Deciphering Differences in Microbial Community Diversity between Clubroot-Diseased and Healthy Soils
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description and Sample Collection
2.2. Quantification of P. brassicae
2.3. Soil Chemical Properties
2.4. Amplicon Sequencing and Data Processing
2.5. Network Analysis
2.6. Statistics Analysis
3. Results
3.1. P. brassicae Abundance in Different Fields
3.2. Soil Chemical Properties
3.3. Microbial Community Diversity
3.4. Microbial Composition
3.5. Effects of Soil Variables and Microbial Species on Clubroot Disease
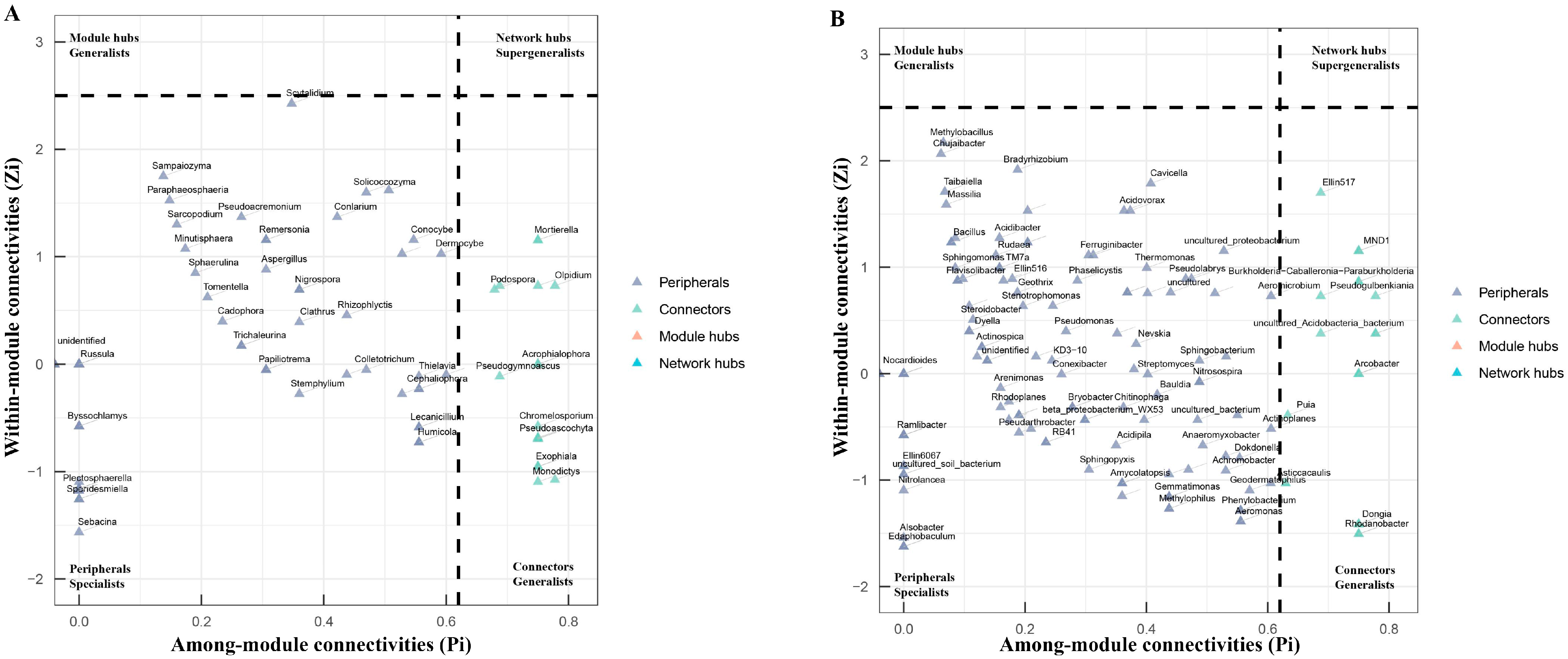
3.6. Network Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kowata-Dresch, L.S.; May-De Mio, L.L. Clubroot management of highly infested soils. Crop Prot. 2012, 35, 47–52. [Google Scholar] [CrossRef]
- Donald, C.; Porter, I. Integrated control of clubroot. J. Plant Growth Regul. 2009, 28, 289–303. [Google Scholar] [CrossRef]
- Struck, C.; Rüsch, S.; Strehlow, B. Control strategies of clubroot disease caused by Plasmodiophora brassicae. Microorganisms 2022, 10, 620. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.; Bakker, P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, J.; Riera, N.; Jin, T.; Li, J.; Wang, N. Huanglongbing impairs the rhizosphere-to-rhizoplane enrichment process of the citrus root-associated microbiome. Microbiome 2017, 5, 97. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, R.L.; Vismans, G.; Yu, K.; Song, Y.; de Jonge, R.; Burgman, W.P.; Burmølle, M.; Herschend, J.; Bakker, P.A.H.M.; Pieterse, C.M.J. Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 2018, 12, 1496–1507. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; van der Heijden, M.G. Soil microbiomes and one health. Nat. Rev. Microbiol. 2023, 21, 6–20. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Tan, L.; Gu, S.; Xiao, Y.; Xiong, X.; Zeng, W.; Feng, K.; Wei, Z.; Deng, Y. Network analysis infers the wilt pathogen invasion associated with non-detrimental bacteria. NPJ Biofilms Microb. 2020, 6, 8. [Google Scholar] [CrossRef]
- Sun, X.; Xu, Z.; Xie, J.; Hesselberg-Thomsen, V.; Tan, T.; Zheng, D.; Strube, M.L.; Dragoš, A.; Shen, Q.; Zhang, R.; et al. Bacillus velezensis stimulates resident rhizosphere Pseudomonas stutzeri for plant health through metabolic interactions. ISME J. 2022, 16, 774–787. [Google Scholar] [CrossRef]
- Poppeliers, S.W.M.; Sánchez-Gil, J.J.; de Jonge, R. Microbes to support plant health: Understanding bioinoculant success in complex conditions. Curr. Opin. Microbiol. 2023, 73, 102286. [Google Scholar] [CrossRef]
- Qian, J.J.; Akçay, E. The balance of interaction types determines the assembly and stability of ecological communities. Nat. Ecol. Evol. 2020, 4, 356–365. [Google Scholar] [CrossRef]
- Lee, S.M.; Kong, H.G.; Song, G.C.; Ryu, C.M. Disruption of Firmicutes and Actinobacteria abundance in tomato rhizosphere causes the incidence of bacterial wilt disease. ISME J. 2021, 15, 330–347. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Li, R.; Ren, Y.; Liu, C.; Zhao, Q.; Wu, H.; Jousset, A.; Shen, Q. Distinct roles for soil fungal and bacterial communities associated with the suppression of vanilla Fusarium wilt disease. Soil Biol. Biochem. 2017, 107, 198–207. [Google Scholar] [CrossRef]
- Wei, Z.; Gu, Y.; Friman, V.P.; Kowalchuk, G.A.; Xu, Y.; Shen, Q.; Jousset, A. Initial soil microbiome composition and functioning predetermine future plant health. Sci. Adv. 2019, 5, eaaw0759. [Google Scholar] [CrossRef] [PubMed]
- Bonanomi, G.; Capodilupo, M.; Incerti, G.; Gaglione, S.A.; Scala, F. Fungal diversity increases soil fungistasis and resistance to microbial invasion by a nonresident species. Biol. Control 2014, 72, 38–45. [Google Scholar] [CrossRef]
- Wei, Z.; Yang, T.; Friman, V.P.; Xu, Y.; Shen, Q.; Jousset, A. Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat. Commun. 2015, 6, 8413. [Google Scholar] [CrossRef] [PubMed]
- Murakami, H.; Tsushima, S.; Shishido, Y. Soil suppressiveness to clubroot disease of Chinese cabbage caused by Plasmodiophora brassicae. Soil Biol. Biochem. 2000, 32, 1637–1642. [Google Scholar] [CrossRef]
- Li, J.; Wan, X.; Liu, X.; Chen, Y.; Slaughter, L.C.; Weindorf, D.C.; Dong, Y. Changes in soil physical and chemical characteristics in intensively cultivated greenhouse vegetable fields in North China. Soil Till. Res. 2019, 195, 104366. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, H.; Sun, L.; Qi, G.; Chen, S.; Zhao, X. Microbial community composition is related to soil biological and chemical properties and bacterial wilt outbreak. Sci. Rep. 2017, 7, 343. [Google Scholar] [CrossRef]
- Orr, R.; Nelson, P.N. Impacts of soil abiotic attributes on Fusarium wilt, focusing on bananas. Appl. Soil Ecol. 2018, 132, 20–33. [Google Scholar] [CrossRef]
- Meng, T.; Wang, Q.; Abbasi, P.; Ma, Y. Deciphering differences in the chemical and microbial characteristics of healthy and Fusarium wilt-infected watermelon rhizosphere soils. Appl. Microbiol. Biotechnol. 2019, 103, 1497–1509. [Google Scholar] [CrossRef]
- Li, X.; Chen, D.; Carrión, V.J.; Revillini, D.; Yin, S.; Dong, Y.; Zhang, T.; Wang, X.; Delgado-Baquerizo, M. Acidification suppresses the natural capacity of soil microbiome to fight pathogenic Fusarium infections. Nat. Commun. 2023, 14, 5090. [Google Scholar] [CrossRef]
- Kasinathan, H. Influence of pH, Temperature, and Biofungicides on Clubroot of Canola. Master’s Thesis, University of Guelph, Guelph, ON, Canada, 2012. [Google Scholar]
- Gludovacz, T.V. Clubroot in Canola and Cabbage in Relation to Soil Temperature, Plant Growth and Host Resistance. Master’s Thesis, University of Guelph, Guelph, ON, Canada, 2013. [Google Scholar]
- Deora, A.; Gossen, B.D.; Walley, F.; McDonald, M.R. Boron reduces development of clubroot in canola. Can. J. Plant Pathol. 2011, 33, 475–484. [Google Scholar] [CrossRef]
- Webster, M.A.; Dixon, G.R. Calcium, pH and inoculum concentration influencing colonization by Plasmodiophora brassicae. Mycol. Res. 1991, 95, 64–73. [Google Scholar] [CrossRef]
- Gossen, B.D.; Deora, A.; Peng, G.; Hwang, S.F.; McDonald, M.R. Effect of environmental parameters on clubroot development and the risk of pathogen spread. Can. J. Plant Pathol. 2014, 36, 37–48. [Google Scholar] [CrossRef]
- Niu, J.; Chao, J.; Xiao, Y.; Chen, W.; Zhang, C.; Liu, X.; Rang, Z.; Yin, H.; Dai, L. Insight into the effects of different cropping systems on soil bacterial community and tobacco bacterial wilt rate. J. Basic Microbiol. 2007, 57, 3–11. [Google Scholar] [CrossRef]
- Chai, A.L.; Li, J.P.; Xie, X.W.; Shi, Y.X.; Li, B.J. Dissemination of Plasmodiophora brassicae in livestock manure detected by qPCR. Plant Pathol. 2016, 65, 137–144. [Google Scholar] [CrossRef]
- Ciavatta, C.; Govi, M.; Antisari, L.V.; Sequi, P. Determination of organic carbon in aqueous extracts of soils and fertilizers. Commun. Soil Sci. Plant Anal. 1991, 22, 795–807. [Google Scholar] [CrossRef]
- Li, J.G.; Ren, G.D.; Jia, Z.J.; Dong, Y.H. Composition and activity of rhizosphere microbial communities associated with healthy and diseased greenhouse tomatoes. Plant Soil 2004, 380, 337–347. [Google Scholar] [CrossRef]
- Shen, Z.Q.; Zhang, Q.; Liu, L.J.; Qiu, Y. Determination of available phosphorus in soil by sodium bicarbonate extraction Mo-Sb anti-spectrophotometry method. Environ. Monit. Forew. 2001, 3, 12–15. [Google Scholar]
- Mulvaney, R.L.; Yaremych, S.A.; Khan, S.A.; Swiader, J.M.; Horgan, B.P. Use of diffusion to determine soil cation-exchange capacity by ammonium saturation. Commun. Soil Sci. Plant Anal. 2004, 35, 51–67. [Google Scholar] [CrossRef]
- Bao, S.D. Analysis of water soluble salts in soil. In Soil and Agricultural Chemistry Analysis, 3rd ed.; Chinese Agricultural Press: Beijing, China, 2000; pp. 189–191. [Google Scholar]
- Bhering, A.D.S.; do Carmo, M.G.; Matos, T.D.S.; Lima, E.S.; do Amaral Sobrinho, N.M. Soil factors related to the severity of clubroot in Rio de Janeiro, Brazil. Plant Dis. 2007, 101, 1345–1353. [Google Scholar] [CrossRef] [PubMed]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Qi, G.; Ma, G.; Chen, S.; Lin, C.; Zhao, X. Microbial network and soil properties are changed in bacterial wilt-susceptible soil. Appl. Environ. Microbiol. 2019, 85, e00162-19. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.B.; He, J.Z.; Geisen, S.; Han, L.L.; Wang, J.T.; Shen, J.P.; Wei, W.X.; Fang, Y.T.; Li, P.P.; Zhang, L.M. Protist communities are more sensitive to nitrogen fertilization than other microorganisms in diverse agricultural soils. Microbiome 2009, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Jiang, Y.H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular ecological network analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef]
- Ling, N.; Wang, T.; Kuzyakov, Y. Rhizosphere bacteriome structure and functions. Nat. Commun. 2022, 13, 836. [Google Scholar] [CrossRef]
- Banerjee, S.; Kirkby, C.A.; Schmutter, D.; Bissett, A.; Kirkegaard, J.A.; Richardson, A.E. Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol. Biochem. 2016, 97, 188–198. [Google Scholar] [CrossRef]
- R Core Team. A Language and Environment for Statistical Computing, 3.6.0 edn.; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Shi, W.; Li, M.; Wei, G.; Tian, R.; Li, C.; Wang, B.; Lin, R.; Shi, C.; Chi, X.; Zhou, B.; et al. The occurrence of potato common scab correlates with the community composition and function of the geocaulosphere soil microbiome. Microbiome 2019, 7, 14. [Google Scholar] [CrossRef]
- Niwa, R.; Nomura, Y.; Osaki, M.; Ezawa, T. Suppression of clubroot disease under neutral pH caused by inhibition of spore germination of Plasmodiophora brassicae in the rhizosphere. Plant Pathol. 2008, 5, 445–452. [Google Scholar] [CrossRef]
- Bhering, A.S.; Carmo, M.G.F.; Coelho, I.S.; Lima, E.S.A.; de Carvalho, C.F.; Saraiva, A.L.R.F.; Passos, S.R.; Sobrinho, N.M.B.A. Soil management in a mountain agroecosystem and clubroot disease. Plant Pathol. 2020, 69, 302–309. [Google Scholar] [CrossRef]
- Gossen, B.D.; Kasinathan, H.; Cao, T.; Manolii, V.P.; Strelkov, S.E.; Hwang, S.F.; McDonald, M.R. Interaction of pH and temperature affect infection and symptom development of Plasmodiophora brassicae in canola. Can. J. Plant Pathol. 2013, 35, 294–303. [Google Scholar] [CrossRef]
- Shi, H.; Xu, P.; Wu, S.; Yu, W.; Cheng, Y.; Chen, Z.; Yang, X.; Yu, X.; Li, B.; Ding, A.; et al. Analysis of rhizosphere bacterial communities of tobacco resistant and non-resistant to bacterial wilt in different regions. Sci. Rep. 2022, 12, 18309. [Google Scholar] [CrossRef] [PubMed]
- Rousk, J.; Bååth, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef]
- Fox, N.M.; Hwang, S.F.; Manolii, V.P.; Turnbull, G.; Strelkov, S.E. Evaluation of lime products for clubroot (Plasmodiophora brassicae) management in canola (Brassica napus) croping systems. Can. J. Plant Pathol. 2022, 44, 21–38. [Google Scholar] [CrossRef]
- Dixon, G.R. Managing clubroot disease (caused by Plasmodiophora brassicae Wor.) by exploiting the interactions between calcium cyanamide fertilizer and soil microorganisms. J. Agric. Sci. 2017, 155, 527–543. [Google Scholar] [CrossRef]
- Saraiva, A.L.R.F.; Bhering, A.S.; do Carmo, M.G.F.; Andreote, F.D.; Dias, A.C.F.; Coelho, I.D.S. Bacterial composition in brassica-cultivated soils with low and high severity of clubroot. J. Phytopathol. 2020, 168, 613–619. [Google Scholar] [CrossRef]
- Jia, M.; Sun, X.; Chen, M.; Liu, S.; Zhou, J.; Peng, X. Deciphering the microbial diversity associated with healthy and wilted Paeonia suffruticosa rhizosphere soil. Front. Microbiol. 2022, 13, 967601. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Jing, T.; Chen, Y.; Wang, F.; Qi, D.; Feng, R.; Xie, J.; Li, H. Deciphering microbial diversity associated with Fusarium wilt-diseased and disease-free banana rhizosphere soil. BMC Microbiol. 2019, 19, 161. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, R.; Xue, C.; Xun, W.; Sun, L.; Xu, Y.; Shen, Q. Pyrosequencing reveals contrasting soil bacterial diversity and community structure of two main winter wheat cropping systems in China. Microb. Ecol. 2004, 67, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Ruan, Y.; Xue, C.; Zhong, S.; Li, R.; Shen, Q. Soils naturally suppressive to banana Fusarium wilt disease harbor unique bacterial communities. Plant Soil 2015, 393, 21–33. [Google Scholar] [CrossRef]
- Van Elsas, J.D.; Chiurazzi, M.; Mallon, C.A.; Elhottovā, D.; Krištůfek, V.; Salles, J.F. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. USA 2022, 109, 1159–1164. [Google Scholar] [CrossRef]
- Liu, L.; Yan, Y.; Ding, H.; Zhao, J.; Cai, Z.; Dai, C.; Huang, X. The fungal community outperforms the bacterial community in predicting plant health status. Appl. Microbiol. Biotechnol. 2021, 105, 6499–6513. [Google Scholar] [CrossRef]
- Ou, Y.; Penton, C.R.; Geisen, S.; Shen, Z.; Sun, Y.; Lv, N.; Wang, B.; Ruan, Y.; Xiong, W.; Li, R.; et al. Deciphering underlying drivers of disease suppressiveness against pathogenic Fusarium oxysporum. Front. Microbiol. 2019, 10, 2535. [Google Scholar] [CrossRef]
- Cha, J.Y.; Han, S.; Hong, H.J.; Cho, H.; Kim, D.; Kwon, Y.; Crüsemann, M.; Lee, Y.B.; Kim, J.F.; Giaever, G.; et al. Microbial and biochemical basis of a Fusarium wilt-suppressive soil. ISME J. 2016, 10, 119–129. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Diamond, S.; Butterfield, C.N.; Thomas, B.C.; Banfield, J.F. Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature 2018, 558, 440–444. [Google Scholar] [CrossRef]
- Prieto, C. Characterization of nonribosomal peptide synthetases with NRPSsp. In Nonribosomal Peptide and Polyketide Biosynthesis; Bradley, S.E., Ed.; Springer: New York, NY, USA, 2016; Volume 1401, pp. 273–278. [Google Scholar]
- Raaijmakers, J.M.; De Bruijn, I.; Nybroe, O.; Ongena, M. Natural functions of lipopeptides from Bacillus and Pseudomonas: More than surfactants and antibiotics. FEMS Microbiol. Rev. 2010, 34, 1037–1062. [Google Scholar] [CrossRef] [PubMed]
- Soytong, K.; Kanokmedhakul, S.; Kukongviriyapa, V.; Isobe, M. Application of Chaetomium species (Ketomium) as a new broad spectrum biological fungicide for plant disease control. Fungal Divers. 2001, 7, 1–15. [Google Scholar]
- Pothiraj, G.; Hussain, Z.; Singh, A.K.; Solanke, A.U.; Aggarwal, R.; Ramesh, R.; Shanmugam, V. Characterization of Fusarium spp. inciting vascular wilt of tomato and its management by a Chaetomium-based biocontrol consortium. Front. Plant Sci. 2021, 12, 748013. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Y.; Wei, F.; Zhao, L.; Zhou, J.; Qi, G.; Ma, Z.; Zhu, H.; Feng, H.; Feng, Z. Applications of Chaetomium globosum CEF-082 improve soil health and mitigate the continuous croping obstacles for Gossypium hirsutum. Ind. Crops Prod. 2023, 197, 116586. [Google Scholar] [CrossRef]
- Nerva, L.; Zanzotto, A.; Gardiman, M.; Gaiotti, F.; Chitarra, W. Soil microbiome analysis in an ESCA diseased vineyard. Soil Biol. Biochem. 2019, 135, 60–70. [Google Scholar] [CrossRef]
- Chu, C.; Liu, B.; Liu, J.; He, J.; Lv, L.; Wang, H.; Xie, X.; Tao, Q.; Chen, Q. Phytoremediation of acetochlor residue by transgenic Arabidopsis expressing the acetochlor N-dealkylase from Sphingomonas wittichii DC-6. Sci. Total Environ. 2020, 728, 138687. [Google Scholar] [CrossRef] [PubMed]
- Lombardino, J.; Bijlani, S.; Singh, N.K.; Wood, J.M.; Barker, R.; Gilroy, S.; Wang, C.C.C.; Venkateswaran, K. Genomic characterization of potential plant growth-promoting features of Sphingomonas strains isolated from the International Space Station. Microbiol. Spectr. 2022, 10, e01994-21. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wei, Y.; Yan, T.; Wang, C.; Chao, Y.; Jia, M.; An, L.; Sheng, H. Sphingomonas sp. Hbc-6 alters physiological metabolism and recruits beneficial rhizosphere bacteria to improve plant growth and drought tolerance. Front. Plant Sci. 2022, 13, 1002772. [Google Scholar] [CrossRef] [PubMed]
- Mazoyon, C.; Hirel, B.; Pecourt, A.; Catterou, M.; Gutierrez, L.; Sarazin, V.; Dubois, F.; Duclercq, J. Sphingomonas sediminicola is an endosymbiotic bacterium able to induce the formation of root nodules in Pea (Pisum sativum L.) and to enhance plant biomass production. Microorganisms 2023, 11, 199. [Google Scholar] [CrossRef]
- Asaf, S.; Numan, M.; Khan, A.L.; Al-Harrasi, A. Sphingomonas: From diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 2020, 40, 138–152. [Google Scholar] [CrossRef]
- Morrien, E.; Hannula, S.E.; Snoek, L.B.; Helmsing, N.R.; Zweers, H.; de Hollander, M.; Soto, R.L.; Bouffaud, M.L.; Buée, M.; Dimmers, W.; et al. Soil networks become more connected and take up more carbon as nature restoration progresses. Nat. Commun. 2017, 8, 14349. [Google Scholar] [CrossRef]
- Ratzke, C.; Barrere, J.; Gore, J. Strength of species interactions determines biodiversity and stability in microbial communities. Nat. Ecol. Evol. 2020, 4, 376–383. [Google Scholar] [CrossRef]
- Schmitt, S.; Tsai, P.; Bell, J.; Fromont, J.; Ilan, M.; Lindquist, N.; Perez, T.; Rodrigo, A.; Schupp, P.J.; Vacelet, J.; et al. Assessing the complex sponge microbiota: Core, variable and species-specific bacterial communities in marine sponges. ISME J. 2012, 6, 564–576. [Google Scholar] [CrossRef]
- Wagg, C.; Schlaeppi, K.; Banerjee, S.; Kuramae, E.E.; van der Heijden, M.G. Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 2019, 10, 4841. [Google Scholar] [CrossRef]
- Coyte, K.Z.; Schluter, J.; Foster, K.R. The ecology of the microbiome: Networks, competition, and stability. Science 2015, 350, 663–666. [Google Scholar] [CrossRef]
- Palmer, J.D.; Foster, K.R. Bacterial species rarely work together. Science 2022, 376, 581–582. [Google Scholar] [CrossRef]
- Price, G.W.; Langille, M.G.; Yurgel, S.N. Microbial co-occurrence network analysis of soils receiving short-and long-term applications of alkaline treated biosolids. Sci. Total Environ. 2021, 751, 141687. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.; Weisenhorn, P.; Gilbert, J.A.; Chu, H. Wheat rhizosphere harbors a less complex and more stable microbial co-occurrence pattern than bulk soil. Soil Biol. Biochem. 2018, 125, 251–260. [Google Scholar] [CrossRef]
- Jäschke, D.; Dugassa-Gobena, D.; Karlovsky, P.; Vidal, S.; Ludwig-Müller, J. Suppression of clubroot (Plasmodiophora brassicae) development in Arabidopsis thaliana by the endophytic fungus Acremonium alternatum. Plant Pathol. 2010, 59, 100–111. [Google Scholar] [CrossRef]
- Koch, H.; Lücker, S.; Albertsen, M.; Kitzinger, K.; Herbold, C.; Spieck, E.; Nielsen, P.H.; Wagner, M.; Daims, H. Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc. Natl. Acad. Sci. USA 2015, 112, 11371–11376. [Google Scholar] [CrossRef] [PubMed]
- Martikainen, P.J. Heterotrophic nitrification-An eternal mystery in the nitrogen cycle. Soil Biol. Biochem. 2022, 168, 108611. [Google Scholar] [CrossRef]
- Kuypers, M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef]
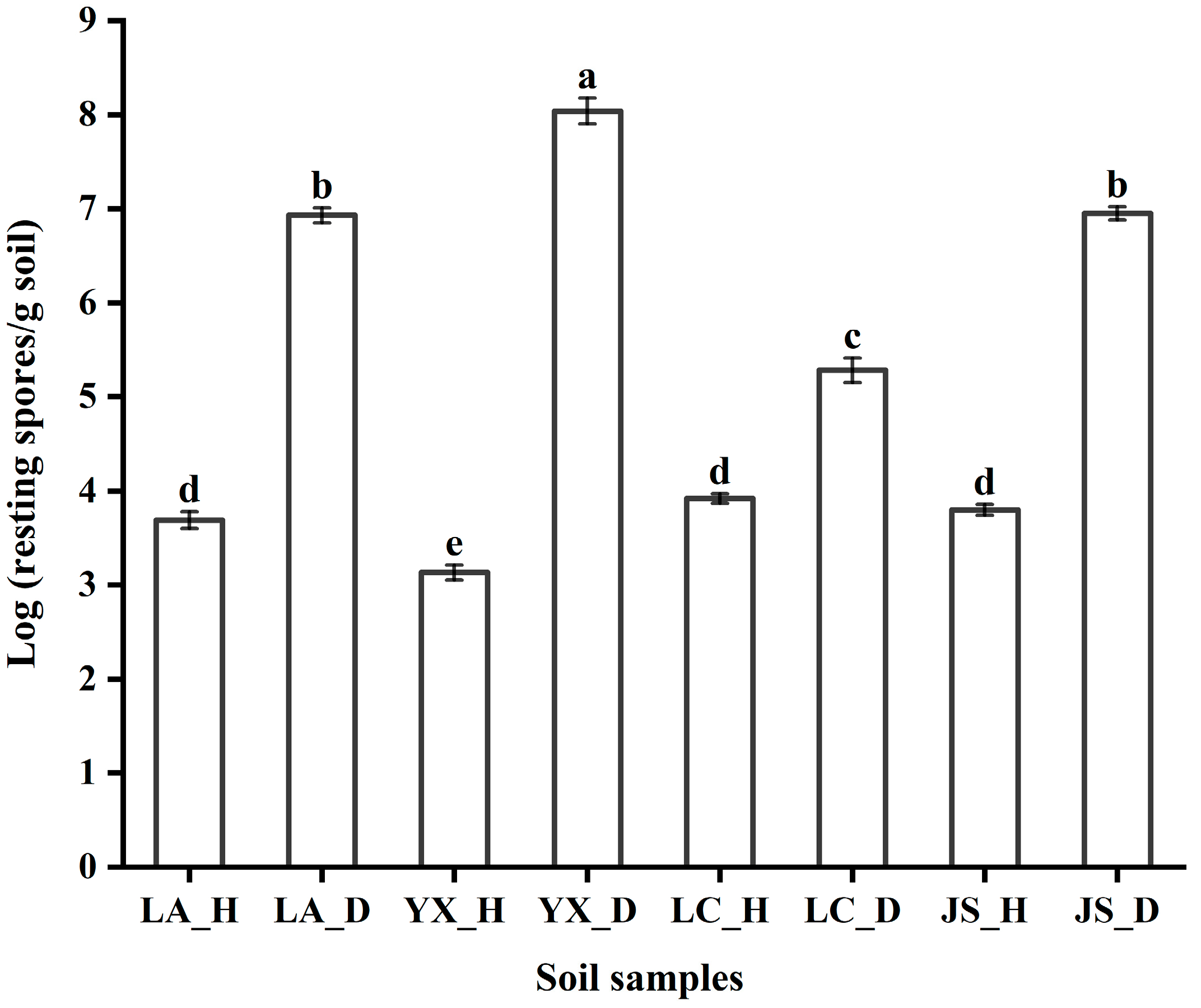
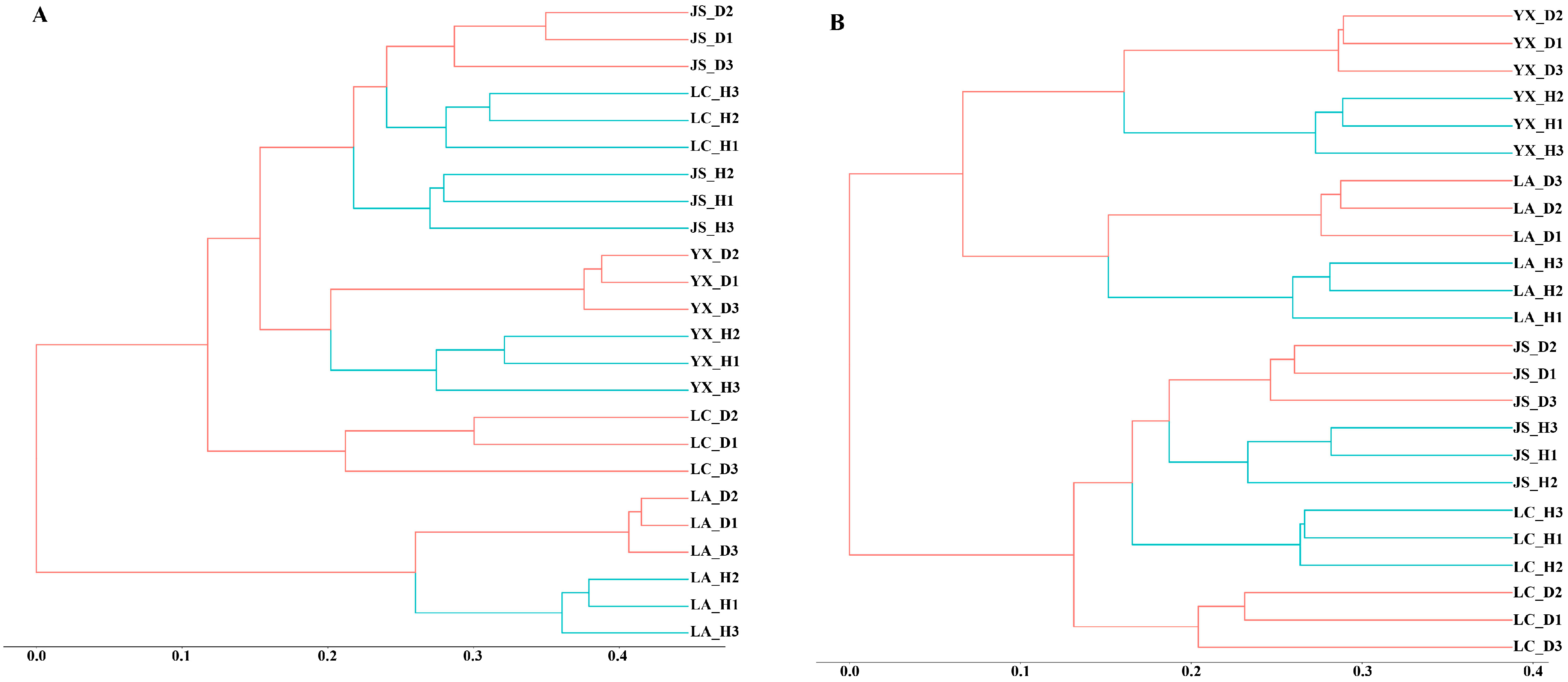

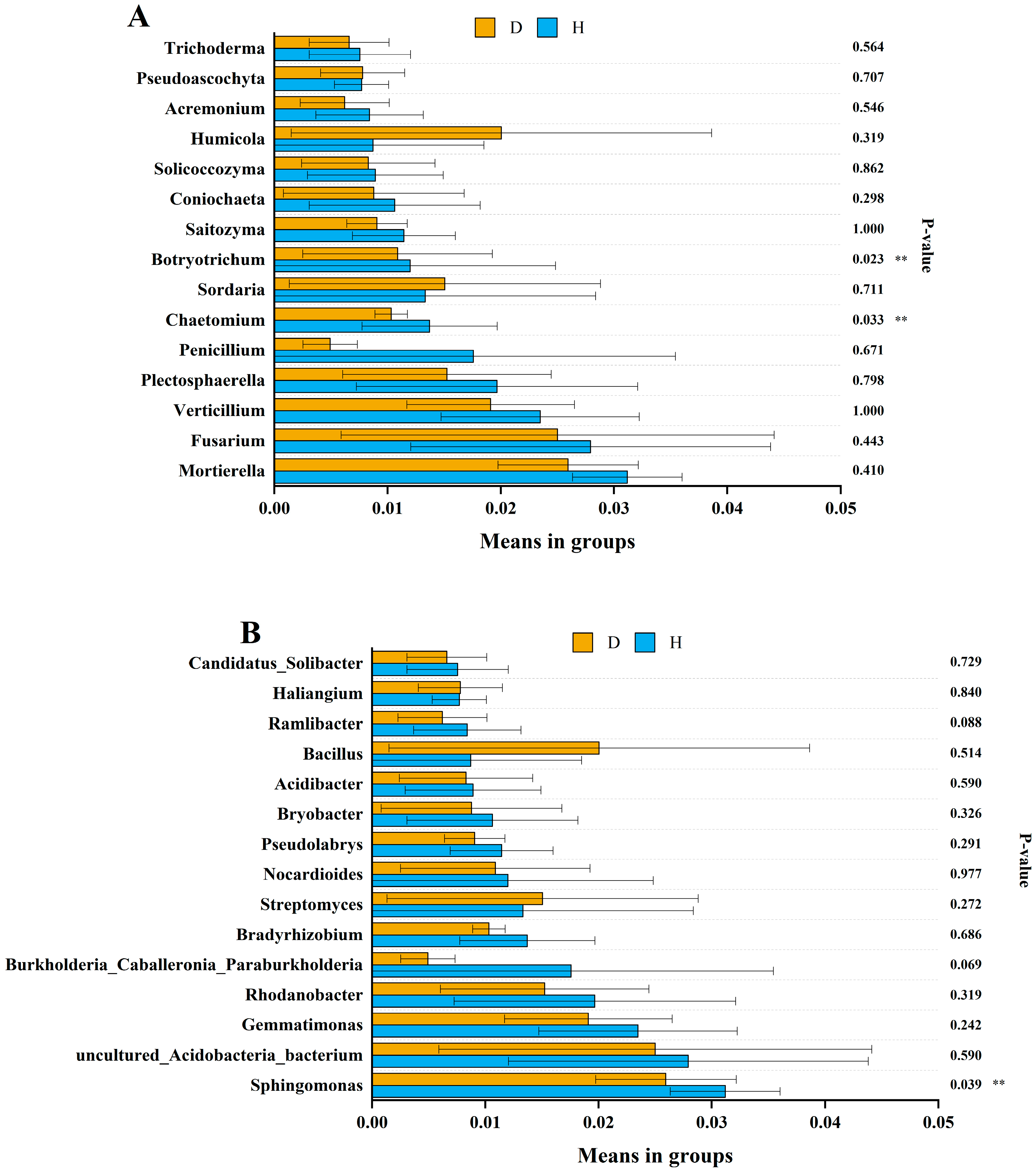
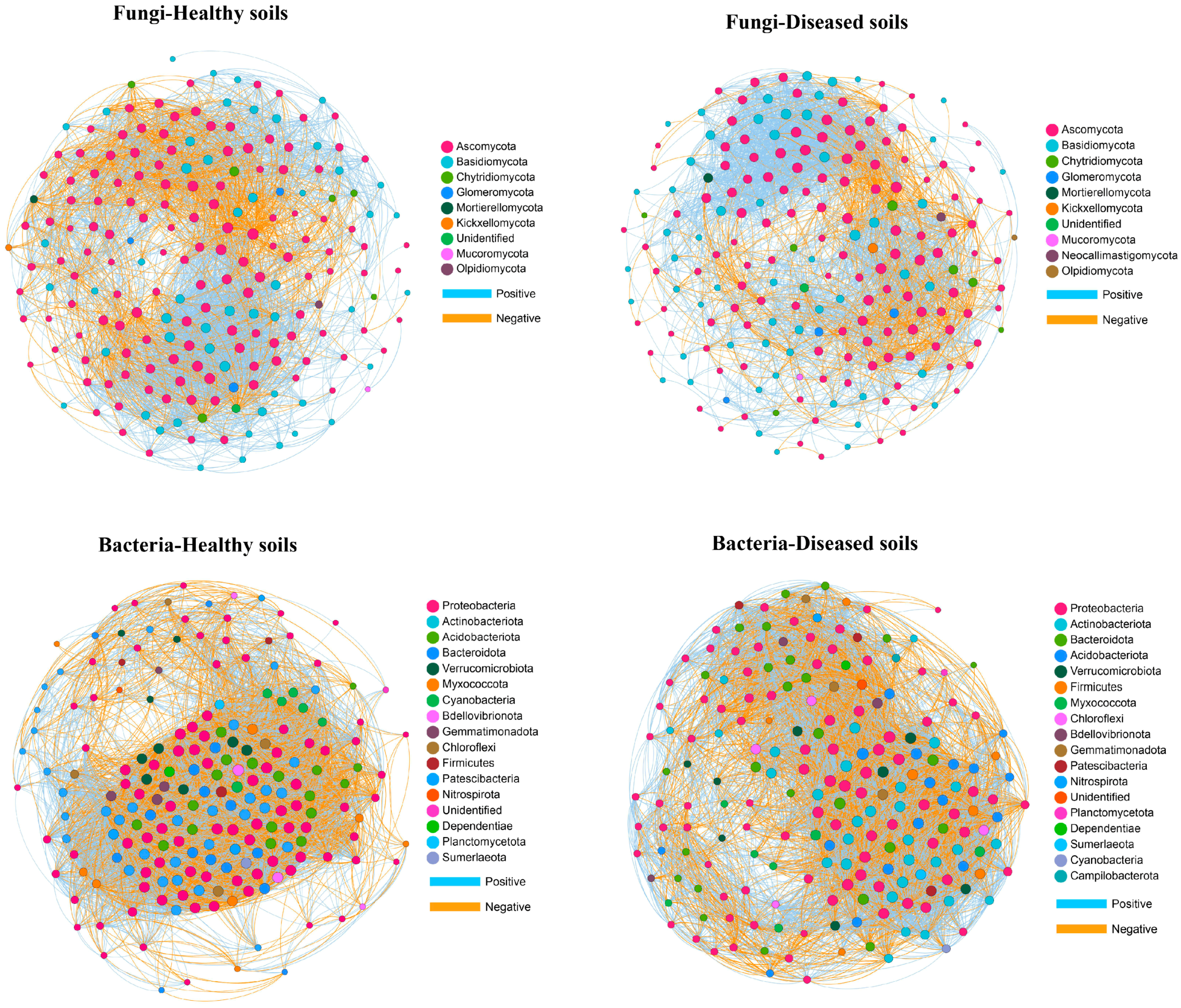
| Sample Location | Clubroot Infection, % | Coordinates | Altitude (m) | Mean Annual Temperature (°C) | Mean Annual Precipitation (mm) | Year | Soil Type | Crops |
|---|---|---|---|---|---|---|---|---|
| Licang, Shandong Province | 55.38% | 36.1511° N 120.4267° E | 99 | 14.5 | 732.0 | 2020 | Luvisols | Chinese cabbage |
| Lichuan, Hubei Province | 77.46% | 30.3212° N 108.6638° E | 1595 | 12.3 | 1200.0 | 2020 | Cambisols | Chinese cabbage |
| Jianshi, Hubei Province | 72.29% | 30.7344° N 109.7990° E | 726 | 16.0 | 1480.0 | 2020 | Cambisols | Chinese cabbage |
| Youxian, Sichuan Province | 85.73% | 31.5959° N 104.8624° E | 568 | 16.4 | 969.6 | 2020 | Cambisols | Cabbage |
| Sample Location | Soil Trait | pH | SOM (g/kg) | TN (g/kg) | AN (mg/kg) | AP (mg/kg) | AK (mg/kg) | CEC (col (+)/kg) | EC (ms/cm) | Ca (g/kg) | B (mg/kg) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Licang, Shandong Province | Healthy | 7.52 ± 0.01 a | 19.97 ± 0.94 c | 0.89 ± 0.01 d | 94.73 ± 2.38 e | 99.20 ± 0.42 d | 113.83 ± 0.47 f | 6.63 ± 0.41 d | 233.67 ± 5.44 c | 2.57 ± 0.04 c | 0.31 ± 0.04 ab |
| Diseased | 7.54 ± 0.00 a | 15.56 ± 0.79 d | 0.82 ± 0.00 d | 102.20 ± 1.14 e | 97.61 ± 0.54 d | 120.83 ± 0.47 f | 6.57 ± 0.25 d | 330.67 ± 17.25 b | 2.36 ± 0.03 d | 0.34 ± 0.02 ab | |
| Youxian, Sichuan Province | Healthy | 7.25 ± 0.02 b | 22.55 ± 0.30 bc | 1.44 ± 0.03 c | 144.67 ± 0.66 d | 48.10 ± 0.56 e | 139.17 ± 4.92 e | 12.97 ± 0.25 ab | 66.53 ± 2.99 d | 4.27 ± 0.04 a | 0.41 ± 0.03 a |
| Diseased | 6.11 ± 0.04 c | 24.15 ± 0.38 b | 1.62 ± 0.03 b | 203.93 ± 0.66 c | 44.22 ± 0.55 e | 247.50 ± 2.83 d | 14.17 ± 0.34 a | 43.37 ± 4.29 de | 3.90 ± 0.02 b | 0.39 ± 0.05 a | |
| Lichuan, Hubei Province | Healthy | 5.03 ± 0.05 f | 35.87 ± 0.04 a | 2.29 ± 0.01 a | 245.00 ± 1.14 b | 153.05 ± 1.80 a | 389.50 ± 3.27 b | 12.77 ± 0.41 ab | 54.35 ± 10.30 de | 1.53 ± 0.02 h | 0.22 ± 0.01 c |
| Diseased | 5.20 ± 0.03 e | 37.72 ± 0.93 a | 2.31 ± 0.04 a | 257.60 ± 1.14 ab | 119.28 ± 0.85 c | 533.17 ± 7.76 a | 12.50 ± 1.02 b | 66.36 ± 3.16 d | 2.26 ± 0.02 e | 0.25 ± 0.02 bc | |
| Jianshi, Hubei Province | Healthy | 5.50 ± 0.01 d | 36.50 ± 1.45 a | 2.27 ± 0.04 a | 260.40 ± 2.29 a | 143.63 ± 2.75 b | 333.83 ± 2.62 c | 10.77 ± 0.09 c | 761.00 ± 25.15 a | 1.68 ± 0.02 g | 0.34 ± 0.02 ab |
| Diseased | 5.13 ± 0.02 e | 36.64 ± 1.19 a | 2.23 ± 0.02 a | 259.93 ± 9.86 a | 145.19 ± 0.58 b | 340.83 ± 2.05 c | 12.30 ± 0.33 bc | 25.20 ± 1.26 e | 1.82 ± 0.02 f | 0.35 ± 0.04 ab |
| Sample Location | Sample Trait | Fungi | Bacteria | ||||
|---|---|---|---|---|---|---|---|
| OTU Number | Chao1 | Shannon | OTU Number | Chao1 | Shannon | ||
| Licang, Shandong Province | Healthy | 437 ± 13.77 cd | 697 ± 56.21 c | 4.30 ± 0.09 bc | 3237 ± 57.12 b | 4204 ± 137.42 b | 10.07 ± 0.07 a |
| Diseased | 361 ± 71.30 d | 609 ± 156.53 c | 2.39 ± 0.09 d | 2860 ± 45.15 c | 3908 ± 43.01 b | 9.63 ± 0.11 c | |
| Youxian, Sichuan Province | Healthy | 852 ± 48.24 a | 1235± 49.45 a | 5.89 ± 0.24 a | 3409 ± 38.58 a | 4703 ± 146.66 a | 9.99 ± 0.01 b |
| Diseased | 725 ± 56.10 ab | 1136 ± 44.29 ab | 5.05 ± 0.23 ab | 2703 ± 32.07 c | 3954 ± 85.93 b | 9.31 ± 0.01 d | |
| Lichuan, Hubei Province | Healthy | 574 ± 1.63 bc | 809 ± 19.93 bc | 4.53 ± 0.26 bc | 1918 ± 48.19 f | 2690 ± 75.72 d | 8.84 ± 0.16 e |
| Diseased | 483 ± 12.23 bc | 775 ± 27.53 bc | 3.99 ± 0.45 c | 2307 ± 45.11 d | 3117 ± 82.60 c | 9.24 ± 0.04 dc | |
| Jianshi, Hubei Province | Healthy | 641 ± 173.36 ab | 938 ± 209.11 ab | 4.82 ± 0.46 bc | 2124 ± 28.16 e | 2968 ± 143.29 cd | 9.11 ± 0.09 dc |
| Diseased | 630 ± 68.78 ab | 944 ± 119.06 ab | 4.71 ± 0.12 bc | 2271 ± 71.98 de | 3265 ± 67.68 c | 8.96 ± 0.09 de | |
| Sample Trait | OTU Number | Chao1 | Shannon | |
|---|---|---|---|---|
| Fungi | Healthy | 626 ± 174.66 a | 920 ± 229.74 a | 4.88 ± 0.67 a |
| Diseased | 550 ± 150.48 a | 866 ± 220.62 a | 4.03 ± 1.06 b | |
| Bacteria | Healthy | 2672 ± 659.28 a | 3641 ± 847.00 a | 9.50 ± 0.55 a |
| Diseased | 2535 ± 257.83 a | 3560 ± 380.82 a | 9.29 ± 0.25 a |
| pH | SOM | TON | AN | AP | AK | CEC | EC | Ca | B | |
|---|---|---|---|---|---|---|---|---|---|---|
| Pearson | −0.49 | 0.421 | 0.577 | 0.565 | −0.302 | 0.43 | 0.983 | −0.629 | 0.456 | 0.257 |
| p-value | 0.51 | 0.579 | 0.423 | 0.435 | 0.698 | 0.57 | 0.017 * | 0.371 | 0.544 | 0.743 |
| Fungal Genera | Pearson | p-Value | Bacterial Genera | Pearson | p Value |
|---|---|---|---|---|---|
| Mortierella | −0.006 | 0.978 | Sphingomonas | −0.245 | 0.249 |
| Fusarium | 0.178 | 0.407 | uncultured_Acidobacteria_bacterium | −0.098 | 0.649 |
| Verticillium | 0.176 | 0.41 | Gemmatimonas | −0.323 | 0.123 |
| Plectosphaerella | −0.157 | 0.465 | Rhodanobacter | −0.097 | 0.652 |
| Penicillium | −0.022 | 0.919 | Burkholderia_Caballeronia_Paraburkholderia | −0.389 | 0.06 |
| Chaetomium | −0.507 | 0.011 * | Bradyrhizobium | −0.236 | 0.267 |
| Sordaria | −0.256 | 0.227 | Streptomyces | 0.172 | 0.423 |
| Botryotrichum | −0.407 | 0.049 * | Nocardioides | −0.002 | 0.991 |
| Saitozyma | −0.077 | 0.719 | Pseudolabrys | −0.240 | 0.258 |
| Coniochaeta | −0.248 | 0.243 | Bryobacter | −0.078 | 0.719 |
| Solicoccozyma | 0.009 | 0.966 | Acidibacter | −0.113 | 0.598 |
| Humicola | −0.250 | 0.238 | Bacillus | 0.527 | 0.008 ** |
| Acremonium | −0.453 | 0.026 * | Ramlibacter | −0.358 | 0.086 |
| Pseudoascochyta | −0.004 | 0.987 | Haliangium | −0.264 | 0.213 |
| Trichoderma | 0.111 | 0.605 | Candidatus_Solibacter | −0.150 | 0.485 |
| Fungal Networks | Bacterial Networks | |||
|---|---|---|---|---|
| Healthy Soils | Diseased Soils | Healthy Soils | Diseased Soils | |
| Number of positive links | 2497 | 2238 | 4721 | 3852 |
| Number of negative links | 1160 | 742 | 4068 | 2898 |
| Positive link/negative link ratio | 2.15 | 3.02 | 1.16 | 1.33 |
| Network density | 0.19 | 0.15 | 0.44 | 0.34 |
| Clustering coefficient | 0.57 | 0.53 | 0.75 | 0.68 |
| Modularity | 0.39 | 0.40 | 0.18 | 0.31 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, H.; Chai, A.; Lin, Z.; Shi, Y.; Xie, X.; Li, L.; Fan, T.; Xiang, S.; Xie, J.; Li, B. Deciphering Differences in Microbial Community Diversity between Clubroot-Diseased and Healthy Soils. Microorganisms 2024, 12, 251. https://doi.org/10.3390/microorganisms12020251
Kang H, Chai A, Lin Z, Shi Y, Xie X, Li L, Fan T, Xiang S, Xie J, Li B. Deciphering Differences in Microbial Community Diversity between Clubroot-Diseased and Healthy Soils. Microorganisms. 2024; 12(2):251. https://doi.org/10.3390/microorganisms12020251
Chicago/Turabian StyleKang, Huajun, Ali Chai, Zihan Lin, Yanxia Shi, Xuewen Xie, Lei Li, Tengfei Fan, Sheng Xiang, Jianming Xie, and Baoju Li. 2024. "Deciphering Differences in Microbial Community Diversity between Clubroot-Diseased and Healthy Soils" Microorganisms 12, no. 2: 251. https://doi.org/10.3390/microorganisms12020251
APA StyleKang, H., Chai, A., Lin, Z., Shi, Y., Xie, X., Li, L., Fan, T., Xiang, S., Xie, J., & Li, B. (2024). Deciphering Differences in Microbial Community Diversity between Clubroot-Diseased and Healthy Soils. Microorganisms, 12(2), 251. https://doi.org/10.3390/microorganisms12020251






