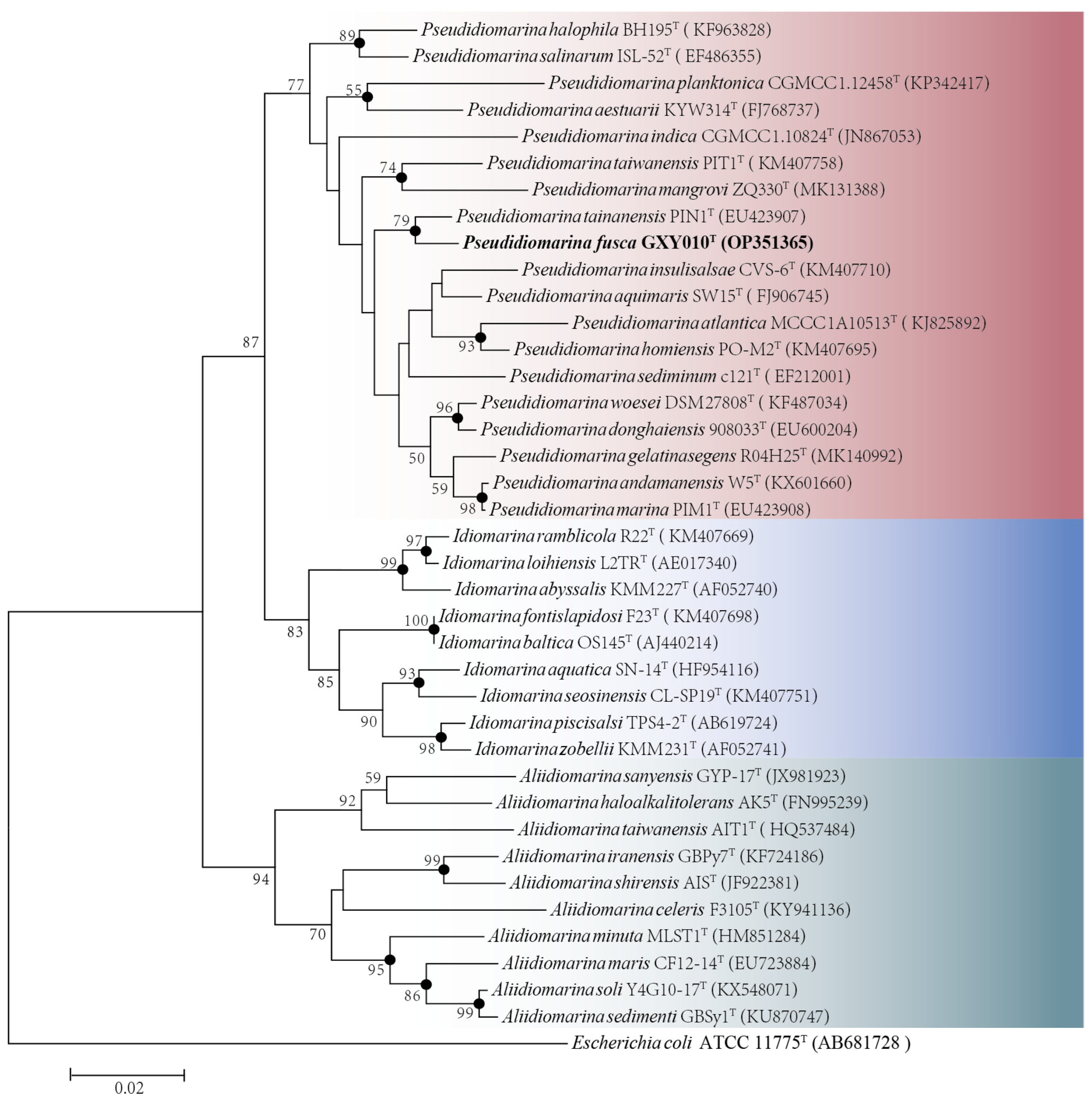
Based on the complete sequence of the 16S rRNA gene (1505 bp) of the strain GXY010T, pairwise alignment exhibited the largest sequence similarity of 98.15% and 97.81% to P. tainanensis MCCC 1A02633T and P. marina PIM1T, respectively. Phylogenetic analysis on the basis of the NJ (Figure 1), ML (Figure S2) and MP (Figure S3) algorithms indicated that strain GXY010T formed a close branch with P. tainanensis MCCC 1A02633T. From the 16S rRNA gene identification and phylogenetic analysis, parallel experiments were conducted using P. tainanensis MCCC 1A02633T and P. taiwanensis MCCC 1A00163T from the MCCC as reference strains. In the 16S rRNA gene-based tree of the family Idiomarinaceae, the genera Pseudidiomarina and Idiomarina formed sister groups, with the genus Aliidiomarina being more closely related to them.
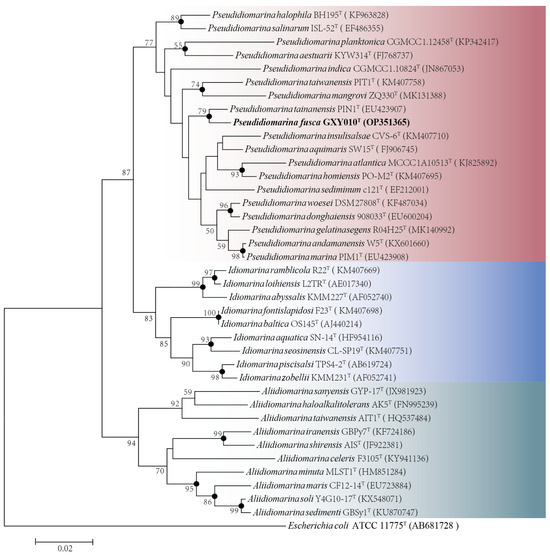
Figure 1.
Neighbor-joining phylogenetic tree based on 16S rRNA gene sequences (1505 bp) showing the phylogenetic position of strain GXY010T and other closely related species. Percentage bootstrap values above 50% (1000 replicates) are shown at branch nodes. Closed circles indicate that the corresponding nodes were also recovered in trees generated with the neighbor-joining, maximum-likelihood and maximum-parsimony algorithms. Red, Pseudidiomarina genus. Blue, Idiomarina genus. Green, Aliidiomarina genus. Escherichia coli ATCC 11775T (GenBank accession: AB681728) was used as the outgroup. The putative novel genospecies are highlighted in bold. Bar, 0.02 nucleotide substitutions per nucleotide position.
3.4. Genome Analysis
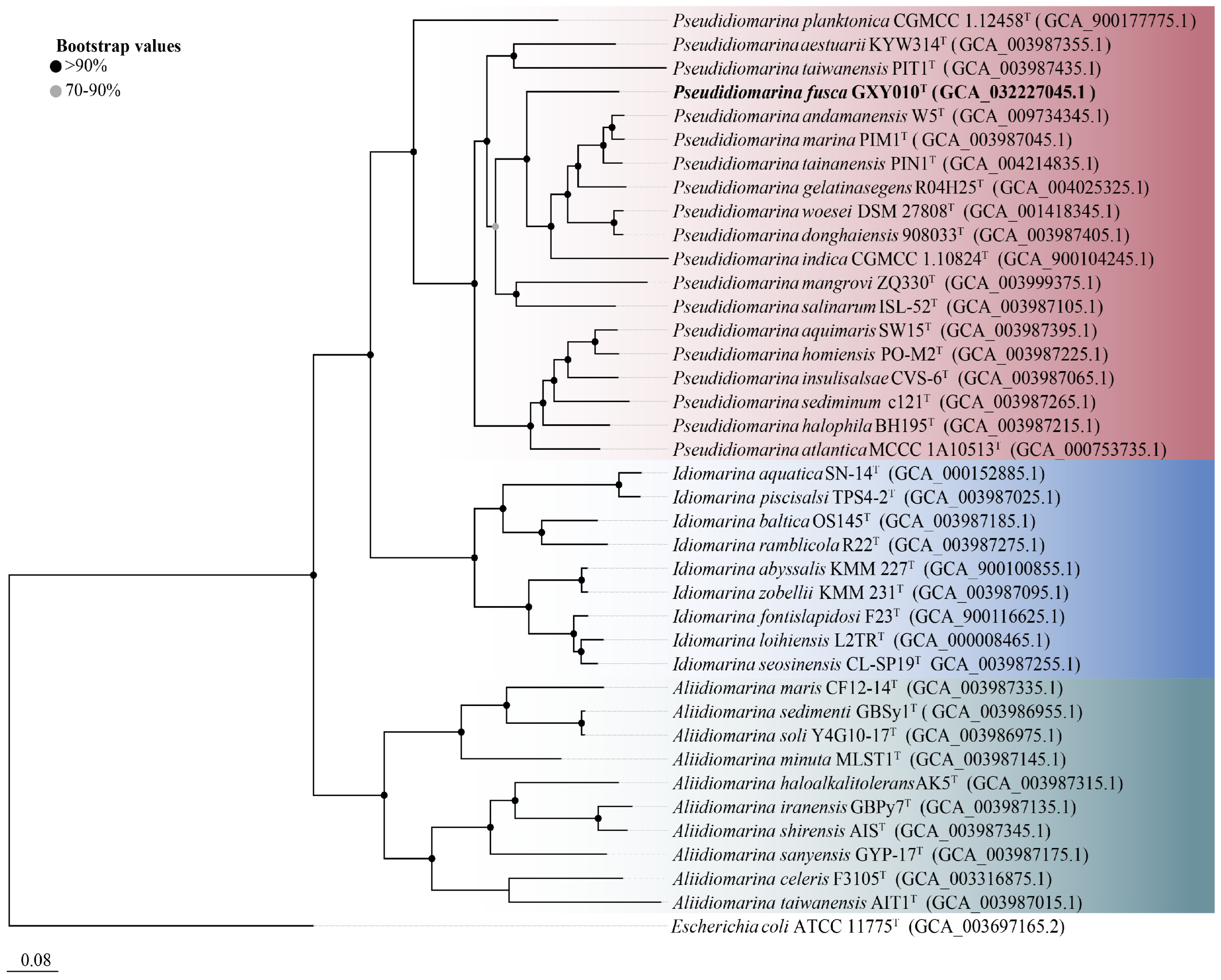
The genome of strain GXY010T is composed of 2,766,857 bp, housing 2664 ORFs, which encompass 2586 CDSs and 78 RNAs (71 tRNAs, 3 rRNAs and 4 ncRNAs). Detailed draft genomic characteristics of strain GXY010T and reference strains can be found in Table S1. The core genome-based phylogenetic tree aligns with the relationships observed in the 16S rRNA gene-based tree, placing GXY010T within the genus Pseudidiomarina (Figure 2). WGS results indicate that the genomic DNA of strain GXY010T has a G+C content of 48.0% and is classified within the genus Pseudidiomarina (47.2–53.0%). The ANI values between strain GXY010T and reference species P. tainanensis MCCC 1A02633T and P. taiwanensis MCCC 1A00163T were 73.4% and 70.6%, respectively, which were designated below the threshold of 95–96% [34]. Similarly, the digital DDH values between strain GXY010T and reference species P. tainanensis MCCC 1A02633T and P. taiwanensis MCCC 1A00163T were 19.2% and 14.5%, respectively, which were well below the recognized threshold of 70% for establishing a novel species [35].
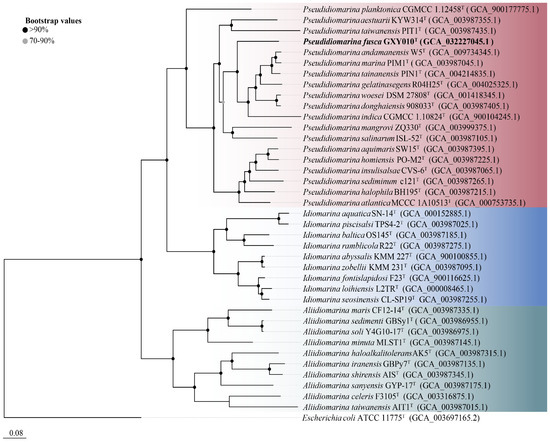
Figure 2.
Core genome-based phylogenetic tree of the family Idiomarinaceae inferred using FastTree 2.1.10 with JTT+CAT parameters and 1000 bootstrap replicates and rooted using Escherichia coli ATCC 11775T. Bootstrap values are indicated at the nodes of the solid circle filled with different colors. The putative novel genospecies are highlighted in bold. Red, Pseudidiomarina genus. Blue, Idiomarina genus. Green, Aliidiomarina genus. Genome accession numbers are indicated in parentheses. Bar, 0.08 substitutions per position.
Strain GXY010T, along with P. tainanensis MCCC 1A02633T and P. taiwanensis MCCC 1A00163T, underwent annotation using the COG database. As shown in Figure S4, strain GXY010T had a higher relative abundance of K (Transcription), L (Replication, recombination and repair), V (Defense mechanisms), T (Signal transduction mechanisms) and S (Function unknown), and lower relative abundance of A (RNA processing and modification), J (Translation, ribosomal structure and biogenesis), D (Cell cycle control, cell division, chromosome partitioning), O (Posttranslational modification, protein turnover, chaperones), C (Energy production and conversion), E (Amino acid transport and metabolism), F (Nucleotide transport and metabolism), H (Coenzyme transport and metabolism) and I (Lipid transport and metabolism) than its reference strains. Based on the KEGG database, glutamate synthase (NADPH/NADH) (EC 1.4.1.13) and alcohol dehydrogenase (EC 1.1.1.1) were found in strain GXY010T, whilst it was not found in P. tainanensis MCCC 1A02633T and P. taiwanensis MCCC 1A00163T. The genome of strain GXY010T features a gene encoding octaprenyl diphosphate synthase (EC 2.5.1.90), indicating its capacity for synthesizing ubiquinone-8 (Q-8), the most abundant respiratory quinone in the family Idiomarinaceae. Additionally, it harbors genes responsible for cardiolipin synthase (EC 2.7.8.41), facilitating DPG production and phosphatidylserine decarboxylase (EC 4.1.1.65), enabling PE synthesis. Along with PG, these constitute the major identified polar lipids of the genus Pseudidiomarina.
4. Conclusions
The enzyme activities of strain GXY010T share similarities with other reference bacteria, as all strains exhibit positive results for oxidase and catalase tests. However, a notable point of distinction is that only GXY010T demonstrates the ability to hydrolyze gelatin. While all strains share the same major fatty acid types, there are variations in their respective content percentages. Strain GXY010T possesses a unique, unidentifiable glycolipid (GL) absent in other strains, further distinguishing it from other members of the genus Pseudidiomarina in terms of polar lipid composition. In summary, the GXY010T strain can be distinguished from the reference strains by several phenotypic features, including both morphological and chemical taxonomic markers. The 16S rRNA phylogenetic tree, core genome-based phylogenetic tree and whole-genome indices (ANI and DDH) collectively validate that strain GXY010T is affiliated with the genus Pseudidiomarina and exhibits noticeable distinctions from other species within the genus. The alignment of evidence from phenotypic, phylogenetic and genetic data unequivocally designates strain GXY010T as a novel species within the genus Pseudidiomarina, leading to the formal proposal of the name Pseudidiomarina fusca sp. nov. A description of Pseudidiomarina fusca sp. nov. can be found below.
Pseudidiomarina fusca (fus’ca. L. fem. adj. fusca, brown): The strain grew aerobically on 2216E agar plates, the colonies were light brown, smooth and rounded, the cell was non-motile and without flagellum, and the cell Gram staining result was negative. Strain GXY010T grew most optimally at 37 °C, pH 7.5 and salinity 5.0%. It showed positive activities for oxidase and catalase. Hydrolysis activities were observed for Tween 20, Tween 40, gelatin and DNA, but not for Tween 80, chitin, sodium alginate, cellulose, casein and starch. The principal respiratory quinone was ubiquinone-8 (Q-8). The dominant fatty acids (>10%) of strain GXY010T were iso-C15:0, summed feature 9 (iso-C17:1 ω9c and/or 10-methyl C16:0), iso-C17:0 and summed feature 3 (C16:1 ω7c/C16:1 ω6c). The polar lipids consisted mainly of PE, PG, DPG, unidentified GL and four unidentified ALs. The species is affiliated with the genus Pseudidiomarina of the family Idiomarinaceae. The DNA G+C content of the type strain is 48.0%.
The type strain, GXY010T (=JCM 35760T = MCCC M28199T = KCTC 92693T), was isolated from the surface seawater of the western Pacific Ocean. The GenBank accession number for the 16S rRNA gene sequence of GXY010T is OP351365. The WGS project of strain GXY010T has been deposited in GenBank under the accession number JANFPJ000000000. The version presented in this paper is the JANFPJ000000000 version.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms12020408/s1, Table S1: Genome features of strain GXY010T and strains of the genus Pseudidiomarina; Figure S1: Transmission electron micrograph of a negatively stained cell of GXY010T; Figure S2: Maximum-likelihood phylogenetic tree based on 16S rRNA gene sequences (1505 bp) showing the phylogenetic position of strain GXY010T and other closely related species; Figure S3: Maximum-parsimony phylogenetic tree based on 16S rRNA gene sequences (1505 bp) showing the phylogenetic positions of strain GXY010T and other closely related species; Figure S4: Comparison of gene content between GXY010T and their reference strains; Figure S5: Total polar lipids of strain GXY010T and the reference strain were separated by two-dimensional TLC and detected with 10% ethanolic molybdophosphoric acid.
Author Contributions
Conceptualization, X.S.; Data curation, Y.W., X.W., X.G., J.H., X.Y. and Y.Z.; Formal analysis, Y.W. and X.W.; Funding acquisition, X.S.; Investigation, X.Z.; Project administration, X.Z. and X.S.; Software, X.S.; Supervision, X.Z.; Writing—original draft, Y.W.; Writing—review and editing, X.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by projects from the National Natural Science Foundation of China (NSFC; No. 42230411) and the China Ocean Mineral Resources R and D Association (COMRA) Special Foundation (DY135-B2-10).
Data Availability Statement
All data generated or analyzed during this study are included.
Acknowledgments
We are thankful to the kind help of Aharon Oren (The Hebrew University of Jerusalem, Israel) for the naming of this novel strain.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Jean, W.D.; Shieh, W.Y.; Chiu, H.-H. Pseudidiomarina taiwanensis gen. nov., sp. nov., a marine bacterium isolated from shallow coastal water of An-Ping Harbour, Taiwan, and emended description of the family Idiomarinaceae. Int. J. Syst. Evol. Microbiol. 2006, 56, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Taborda, M.; Antunes, A.; Tiago, I.; Veríssimo, A.; Nobre, M.F.; da Costa, M.S. Description of Idiomarina insulisalsae sp. nov., isolated from the soil of a sea salt evaporation pond, proposal to transfer the species of the genus Pseudidiomarina to the genus Idiomarina and emended description of the genus Idiomarina. Syst. Appl. Microbiol. 2009, 32, 371–378. [Google Scholar] [CrossRef]
- Liu, Y.; Lai, Q.; Shao, Z. Genome-Based Analysis Reveals the Taxonomy and Diversity of the Family Idiomarinaceae. Front. Microbiol. 2018, 9, 2453. [Google Scholar] [CrossRef]
- Jean, W.D.; Leu, T.-Y.; Lee, C.-Y.; Chu, T.-J.; Lin, S.Y.; Shieh, W.Y. Pseudidiomarina marina sp. nov. and Pseudidiomarina tainanensis sp. nov. and reclassification of Idiomarina homiensis and Idiomarina salinarum as Pseudidiomarina homiensis comb. nov. and Pseudidiomarina salinarum comb. nov., respectively. Int. J. Syst. Evol. Microbiol. 2009, 59, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.-W.; Kim, B.-Y.; Weon, H.-Y.; Baek, Y.-K.; Koo, B.-S.; Go, S.-J. Idiomarina homiensis sp. nov., isolated from seashore sand in Korea. Int. J. Syst. Evol. Microbiol. 2006, 56, 2229–2233. [Google Scholar] [CrossRef]
- Wu, Y.-H.; Shen, Y.-Q.; Xu, X.-W.; Wang, C.-S.; Oren, A.; Wu, M. Pseudidiomarina donghaiensis sp. nov. and Pseudidiomarina maritima sp. nov., isolated from the East China Sea. Int. J. Syst. Evol. Microbiol. 2009, 59, 1321–1325. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Jung, S.-Y.; Jung, Y.-T.; Oh, T.-K. Idiomarina salinarum sp. nov., isolated from a marine solar saltern in Korea. Int. J. Syst. Evol. Microbiol. 2007, 57, 2503–2506. [Google Scholar] [CrossRef]
- Parte, A.C.; Carbasse, J.S.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Poddar, A.; Lepcha, R.T.; Mukherjee, D.; Bhattacharyya, D.; Das, S.K. Comparative analysis of 16S rRNA signature sequences of the genus Idiomarina and Idiomarina woesei sp. nov., a novel marine bacterium isolated from the Andaman Sea. Res. Microbiol. 2014, 165, 501–507. [Google Scholar] [CrossRef]
- Moore, E.R.B.; Arnscheidt, A.; Krüger, A.; Strömpl, C.; Mau, M. Simplified protocols for the preparation of genomic DNA from bacterial cultures. Mol. Microb. Ecol. Man. 1999, 1, 1–15. [Google Scholar]
- Lane, D.J. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematics; John Wiley and Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Fitch, W.M. Towards defining the course of evolution: Minimum change for a specific tree topology. Syst. Zool. 1971, 20, 406–416. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience 2012, 1, 18. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Beveridge, T.J.; Lawrence, J.R.; Murray, R.G. Sampling and staining for light microscopy. In Methods for General and Molecular Microbiology; Reddy, C.A., Beveridge, T.J., Breznak, T.A., Marzluf, G., Schmidt, T.M., Snyder, L.R., Eds.; American Society for Microbiology: Washington, DC, USA, 2007; pp. 19–33. [Google Scholar]
- Lyman, J.; Fleming, R.H. Composition of sea water. J. Mar. Res. 1940, 3, 134–146. [Google Scholar]
- Li, A.Z.; Lin, L.Z.; Zhang, M.X.; Zhu, H.H. Arenibacter antarcticus sp. nov., isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 2017, 67, 4601–4605. [Google Scholar] [CrossRef]
- Taha, M. Buffers for the Physiological pH Range: Acidic Dissociation Constants of Zwitterionic Compounds in Various Hydroorganic Media. Ann. Chim. 2005, 95, 105–109. [Google Scholar] [CrossRef]
- Tindall, B.J.; Sikorski, J.; Smibert, R.A.; Krieg, N.R. Phenotypic characterization and the principles of comparative systematics. In Methods for General and Molecular Microbiology; Reddy, C.A., Beveridge, T.J., Breznak, J.A., Marzluf, G., Schmidt, T.M., Eds.; American Society for Microbiology: Washington, DC, USA, 2007; pp. 330–393. [Google Scholar]
- Komagata, K.; Suzuki, K.I. 4 Lipid and Cell-Wall Analysis in Bacterial Systematics. Methods Microbiol. 1988, 19, 161–207. [Google Scholar]
- ResearchGate home page. Available online: https://www.researchgate.net/publication/284025789 (accessed on 16 February 2024).
- Xie, C.-H.; Yokota, A. Phylogenetic analyses of Lampropedia hyalina based on the 16S rRNA gene sequence. J. Gen. Appl. Microbiol. 2003, 49, 345–349. [Google Scholar] [CrossRef]
- Minnikin, D.; O’Donnell, A.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, A.; Parlett, J. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Collins, M.D.; Shah, H.N. Fatty acid, menaquinone and polar lipid composition of Rothia dentocariosa. Arch. Microbiol. 1984, 137, 247–249. [Google Scholar] [CrossRef]
- Wayne, L.G.; Moore, W.E.C.; Stackebrandt, E.; Kandler, O.; Colwell, R.R.; Krichevsky, M.I.; Truper, H.G.; Murray, R.G.E.; Grimont, P.A.D.; Brenner, D.J.; et al. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int. J. Syst. Evol. Microbiol. 1987, 37, 463–464. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).