The Antimicrobial Resistance of Enterotoxigenic Escherichia coli from Diarrheal Patients and the Environment in Two Geographically Distinct Rural Areas in Bangladesh over the Years
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Processing in the Surveillance Study
2.2. Antibiotic Susceptibility Testing
2.3. Data Analysis
2.4. Ethics Statement
3. Results
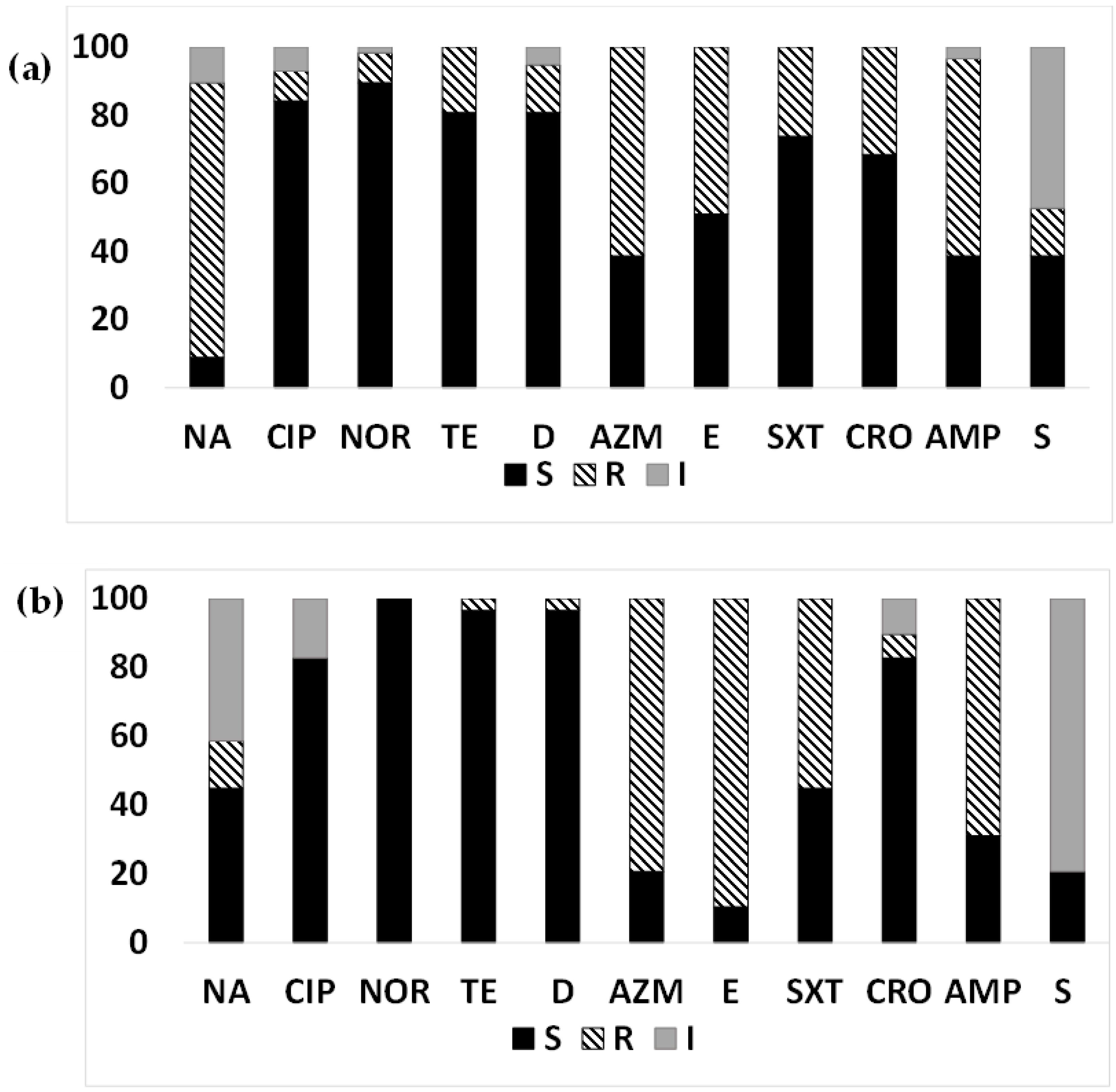
3.1. AMR in the ETEC Strains Isolated from Diarrheal Patients
3.2. MDR-ETEC from Diarrheal Patients
3.3. AMR Pattern of ETEC Strains Based on Toxin Gene Profiles Isolated from Diarrheal Patients
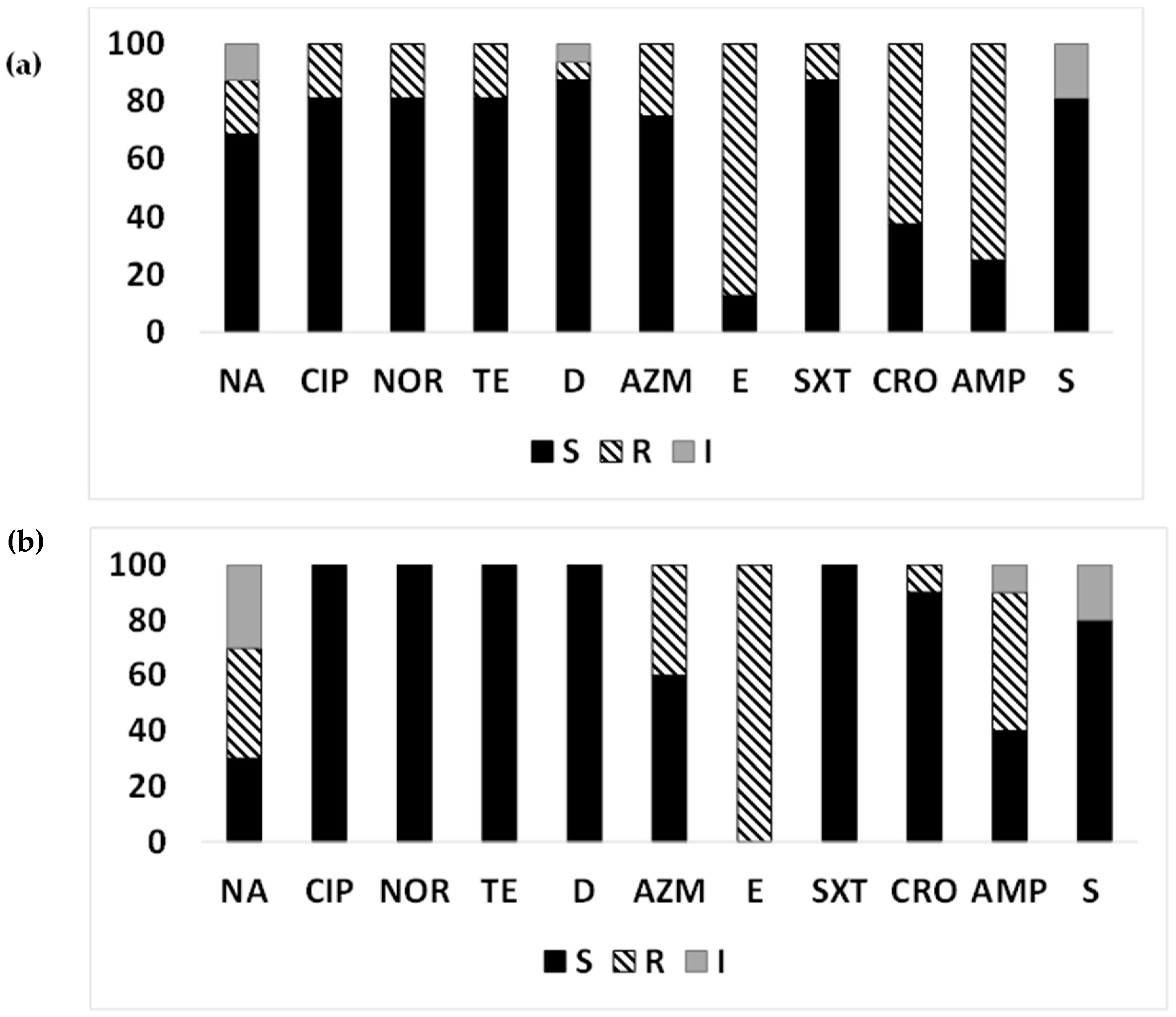
3.4. AMR in the ETEC Strains from the Aquatic Environment
3.5. MDR-ETEC from Surface Water
3.6. AMR Pattern of ETEC Strains Based on Toxin Gene Profiles Isolated from Surface Water
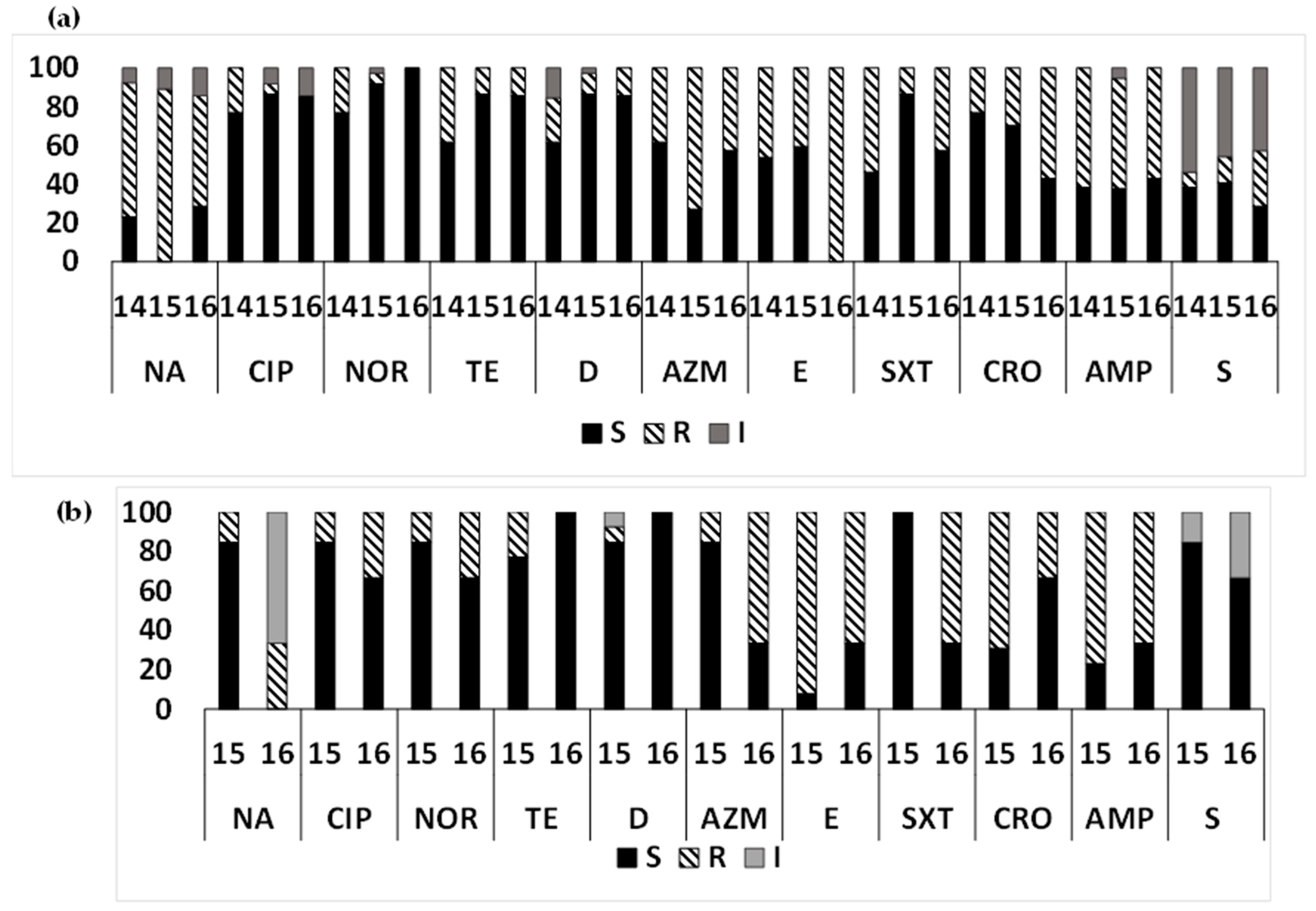
3.7. Year-Wise AMR Pattern of ETEC Strains
3.8. AMR Pattern in Non-ETEC Strains
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khalil, I.A.; Troeger, C.; Blacker, B.F.; Rao, P.C.; Brown, A.; Atherly, D.E.; Brewer, T.G.; Engmann, C.M.; Houpt, E.; Kang, G.; et al. Morbidity and mortality due to shigella and enterotoxigenic Escherichia coli diarrhoea: The Global Burden of Disease Study 1990–2016. Lancet Infect. Dis. 2018, 18, 1229–1240. [Google Scholar] [CrossRef]
- Chakraborty, S.; Johura, F.T.; Sultana, M.; Zhang, X.; Sadique, A.; George, C.; Monira, S.; Sack, D.A.; Sack, R.B.; Alam, M. Epidemiology of enterotoxigenic Escherichia coli among children and adults seeking care at hospitals in two geographically distinct rural areas in Bangladesh. Microorganisms 2023. [Google Scholar]
- Kotloff, K.L.; Nataro, J.P.; Blackwelder, W.C.; Nasrin, D.; Farag, T.H.; Panchalingam, S.; Wu, Y.; Sow, S.O.; Sur, D.; Breiman, R.F.; et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet 2013, 382, 209–222. [Google Scholar] [CrossRef]
- World Health Organization. Vaccine against enterotoxigenic Escherichia coli. 2021. Available online: https://cdn.who.int/media/docs/default-source/immunization/final-etec-ppc-2021eng.pdf? (accessed on 19 May 2023).
- Goldsmid, J.M.; Leggat, P.A. The returned traveler with diarrhea. Aust. Fam. Physician 2007, 36, 322–327. [Google Scholar]
- Ahmed, S.; Korpe, P.; Ahmed, T.; Chisti, M.J.; Faruque, A.S.G. Burden and Risk Factors of Antimicrobial Use in Children Less Than 5 Years of Age with Diarrheal Illness in Rural Bangladesh. Am. J. Trop. Med. Hyg. 2018, 98, 1571–1576. [Google Scholar] [CrossRef] [PubMed]
- O’Ryan, G.M.; Ashkenazi-Hoffnung, L.; O’Ryan-Soriano, M.A.; Ashkenazi, S. Management of acute infectious diarrhea for children living in resource-limited settings. Expert Rev. Anti Infect. Ther. 2014, 12, 621–632. [Google Scholar] [CrossRef] [PubMed]
- Becker, S.L.; Vogt, J.; Knopp, S.; Panning, M.; Warhurst, D.C.; Polman, K.; Marti, H.v.M.L.; Yansouni, C.P.; Jacobs, J.; Bottieau, E.; et al. Persistent digestive disorders in the tropics: Causative infectious pathogens and reference diagnostic tests. BMC Infect. Dis. 2013, 13, 37. [Google Scholar] [CrossRef] [PubMed]
- Sack, R.B. Travelers’ diarrhea: Microbiologic bases for prevention and treatment. Rev. Infect. Dis. 1990, 12 (Suppl. 1), S59–S63. [Google Scholar] [CrossRef]
- Diptyanusa, A.; Ngamprasertchai, T.; Piyaphanee, W. A review of antibiotic prophylaxis for traveler’s diarrhea: Past to present. Trop. Dis. Travel Med. Vaccines 2018, 4, 14. [Google Scholar] [CrossRef]
- Diemert, D.J. Prevention and self-treatment of traveler’s diarrhea. Clin. Microbiol. Rev. 2006, 19, 583–594. [Google Scholar] [CrossRef]
- Rademaker, C.M.; Hoepelman, I.M.; Wolfhagen, M.J.; Beumer, H.; Rozenberg-Arska, M.; Verhoef, J. Results of a double-blind placebo-controlled study using ciprofloxacin for prevention of travelers’ diarrhea. Eur. J. Clin. Microbiol. Infect. Dis. 1989, 8, 690–694. [Google Scholar] [CrossRef]
- DuPont, H.L.; Jiang, Z.D.; Okhuysen, P.C.; Ericsson, C.D.; de la Cabada, F.J.; Ke, S.; DuPont, M.W.; Martinez-Sandoval, F. A randomized, double-blind, placebo-controlled trial of rifaximin to prevent travelers’ diarrhea. Ann. Intern. Med. 2005, 142, 805–812. [Google Scholar] [CrossRef]
- Steffen, R.; Hill, D.R.; DuPont, H.L. Traveler’s diarrhea: A clinical review. JAMA 2015, 313, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Vila, J. Fluooroquinolones. In Frontiers in Antimicrobial Resistance: A Tribute to Stuart B. Levy; White, D.G., Alekshun, M.N., McDermott, P.F., Eds.; ASM Press: Washington, DC, USA, 2005; pp. 41–52. [Google Scholar]
- DuPont, H.L.; Steffen, R. Use of antimicrobial agent for treatment and prevention of travellers’ diarrhoea in the face of enhanced risk of transient fecal carriage of multidrug resistant Enterobacteriaceae: Setting the stage for consensus recommendations. J. Travel. Med. 2017, 24, S57–S62. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.; Baron, E.; Pfaller, M.; Tenover, F.; Yolken, R. (Eds.) Manual of Clinical Microbiology, 7th ed.; American Society for Microbiology: Washington, DC, USA, 1999; pp. 497–506. [Google Scholar]
- World Health Organization. WHO Guidelines for the Laboratory Diagnosis of Cholera; Bacterial Disease Unit, World Health Organization: Geneva, Switzerland, 1974. [Google Scholar]
- Alam, M.; Hasan, N.A.; Sadique, A.; Bhuiyan, N.A.; Ahmed, K.U.; Nusrin, S.; Nair, G.B.; Siddique, A.K.; Sack, R.B.; Sack, D.A.; et al. Seasonal cholera caused by Vibrio cholerae serogroups O1 and O139 in the coastal aquatic environment of Bangladesh. Appl. Environ. Microbiol. 2006, 72, 4096–4104. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk Susceptibility Tests: Approved Standard, 10th ed.; CLSI: Wayne, PA, USA, 2009; Volume M02-A11. [Google Scholar]
- Chakraborty, S.; Deokule, J.S.; Garg, P.; Bhattacharya, S.K.; Nandy, R.K.; Nair, G.B.; Yamasaki, S.; Takeda, Y.; Ramamurthy, T. Concomitant infection of enterotoxigenic Escherichia coli in an outbreak of cholera caused by Vibrio cholerae O1 and O139 in Ahmedabad. India J. Clin. Microbiol. 2001, 39, 3241–3246. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Wu, F.; Chai, Y.; Xu, X.; Yang, L.; Tian, S.; Zhang, H.; Li, Y.; Yang, C.; Liu, H.; et al. A new plasmid carrying mphA causes prevalence of azithromycin resistance in enterotoxigenic Escherichia coli serogroup O6. BMC Microbiol. 2020, 20, 247. [Google Scholar] [CrossRef]
- Das, R.; Nasrin, S.; Palit, P.; Sobi, R.A.; Sultana, A.A.; Khan, S.H.; Haque, M.A.; Nuzhat, S.; Ahmed, T.; Faruque, A.S.G.; et al. Vibrio cholerae in rural and urban Bangladesh, findings from hospital-based surveillance, 2000–2021. Sci. Rep. 2023, 13, 6411. [Google Scholar] [CrossRef]
- Rashed, S.M.; Hasan, N.A.; Alam, M.; Sadique, A.; Sultana, M.; Hoq, M.M.; Sack, R.B.; Colwell, R.R.; Huq, A. Vibrio cholerae O1 with Reduced Susceptibility to Ciprofloxacin and Azithromycin Isolated from a Rural Coastal Area of Bangladesh. Front. Microbiol. 2017, 8, 252. [Google Scholar] [CrossRef]
- Begum, Y.A.; Talukder, K.A.; Azmi, I.J.; Shahnaij, M.; Sheikh, A.; Sharmin, S.; Svennerholm, A.M.; Qadri, F. Resistance Pattern and Molecular Characterization of Enterotoxigenic Escherichia coli (ETEC) Strains Isolated in Bangladesh. PLoS ONE 2016, 11, e0157415. [Google Scholar] [CrossRef]
- Matsushita, S.; Kawamura, M.; Takahashi, M.; Yokoyama, K.; Konishi, N.; Hatakeyama, K.; Kai, A.; Morozumi, S.; Morita, K.; Watanabe, N.; et al. Increasing fluoroquinolone low sensitivity in enterotoxigenic Escherichia coli isolated from diarrhea of overseas travelers in Tokyo. Kansenshogaku Zasshi 2001, 75, 785–791. [Google Scholar] [CrossRef][Green Version]
- Shaheen, H.I.; Khalil, S.B.; Rao, M.R.; Abu, E.R.; Wierzba, T.F.; Peruski, L.F., Jr.; Putnam, S.; Navarro, A.; Morsy, B.Z.; Cravioto, A.; et al. Phenotypic profiles of enterotoxigenic Escherichia coli associated with early childhood diarrhea in rural Egypt. J. Clin. Microbiol. 2004, 47, 5588–5595. [Google Scholar] [CrossRef]
- Medina, A.M.; Rivera, F.P.; Pons, M.J.; Riveros, M.; Gomes, C.; Bernal, M.; Meza, R.; Maves, R.C.; Huicho, L.; Chea-Woo, E.; et al. Comparative analysis of antimicrobial resistance in enterotoxigenic Escherichia coli isolates from two paediatric cohort studies in Lima, Peru. Trans. R Soc. Trop. Med. Hyg. 2015, 109, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Pal, B.B.; Nayak, S.R.; Khuntia, H.K. Epidemiology and Antibiogram Profile of Vibrio cholerae Isolates between 2004–2013 from Odisha, India. Jpn. J. Infect. Dis. 2018, 71, e99–e103. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Song, D.; Zhou, B.; Hua, Q.; Lin, X.; Wang, Y. Persistence of Salmonella Typhimurium and antibiotic resistance genes in different types of soil influenced by flooding and soil properties. Ecotoxicol. Environ. Saf. 2022, 248, 114330. [Google Scholar] [CrossRef] [PubMed]
- Talukdar, P.K.; Rahman, M.; Rahman, M.; Nabi, A.; Islam, Z.; Hoque, M.M.; Endtz, H.P.; Islam, M.A. Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PLoS ONE 2013, 8, e61090. [Google Scholar] [CrossRef] [PubMed]



| Mathbaria | Chhatak | |
|---|---|---|
| Patients | 57 | 29 |
| Surface water | 15 | 10 |
| Pond sand filter (drinking water) | 8 | |
| Sediment | 3 | |
| Plankton | 0 | 2 |
| Totals | 83 | 41 |
| Location | Type | Resistance Profile | % Of Strains |
|---|---|---|---|
| Mathbaria | I | T, M, SXT, CRO, AMP, S | 14.7 |
| II | Q, T, SXT, CRO, AMP, S | 2.9 | |
| III | Q, T, SXT, CRO, AMP | 5.88 | |
| IV | Q, T, M, SXT, AMP | 2.9 | |
| V | Q, M, CRO, AMP | 23.53 | |
| VI | M, SXT, CRO, AMP | 5.88 | |
| VII | Q, M, AMP | 29.41 | |
| VIII | M, SXT, AMP | 11.76 | |
| IX | Q, M, S | 2.9 | |
| Chhatak | I | Q, T, M | 4.76 |
| II | M, CRO, AMP | 9.5 | |
| III | M, SXT, AMP | 71.43 | |
| IV | Q, M, AMP | 14.3 |
| Location | Type | Resistance Profile | % of Strains |
|---|---|---|---|
| Mathbaria | I | T, M, AMP | 9.1 |
| II | M, CRO, AMP | 63.6 | |
| III | Q, M, CRO, AMP | 9.1 | |
| IV | T, M, CRO, AMP | 9.1 | |
| V | Q, M, SXT, AMP | 9.1 | |
| Chhatak | I | M, CRO, AMP | 33.33 |
| II | Q, M, AMP | 66.67 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Johura, F.-T.; Sultana, M.; Sadique, A.; Monira, S.; Sack, D.A.; Sack, R.B.; Alam, M.; Chakraborty, S. The Antimicrobial Resistance of Enterotoxigenic Escherichia coli from Diarrheal Patients and the Environment in Two Geographically Distinct Rural Areas in Bangladesh over the Years. Microorganisms 2024, 12, 301. https://doi.org/10.3390/microorganisms12020301
Johura F-T, Sultana M, Sadique A, Monira S, Sack DA, Sack RB, Alam M, Chakraborty S. The Antimicrobial Resistance of Enterotoxigenic Escherichia coli from Diarrheal Patients and the Environment in Two Geographically Distinct Rural Areas in Bangladesh over the Years. Microorganisms. 2024; 12(2):301. https://doi.org/10.3390/microorganisms12020301
Chicago/Turabian StyleJohura, Fatema-Tuz, Marzia Sultana, Abdus Sadique, Shirajum Monira, David A. Sack, Richard Bradley Sack, Munirul Alam, and Subhra Chakraborty. 2024. "The Antimicrobial Resistance of Enterotoxigenic Escherichia coli from Diarrheal Patients and the Environment in Two Geographically Distinct Rural Areas in Bangladesh over the Years" Microorganisms 12, no. 2: 301. https://doi.org/10.3390/microorganisms12020301
APA StyleJohura, F.-T., Sultana, M., Sadique, A., Monira, S., Sack, D. A., Sack, R. B., Alam, M., & Chakraborty, S. (2024). The Antimicrobial Resistance of Enterotoxigenic Escherichia coli from Diarrheal Patients and the Environment in Two Geographically Distinct Rural Areas in Bangladesh over the Years. Microorganisms, 12(2), 301. https://doi.org/10.3390/microorganisms12020301






