Abstract
Gut luminal dysbiosis and pathobiosis result in compositional and biodiversified alterations in the microbial and host co-metabolites. The primary mechanism of bacterial evolution is horizontal gene transfer (HGT), and the acquisition of new traits can be achieved through the exchange of mobile genetic elements (MGEs). Introducing genetically engineered microbes (GEMs) might break the harmonized balance in the intestinal compartment. The present objectives are: 1. To reveal the role played by the GEMs’ horizontal gene transfers in changing the landscape of the enteric microbiome eubiosis 2. To expand on the potential detrimental effects of those changes on the human genome and health. A search of articles published in PubMed/MEDLINE, EMBASE, and Scielo from 2000 to August 2023 using appropriate MeSH entry terms was performed. The GEMs’ horizontal gene exchanges might induce multiple human diseases. The new GEMs can change the long-term natural evolution of the enteric pro- or eukaryotic cell inhabitants. The worldwide regulatory authority’s safety control of GEMs is not enough to protect public health. Viability, biocontainment, and many other aspects are only partially controlled and harmful consequences for public health should be avoided. It is important to remember that prevention is the most cost-effective strategy and primum non nocere should be the focus.
1. Introduction
Many essential functions of the human body depend on the enteric symbiotic microbiota composition and biodiversity, essential components for human health. This intricate host–taxa relationship is a dynamic result of their long-term coevolution. This eubiosis harmonically maintains the host’s nutrition, metabolic passways, physiology, protective immune system and even behavior to the extent that we need them and cannot live without them. Greater phyla diversity is associated with microbiota resilience, sustained stability and greater ability to perform metabolic functions. The loss of microbiota phylogenic diversity and enhanced gut dysbiotic composition were associated with the Western lifestyle and several inflammatory, neurodegenerative, neurodevelopmental, infectious, metabolic, cancer and autoimmune diseases (ADs) that put human health at risk [1,2,3,4,5].
The primary mechanism of bacterial evolution is horizontal gene transfer (HGT), and new traits can be acquired through this mobile element exchange. Introducing GEMs might break the harmonized balance in the intestinal compartment [1,6]. The stable temperature, constant physicochemical conditions, continuous food supply, extremely high concentration of prokaryotic cells and phages, and plenty of opportunities for conjugation on the surfaces of host tissues and food particles represent one of the most favorable ecological niches for GEM-originated horizontal gene exchange of detrimental and harmful genetic sequences. Newly developed techniques of bacterial-mediated drug delivery have recently emerged using genetically engineered microbes aiming to locally deliver recombinant therapeutic proteins to the human gut. They are often called live biotherapeutic products, but they deliberately embed potential risks.
Entry of potentially unsafe GEMs into the human gut lumen can impact and change the selective pressures on gut microbiota and potentially contaminate the human microbiome with harmful genes exchanged horizontally. Presumably, the gut microbiome responds to these changes by genetic restructuring of gut populations, driven mainly via horizontal gene exchange. The objectives of the present narrative review are: 1. To reveal the role played by GEMs’ horizontal gene transfers in the changing landscape of the enteric microbiome eubiosis; 2. To expand on the potential detrimental effects of those changes on human health in general and autoimmune diseases in particular; 3. To warn against the impact of the new GEMs intestinal dwellers on many other pro- or eukaryotic cells, including changing the human genome. The following are some of the scientifically reported examples.
2. Numerous Harmful Mobile Genetic Elements (MGEs) Can Be Transferred to the Human Microbiome
Through their genomes, bacteria are subjected to rapid mutations and numerous rearrangements or HGT among and/or within bacterial species. Those MGEs, represented by bacteriophages, transposons, plasmids, and other pathogenic islands, represent a substantial amount of the microbial genome. Applying GEMs to the intestinal lumen can annulate the expression of beneficial genes while inducing the secretion of detrimental proteins. Alternatively, the GEMs can acquire the MGEs in the gut lumen. The following are various major and harmful clinical examples (Figure 1, Table 1).
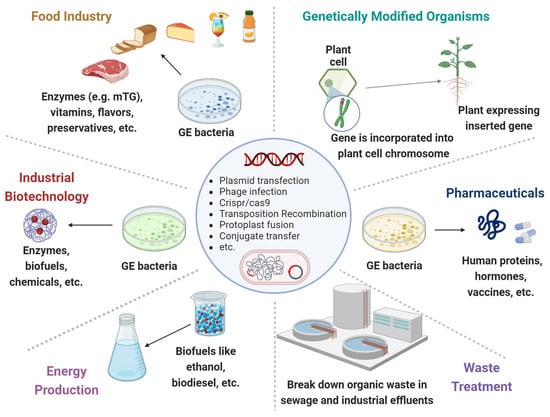
Figure 1.
Genetically engineered microorganisms (GEMs) applications. GEMs have a wide range of applications across various fields due to their versatility and the precision of genetic engineering techniques. Food Industry: Production of vitamins, flavors, enzymes, and preservatives. They can help in improving the nutritional value, taste, and shelf-life of food products. Agriculture: Promote plant growth, increase nutrient uptake, and protect plants from pests and diseases. Medicine and Health Care: Cost-effective production of pharmaceuticals, including insulin, growth hormones and vaccines. Waste Treatment: Break down hazardous substances like oil spills, heavy metals and other toxic chemicals. Energy Production: The production of biofuels like ethanol and biodiesel. Industrial Biotechnology: Improve chemical production to increase yields and reduce environmental impacts.
- Antibiotic resistance genes (ARGs) and multidrug resistance (MDR) genes are the most reported [1,7,8,9]. Less reported but not less important is the development of resistance of bacteria to phages [10,11], drug resistance to cancer therapy [12], resilience against antimicrobial defensive factors [13] and the MDR genes transfer along the food chain, including by contaminated and industrially processed nutrients [14]. The emergence of the resistome represents a worldwide health threat driven by the increasing unnecessary use of antibiotics and anticancer therapy. It occurs mainly by accumulating ARGs and MDR genes on MGEs, which is made possible by HGT [1,15]. Even the frequently consumed Lactobacillus reuteri was reported to carry ARGs [16,17]. The ARG does not originate only from human antibiotic consumption—antibiotic residue in food from animal sources can also drive the resistome [14,18]. Most recently, a high rate of ARG carried by Enterobacterales and diarrheagenic Escherichia coli in healthy donors screened for human fecal transplantation was noted [19]. The authors recommended multiplex PCR panels for stool donor screening. One wonders if the GEMs, said to benefit human health, are screened for ARGs or MDR genes.
- Microbial-engineered enzymes are an exponentially growing area that has become indispensable to processed food production, pharmaceuticals, and numerous other commercial goods [20]. Despite their beneficial effects on the processed food industries with increased production yields and “enhancing quality and sustainability” [21], multiple scientific publications are calling for a reassessment of their safety [22,23,24,25,26]. Intriguingly, a recent call was to reevaluate the GRAS definition allocated to various processed food additive ingredients. More reliable and updated approaches are offered to enzyme and other food nutritional categories for a more scientifically rigorous, sound and transparent application of the GRAS concept [27,28,29,30,31,32]. Moreover, a call to label, declare utilization and ensure consumer transparency regarding GEM enzymes is expressed in multiple scientific publications [28,33,34,35].Many nutritional components and nutrients are treated by GEM enzymes, resulting in post-translational modified proteins, turning naïve peptides into immunogenic complexes [2,30,32,36]. There are multiple examples of genetically engineered microbial enzymes; hence, one example will be expanded, namely the microbial transglutaminase (mTG).Microbial transglutaminase is a frequently used processed food additive, and its cross-linked complexes usage is expanding exponentially. The enzyme was classified as a processing aid and was granted the GRAS (generally recognized as safe) definition decades ago, thus avoiding a thorough assessment according to current criteria of toxicity and public health safety [24,25,26,37,38,39].In contrast to the manufacturer’s declarations and claims, mTG and/or its transamidated complexes are proinflammatory, immunogenic, allergenic, pathogenic and potentially toxic, hence compromising public health [24,25,26]. Being a member of the transglutaminase family and functionally imitating the tissue transglutaminase to demidate or transamidate gliadin peptides, it was recently reported as a potential inducer of celiac disease [26,40,41,42]. In addition, its family member, the tissue transglutaminase, is a well-known inflammation inducer, fibrosis mediator and is heavily involved in sepsis [43,44]. Since mTG functionally imitates its endogenous member, one wonders if it contributes to those morbid conditions.Microbial transglutaminase and its docked complexes have numerous detrimental effects. Interestingly, in contrast to many publications showing the positive and beneficial aspects of mTG usage [45,46,47,48,49,50], there is evidence for the negative and harmful aspects of enzyme usage that might impact and compromise public health [25,26,28]. The debate between the GRAS category allocated by the FDA regulatory authorities for safe mTG consumption versus many critical scientific publications is ongoing. Several national regulatory committees have warned the public about the hazardous effects of mTGs [24,25,26]. In the case of mTG, it is possible for the gene responsible for its production to be transferred horizontally between microorganisms and even to eukaryotes [1,51,52]. Indeed, MGEs with mTG activity can potentially be transferred by HGT in between prokaryotes. Their presence in a gut luminal cellular compartment presents new opportunities for HGT, with the risk of inhabiting eukaryotic hosts [1,52,53]. One of the hypothetical scenarios is the acquisition of a classic microbial survival factor, such as a Trojan horse, against host self-defense barriers [1,25,26,54]. This gene exchange can happen through mechanisms like plasmid transfer or the incorporation of the transglutaminase gene into a MGE that can be transferred between bacteria. It is worth noting that the specific mechanisms and frequency of HGT for the mTG gene may depend on the particular microorganisms involved and the environmental conditions. The efforts to improve mTG production, thermostability and pH dependency by genetic engineering may do the opposite by enhancing the detrimental effects of the manipulated enzyme [55]. Finally, the fact that mTG is a bacterial survival factor can represent a significant positive selective pressure in the harsh, overcrowded luminal compartment [1,2], enhancing its HGT to other intestinal prokaryotic dwellers. It can be summarized that the mTG acts as a double-edged sword, protecting the microbes to survive in the gut lumen, hence compromising human health [24,25,26,54].
- The place of probiotic consumption should be highlighted in terms of their side effects. Drug resistance remains a universal threat, and the fad of probiotic consumption, many of which contain antibiotic-resistant elements, is a major and serious health concern [56,57,58,59]. In 2023, emerging issues in probiotic safety arose. Whole-genome sequencing should be implemented to detect virulence factors, toxins, ARGs and other detrimental MGEs [60]. The clear assignment of species and strain identity risks to vulnerable populations and the need for adverse event reporting are important topics to regulate.Engineered probiotics through gene editing is an emerging domain. Despite the reported clinical benefits for inflammatory bowel disease, infectious, tumor and metabolic diseases, tight regulatory measures are lacking [61]. Engineered and naïve probiotics compete with the luminal microbiome for nutrients or ecological niches and thus might affect the diversity and composition of intestinal microbiota. Human health can be more affected by their interaction with the luminal lipid metabolism [62]. Once again, consumer transparency, visible labeling and safety regulations are far from satisfactory.
- Genetically modified (GM) plants might possess beneficial traits like resistance to drought, pests and diseases, fighting climate change, improved agricultural and industrial production and enhanced nutrition. However, it also has a risky side to humans, animals and environmental health that should be regulated by national food security and regulatory authorities [63]. Mobile elements such as modified DNA can be laterally transferred to other recipients, spanning prokaryotes, eukaryotes and even to people [1,63]. More so, delaying tightened regulation risks facing increased GM plants, including genome-edited crops with deliberately altered and potentially harmful sequences [64,65,66]. A call for reconsideration before consumption [67], problematic and insufficient national legislation [68], risk of allergenicity [69] and consumer’s knowledge versus fears [70,71] are increasingly expressed concerning genetically modified food. Table 1 summarizes the harmful MGEs that potentially can compromise public health.

Table 1.
MGEs harmful effect that can compromise public health.
Table 1.
MGEs harmful effect that can compromise public health.
| MGEs | Potential Harmful Effects | References |
|---|---|---|
| Antibiotic resistance or multidrug resistance genes | Microbial antibiotic resistance Bacterial resistance to phages Drug resistance to cancer therapy Resilience against antimicrobial defensive factors Contaminated and industrially processed nutrients Potential entry to the human genome by HGT | [1,2,3,4,5,6,7,8,9,10,11,12,13,14] |
| Microbial-engineered enzymic genes with MTG as an example. | Post-translational modified proteins, turning naïve peptides into immunogenic complexes Complexes are proinflammatory, allergenic, pathogenic and potentially toxic, hence compromising public health Potential inducer of celiac disease | [2,11,24,25,26,28,33,34,36,41,42,72,73,74,75,76,77] |
| MGE presence in a gut lumen presents new opportunities for HGT, with the risk of inhabiting eukaryotic hosts | [1,52,53] | |
| Transfer of microbial survival factors against host self-defense barriers | [1,25,26,54] | |
| Improved enzyme production, thermostability and pH dependency by genetic engineering might enhance the detrimental effects of the manipulated enzyme | [55] | |
| Probiotics containing MGEs | Transfer of drug resistance Transfer of virulence factors, toxins, ARGs and other detrimental MGEs should be implemented | [56,57,58,59,60,61] |
| Interference with the luminal lipid metabolism | [62] | |
| Genetically modified plants | Modified DNA or other MGEs can be laterally transferred to other recipients, spanning prokaryotes, eukaryotes and even people. | [1,63] |
| Genome-edited plants, like crops with deliberately altered and potentially harmful sequences, can invade the human microbiome or genome | [64,65,66,67,68,69,70,71] |
GEMs—genetically engineered microorganisms, HGT—horizontal gene transfer, MGEs—mobile genetic elements, mTG—microbial transglutaminase, ARGs—antibiotic resistance genes.
3. GEMs’ Horizontal Gene Exchanges Might Induce Human Diseases
The engineered bacteria can produce modified proteins, peptides, nucleic acids, and other hazardous bioactive molecules that might drive various human pathologies and affect human health. Their products can potentially perturbate intracellular metabolic pathways, activate or turn off the expression of related genes and induce the synthesis of biologically active harmful molecules. In fact, current knowledge estimates that MGE represents more than one-half of the human genome [78].
The HGT sharing of DNA can unavoidably spread beneficial genes for prokaryotic survival, with mTG activity playing a role as genetic parasites across communities [79,80]. The resulting selective evolutionary pressure of the new dweller creates a large proportion of the variability acted on by luminal natural selection. This lateral gene exchange appears to be more ubiquitous in the human microbiome than previously described [1,80]. Consequently, unregulated GEMs might introduce new deleterious MGEs into the eubiotic gut lumen.
The toxicity associated with GEM cells, which can limit their declared efficacy and enhance rapid clearance driven by the reactive immune responses stimulated by the bacterial load, might represent major drawbacks. Additionally, alteration of the composition, diversity, and disequilibrium in the gut eubiotic state might compromise human health. Autoimmune disease, inflammatory conditions such as diabetes, multiple sclerosis, rheumatoid arthritis, Inflammatory bowel diseases, obesity and even carcinogenesis might be promoted.
The practical translation and implementation of GEMs are still hindered by potential harmful effects, as well as local legislation and regulations that limit clinical studies to use only bacteria without any genetic manipulations. Multiple challenges exist: 1. Limiting the spillage of genetically inserted genes over into the genomes of other microbes, prokaryotes or eukaryotic cells; 2. Ensuring the stability of the colonized engineered bacteria and the continuous production of the expected mobilome in the targeted tissues; 3. Efficient and helpful interactions with the enteric intestinal microbiome, intending to increase the microbiome–dysbiome ratio; 4. Locking the GEMs into the targeted tissues; 5. Clearing them once they accomplish their mission. Those challenges underscore the importance of ensuring the genetic stability of the foreign HGT cargo inside GEMs under laboratory and normal physiological conditions in vitro, ex. and in vivo [81]. Effective biocontainment measures are pivotal to preventing gene transfer in and out of the engineered microbes [82].
It is hoped that the health considerations of bacterial transgene HGTs will be thoroughly investigated and tightly regulated [65].
Following are some examples of the potential involvement of GEMs in chronic human diseases driven by disequilibrated gut homeostasis (Table 2):
- Autoimmune diseases: Various ADs are associated with specific [83,84,85,86] or pathobiont [87]. Type 1 diabetes, multiple sclerosis, celiac disease and psoriasis are some of them [88]. The above-cited mTG is also associated with AD evolution [24,25,26,34,36,54,73,77]. Intriguingly, the cross-reactive antibodies and sequence similarity between microbial transglutaminase and human tissue antigens were recently reported [54]. Six human epitopes were connected to ten different ADs. The newly described molecular mimicry pathways further strengthen the mTG-ADs pathologic interplay.
- Neurodegenerative conditions: Understanding the involvement of gut dysbiosis and pathobiosis is in its infancy; however, increased knowledge is starting to appear, thus strengthening the gut–brain axis [28,89]. By perturbating enteric eubiosis and/or its beneficial secreted metabolome, the GEMs can potentially drive neuro-inflammatory/degenerative diseases [28,73,89,90,91]. Interestingly, those GEMs included transposable elements that might drive neurodevelopmental and neurodegenerative Disorders [92].
- Metabolic diseases: All the components of the metabolic syndrome are related to a perturbated gut microbiome, hazardous mobilome, and disbalance of a fine synergistic luminal homeostasis [93,94,95,96]. Harmful proteinomes and metabolomes, increased intestinal permeability, post-translational modification of naïve peptides to immunogenic ones, cross-reactive autoantibodies, sequence similarity, molecular mimicry, bacterial fragments blood translocation and some other auto-immunogenic pathways might drive GEM involvement in metabolic conditions [2,85,96,97].
- Allergic conditions: Food allergy is highly related to intestinal dysbiosis, and eubiotic equilibrium might protect allergy patients [98]. Natural or GEM probiotics, prebiotics, synbiotics and potentially fecal microbiota transfer are increasingly being investigated to alleviate allergic reactions. Those trials should be controlled and regulated; they impose a variety of challenges, aiming to improve the reliability and predictability of the allergenicity risk assessment. A clear safety objective that addresses new GM biotechnologies is greatly needed as safety assessments to ensure that allergenic risks of foods are avoided [99].
- Cancer induction or therapy: HGT occurs between prokaryotes and eukaryotes [100] and microbes, viruses or fungi are related to human cancer induction [101]. One recent example is the engineered E. coli Nissle 1917 involvement in colorectal cancer [102]. In contrast, prokaryotes are increasingly reported as key actors in cancer immunotherapy, applying engineered biotechnologies to combat spreading by metastases [103,104]. The potential HGT of carcinogenic constituents, from unicellular prokaryotes to multicellular tissues, including human cancer cells, requires urgent tightened control and regulatory measures on GEMs [1,105,106,107]. Recently reported examples of bacterial DNA were confirmed in lung, pancreatic, breast, bone and colorectal cancers and malignant melanomas [107]. Several mechanisms of microbial DNA integration into the human genome and cancer induction were suggested. One of those is to increase proto-oncogenes or suppress tumor suppressor gene expression in the human genome [107]. However, this can be a self-perpetuating vicious cycle, as recently noted by Yangyanqiu and Shuwen: “The damage caused by bacteria to human DNA, such as inducing DNA breaks, regulating gene expression by epigenetic modifications, and causing genome instability, can facilitate the integration of bacterial DNA into the human genome” [107].In addition, microbial enzymes, like recombinases, can facilitate the site-specific insertion of MGEs into bacterial genomes, thus loading the intestinal microbiome and risking human cells for large-payload genome insertion [108]. Even prebiotic oligosaccharides intake might aggravate DNA damage induced by colibactin-producing gut microbes [109,110]. Interestingly, a high-fiber diet and indigestible prebiotic saccharide are offered to prevent colorectal cancer. In contrast, the authors suggested that the enhanced progression of colorectal cancer operating through cellular senescence, double-strand break induction in cultured cells, and chromosomal abnormalities depends on prebiotic oligosaccharides. Future studies are necessary to resolve this discrepancy.Nevertheless, the topic of microbial genes integrated into the human genome is an ongoing hot topic. Its contribution to the evolution of eukaryotic genomes remains high [1,107,111]. Since prevention is the most cost-effective way to fight cancer or other human chronic diseases, tightly regulating and controlling GEMs and avoiding the entry of MGEs into the microbiome or human genome represent the most rewarding means to protect people from those morbid and mortal conditions.
- Neurodevelopment and behavior: Explicit emotion regulation and cognitive control govern executive functions and mental health throughout the entire lifespan. The intestinal microbiota represents a potential biomarker for the risk of mental and behavioral morbidities. Basically, gut eubiotic diversity and synergistic composition affect brain function, thus playing a pivotal role in emotional processing [28,73,89,112,113,114,115,116]. Recently, the following neuropsychiatric conditions were reported to be dysbiotic-dependent: Alzheimer’s disease, attention deficit hyperactivity disorder, amyotrophic lateral sclerosis, anorexia nervosa, bipolar disorder, generalized anxiety disorder, major depressive disorder, multiple sclerosis and schizophrenia [117]. The microbiome–gut–brain axis plays an essential role in regulating neurodevelopment, brain metabolism and behavior. Tryptophan, the precursor to serotonin, short-chain fatty acids, GABA, acetylcholine, histamine, bile acids, 5-amino valeric acid, taurine and spermine are some of the microbiome-originated neurotransmitters and metabolome that affect brain physiology, human behavior or pathology [89,113,118,119]. Introducing less-regulated GEMs or their foreign mobilome to the luminal compartment might disrupt the evolutionary equilibrium of the enteric inhabitants.
- Female and male infertility: Most recently, genetically proxied intestinal microbes were found to have potential causal effects on females and males [120,121]. This additional potential risk might affect future generations of geo-epidemiology and many other public aspects of life worldwide. One could wonder about the potential impact of deleterious MGE entry into the equilibrated intestinal microbiome on the above-cited chronic human diseases. Table 2 summarizes the potential involvement of GEMs in chronic human diseases driven by perturbated gut homeostasis.

Table 2.
The Role of GEMs in Chronic Diseases Linked to Disrupted Gut Homeostasis.
Table 2.
The Role of GEMs in Chronic Diseases Linked to Disrupted Gut Homeostasis.
| Chronic Disease Category | Disease Examples | References |
|---|---|---|
| Autoimmune diseases | Type 1 diabetes, MM, celiac disease, MG, GBS and psoriasis | [24,25,26,77,83,84,85,86,87,88] |
| Neurodegenerative conditions | Alzheimer’s, Parkinson’s, autism, schizophrenia, ALS and MM | [28,73,89,90] |
| Metabolic diseases | Type 1 diabetes, cardiovascular, hyperlipidemia, obesity and liver steatosis | [1,2,11,28,34,36,75,79,80,85,93,94,95,96,97,122] |
| Allergy | Food allergies | [98,99] |
| Cancer | lung, pancreatic, breast, bone and colorectal cancers and malignant melanoma | [1,100,101,102,103,104,105,106,107,108,109,110,111] |
| Neurodevelopment and behavior | Bipolar, depression, anxiety, ADHD, migraines and headaches | [28,73,89,112,113,114,115,116,117] |
| infertility | Female and male infertility | [120,121] |
Abbreviations: MM—multiple sclerosis, MG—myasthenia gravis, GBS—Guillain–Barré syndrome, ALS—amyotrophic lateral sclerosis, ADHD—attention deficit hyperactivity disorder.
4. The New GEMs Can Change the Long-Term Natural Evolution of the Enteric Pro- or Eukaryotic Cells Inhabitants
Genome editing is an indispensable tool for modulating specific functions of individual genes or changing the expression of important genes in the cells and whole organisms. CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated) is a pivotal prokaryotic adaptive immune machinery that protects the microbiome from invading viruses and plasmids [123,124]. The CRISPR/Cas system represents a major driving force and a game-changer in the life science revolution in the 21st century due to its advantages in genome editing and regulation. On the contrary, despite the advantages and the embedded challenges, using this technology for gene editing in GEMs might bring risks and devastating consequences on the enteric prokaryotic inhabitants and the entire human genome [125]. Genetically engineered probiotics or synthetic microbial consortia should be regulated as drugs, not as less-controlled food supplements [126]. Their gut delivery could bring unexpected consequences, causing biosafety problems that should be addressed and overcome by regulatory authorities. The same applies to genetically modified food after entering the human body [127]. Risks to fetuses, plant toxin production, HGT to the enteric microbiome, spreading ARG and MDR delivery and allergenic reaction induction are some of the reported risks [67,69,127]. Intriguingly, a comparable risk exists in the usage of engineered fungi [128]. There is no doubt that improved and validated ways of safely using genetically modified nutrients should be implemented. The labeling of genetically modified ingredients to satisfy public transparency should be adopted. In summary, tightening the regulation of GEMs and engineered plants is urgently needed. Table 1 and Figure 2 summarize the potential involvement of GEMs in chronic human diseases driven by perturbated gut homeostasis.
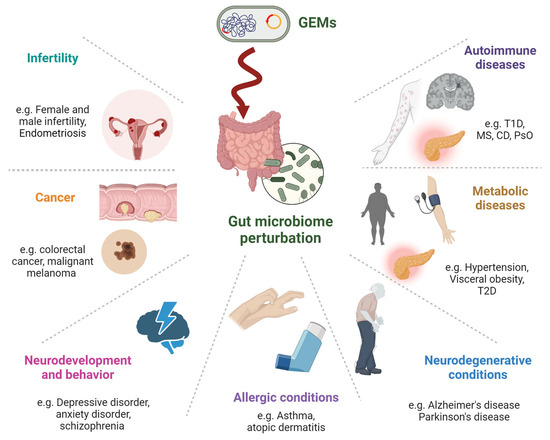
Figure 2.
Potential involvement of GEMs in chronic human diseases related to disrupted gut homeostasis covers a broad spectrum of conditions. Autoimmune Diseases: affecting immune tolerance and increasing inflammation. Metabolic Diseases: Influence the metabolism of lipids, carbohydrates, and other substances, which can lead to obesity, type 2 diabetes, and metabolic syndrome. Neurodegenerative Conditions: Through the gut–brain axis, gut bacteria perturbation can produce neurotoxic substances, affecting neurodegenerative diseases like Alzheimer’s and Parkinson’s. Allergic Conditions: This might influence allergic diseases by modifying the gut microbiome, which is known to play a role in immune responses. Neurodevelopment and Behavior: Via the gut–brain axis, gut microbiome compounds can affect mood, cognition, and behavior. Cancer: Microbial genes that are integrated into the human genome can potentially induce carcinogenic constituents. Infertility: Emerging evidence linking gut health to reproductive health. This might involve the modulation of hormone levels, inflammation, and overall metabolic health.
5. The Worldwide Regulatory Authority’s Safety Control of GEMs Is Not Enough to Protect Public Health
Maximizing the safety of the GEMs is important, necessary and indispensable. Scientific critics are increasingly raising concerns regarding various safety issues in GEM development, clinical applications and usage. The following is a summary of those concerns and warnings.
- Biocontainment should be controlled and regulated for real-world applications [129,130,131,132]. This can be achieved by biocontainment genetic circuits, auxotrophic mechanisms and reliance on synthetic amino acids or protein designs. These means will help to prevent the spread and persistence of GEMs in the environment. Sensors for tight biocontainment will ensure viability control.
- GEM genetic instability to enhance their stability in the gut compartment. This will reduce the probability of loss or gain-of-function mutations [129,130]. Their limited or lack of luminal colonization capacity and easy eradication by routine antibiotic intake administration might limit GEM efficacy.
- Individual inherent microbiome variations may dilute the GEMs’ intestinal functionality [130]. Thus, predicting the long-term engraftment of modified bacteria within any given patient endogenous population might be difficult to achieve, resulting in a kind of personal medicine.
- Competition with stable and long-term eubiotic communities might adversely perturbate the delicate and fragile balance of the gut ecosystem [131].
- Uncontrolled growth of the GEMs within the human gut [130]. Biocontainment strategies are essential prior to the clinical application in order to avoid gut dysfunction, intestinal inflammation [132] or pathogenic infection [81].
- GEM-induced metabolic abnormalities and their toxic effects should be fully evaluated before their in vivo clinical usage [132].
- Controlling and limiting the viability of the inhabitant GEMs is necessary. Live biotherapeutic-engineered microbes can induce unwanted detrimental dysfunction and break gut homeostasis, resulting in microbiome disruption and potential organ pathogenicity [123]. Genetic “kill-switch” strategies designed to lyse the cell when triggered are crucial. Alternatively, GEM pathogenicity should be mitigated by gene knockout or mutated virulence genes [123,132].
- Clearing the foreign-modified bacteria after accomplishing its therapeutic effects is a key task. Alternative selection markers, biocontainment and homologous DNA usage were applied to avoid potential environmental transmission and purge the residual foreign bacteria [132,133].
- Controlling the microbial production pathways. Dynamic regulation is a strategy to control the production of key molecules. Transcription factor-based biosensors for the dynamic regulation of the final product were recently reported [134,135]. By detecting and following the presence of the synthesized molecule and triggering the inhibition or activation of targeted genes in the metabolic pathway, the biosensors might help to tighten the regulation.
In summary, clinical microbial live biotherapy has major safety hurdles to overcome before it can be routinely used, and appropriate regulatory enforcement must be made available. Current safety practices related to GEM evolution, clinical applications and use are far from satisfying public health demands. Several emerging and sophisticated techniques might help track those harmful GEMs, understand the enigma of potential DNA relocation and develop more comprehensive regulatory strategies regarding health benefits.
Indeed, chromosome conformation capture and methylome analyses [136], the bioinformatic pipeline (Xenoseq) application [137], the use of organoids and the microfluidic ‘Gut-on-a-chip’ technique were suggested [138]. Most recently, novel computational strategies were reported to merge theoretical models with experimental methods [139]. Combining those approaches enables numerous strains and GME transfer to be studied, both in vivo and in vitro, thus mimicking the intricacies of luminal-associated dysbiotic and pathobiotic–human morbidity relationships. In 2012, the U.S. Environmental Protection Agency issued a summary on the Regulation of Genetically Engineered Microorganisms Under FIFRA, FFDCA and TSCA [140], which did not address the burning issues of the potentially harmful effects of the HGT of GEMs in the human intestinal compartment. These issues must be addressed. The risk of mutation and the transfer of genetic material are cause for concern and calculated caution [141,142].
6. Conclusions
Along with in-depth analyses, the present information unveils the importance of regulating the GEMs pre-inhabiting our enteric harmonized eubiome. Newly introduced genetic cargo can potentially perturbate the symbiotic and fine, hence, fragile, enteric synergistic homeostasis. The foreign, non-self MGEs represent a potential threat to human physical and mental health.
However, further comprehensive, well-designed and evidence-based studies are required to draw more solid conclusions regarding the tight regulation of GEMs, their mechanisms of action and contemporary and evolutionary potential detrimental impacts that aim to prevent their harmful effects on human beings. Widespread use of natural and genetically engineered intestinal biotics should be halted, and public labeling and clear transparency should be instituted. Regulatory guidelines for gut GEM usage must be backed up by basic and clinical research. A more holistic comprehension of gut HGT-dependent eubiotic–dysbiotic balance, along with multiple environmental and lifestyle factors, is necessary to better manage and prevent the drawbacks of widespread GEM usage. This narrative review encourages regulatory authorities worldwide to take a more holistic and aligned approach to the risk evaluation and regulatory oversight of GEM-produced food ingredients, immune factors, enzymes and any category of food substances that can enable safe and sustainable consumer food choices and consumption.
Despite their proven therapeutic benefits, synthetic microbial biotherapeutics have several safety hurdles to overcome to reach widespread usage and consumer acceptance. Extensive studies are required to explore the multi-directional communication between gut homeostasis and the newly introduced GEMs, which might help researchers understand the newly engineered inhabitant effects on public physical health and mental behavior. It is important to remember that prevention is the most cost-effective strategy and primum non nocere should be the focus.
Author Contributions
A.L. screened the literature, designed and wrote the manuscript, C.B. screened the literature, wrote, edited and revised the manuscript, and designed the figures with BioRender.com’s permission. A.V. designed, edited and supervised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Not applicable.
Conflicts of Interest
Author Aristo Vojdani was employed by the company Immunosciences Lab., Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Lerner, A.; Matthias, T.; Aminov, R. Potential effects of horizontal gene exchange in the human gut. Front. Immunol. 2017, 8, 1630. [Google Scholar] [CrossRef]
- Lerner, A.; Aminov, R.; Matthias, T. Dysbiosis may trigger autoimmune diseases via inappropriate post-translational modification of host proteins. Front. Microbiol. 2016, 7, 84. [Google Scholar] [CrossRef]
- Mosca, A.; Leclerc, M.; Hugot, J.P. Gut Microbiota Diversity and Human Diseases: Should We Reintroduce Key Predators in Our Ecosystem? Front. Microbiol. 2016, 7, 455. [Google Scholar] [CrossRef]
- El Tekle, G.; Garrett, W.S. Bacteria in cancer initiation, promotion and progression. Nat. Rev. Cancer 2023, 23, 600–618. [Google Scholar] [CrossRef]
- Shi, J. Editorial: Reviews in the impact of gut microbiota in health and disease. Front. Microbiol. 2023, 14, 1230925. [Google Scholar] [CrossRef]
- Sitaraman, R. Prokaryotic horizontal gene transfer within the human holobiont: Ecological-evolutionary inferences, implications and possibilities. Microbiome 2018, 6, 163. [Google Scholar] [CrossRef]
- Fredriksen, S.; de Warle, S.; van Baarlen, P.; Boekhorst, J.; Wells, J.M. Resistome expansion in disease-associated human gut microbiomes. Microbiome 2023, 11, 166. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Hallowell, H.A.; Koutouvalis, K.; Suez, J. Good microbes, bad genes? The dissemination of antimicrobial resistance in the human microbiome. Gut Microbes 2022, 14, 2055944. [Google Scholar] [CrossRef]
- Bag, S.; Ghosh, T.S.; Banerjee, S.; Mehta, O.; Verma, J.; Dayal, M.; Desigamani, A.; Kumar, P.; Saha, B.; Kedia, S.; et al. Molecular Insights into Antimicrobial Resistance Traits of Commensal Human Gut Microbiota. Microb. Ecol. 2019, 77, 546–557. [Google Scholar] [CrossRef]
- Suh, G.A.; Patel, R. Clinical phage microbiology: A narrative summary. Clin. Microbiol. Infect. 2023, 29, 710–713. [Google Scholar] [CrossRef]
- Lerner, A.; Ramesh, A.; Matthias, T. The Revival of the Battle between David and Goliath in the Enteric Viruses and Microbiota Struggle: Potential Implication for Celiac Disease. Microorganisms 2019, 7, 173. [Google Scholar] [CrossRef]
- Emran, T.B.; Shahriar, A.; Mahmud, A.R.; Rahman, T.; Abir, M.H.; Siddiquee, M.F.R.; Ahmed, H.; Rahman, N.; Nainu, F.; Wahyudin, E.; et al. Multidrug Resistance in Cancer: Understanding Molecular Mechanisms, Immunoprevention and Therapeutic Approaches. Front. Oncol. 2022, 12, 891652. [Google Scholar] [CrossRef]
- Yadav, M.; Pandey, R.; Chauhan, N.S. Catabolic Machinery of the Human Gut Microbes Bestow Resilience against Vanillin Antimicrobial Nature. Front. Microbiol. 2020, 11, 588545. [Google Scholar] [CrossRef]
- Lerner, A.; Soprun, L.; Benzvi, C. Antimicrobial Resistance along the Food Chain: Contaminated and Industrially Processed Nutrients. J. Food Nutr. Health 2022, 3, 1–11. [Google Scholar] [CrossRef]
- Ondon, B.S.; Li, S.; Zhou, Q.; Li, F. Sources of Antibiotic Resistant Bacteria (ARB) and Antibiotic Resistance Genes (ARGs) in the Soil: A Review of the Spreading Mechanism and Human Health Risks. Rev. Environ. Contam. Toxicol. 2021, 256, 121–153. [Google Scholar]
- Rosander, A.; Connolly, E.; Roos, S. Removal of antibiotic resistance gene-carrying plasmids from Lactobacillus reuteri ATCC 55730 and characterization of the resulting daughter strain, L. reuteri DSM 17938. Appl. Environ. Microbiol. 2008, 74, 6032–6040. [Google Scholar] [CrossRef]
- Egervärn, M.; Lindmark, H.; Olsson, J.; Roos, S. Transferability of a tetracycline resistance gene from probiotic Lactobacillus reuteri to bacteria in the gastrointestinal tract of humans. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2010, 97, 189–200. [Google Scholar] [CrossRef]
- Qamar, M.U.; Aatika; Chughtai, M.I.; Ejaz, H.; Mazhari, B.B.Z.; Maqbool, U.; Alanazi, A.; Alruwaili, Y.; Junaid, K. Antibiotic-Resistant Bacteria, Antimicrobial Resistance Genes, and Antibiotic Residue in Food from Animal Sources: One Health Food Safety Concern. Microorganisms 2023, 11, 161. [Google Scholar] [CrossRef]
- Chuang, C.; Lee, K.C.; Wang, Y.P.; Lee, P.C.; Chang, T.E.; Huang, Y.H.; Lin, Y.T.; Hou, M.C. High carriage rate of extended-spectrum β-lactamase Enterobacterales and diarrheagenic Escherichia coli in healthy donor screening for fecal microbiota transplantation. Eur. J. Clin. Microbiol. Infect. Dis. 2023, 42, 1103–1113. [Google Scholar] [CrossRef]
- Tatta, E.R.; Imchen, M.; Moopantakath, J.; Kumavath, R. Bioprospecting of microbial enzymes: Current trends in industry and healthcare. Appl. Microbiol. Biotechnol. 2022, 106, 1813–1835. [Google Scholar] [CrossRef]
- Jangra, S.; Srivastava, S. Microbial Enzymes in Food Industries: Enhancing Quality and Sustainability. In Food Microbial Sustainability: Integration of Food Production and Food Safety; Springer Nature: Singapore, 2023; pp. 193–221. [Google Scholar] [CrossRef]
- Pariza, M.W.; Johnson, E.A. Evaluating the safety of microbial enzyme preparations used in food processing: Update for a new century. Regul. Toxicol. Pharmacol. 2001, 33, 173–186. [Google Scholar] [CrossRef]
- Deckers, M.; Deforce, D.; Fraiture, M.A.; Roosens, N.H.C. Genetically modified micro-organisms for industrial food enzyme production: An overview. Foods 2020, 9, 326. [Google Scholar] [CrossRef]
- Lerner, A.; Matthias, T. Microbial Transglutaminase is Beneficial to Food Industries but a Caveat to Public Health. Med. One 2019, 4, e190001. [Google Scholar] [CrossRef]
- Lerner, A.; Matthias, T. Processed food additive microbial transglutaminase and its cross-linked gliadin complexes are potential public health concerns in celiac disease. Int. J. Mol. Sci. 2020, 21, 1127. [Google Scholar] [CrossRef]
- Lerner, A.; Benzvi, C. Microbial transglutaminase is a very frequently used food additive and is a potential inducer of autoimmune/neurodegenerative diseases. Toxics 2021, 9, 233. [Google Scholar] [CrossRef]
- Faustman, C.; Aaron, D.; Negowetti, N.; Leib, E.B. Ten years post-GAO assessment, FDA remains uninformed of potentially harmful GRAS substances in foods. Crit. Rev. Food Sci. Nutr. 2021, 61, 1260–1268. [Google Scholar] [CrossRef]
- Lerner, A.; Benzvi, C. “Let food be thy medicine”: Gluten and potential role in neurodegeneration. Cells 2021, 10, 756. [Google Scholar] [CrossRef]
- Sewalt, V.; LaMarta, J.; Shanahan, D.; Gregg, L.; Carrillo, R. Letter to the editor regarding “GRAS from the ground up: Review of the Interim Pilot Program for GRAS notification” by Hanlon et al., 2017. Food Chem. Toxicol. 2017, 107, 520–521. [Google Scholar] [CrossRef]
- Neltner, T.G.; Alger, H.M.; O’Reilly, J.T.; Krimsky, S.; Bero, L.A.; Maffini, M.V. Conflicts of interest in approvals of additives to food: Determined to be generally recognized as safe: Out of balance. JAMA Intern. Med. 2013, 173, 2032–2036. [Google Scholar] [CrossRef]
- Kruger, C. The relevance of international assessments to GRAS determinations. Regul. Toxicol. Pharmacol. 2016, 79, S119–S123. [Google Scholar] [CrossRef]
- Roberts, A.; Haighton, L.A. A hard look at FDA’s review of GRAS notices. Regul. Toxicol. Pharmacol. 2016, 79, S124–S128. [Google Scholar] [CrossRef]
- Lerner, A.; Matthias, T. Possible association between celiac disease and bacterial transglutaminase in food processing: A hypothesis. Nutr. Rev. 2015, 73, 544–552. [Google Scholar] [CrossRef]
- Lerner, A.; Matthias, T. Changes in intestinal tight junction permeability associated with industrial food additives explain the rising incidence of autoimmune disease. Autoimmun. Rev. 2015, 14, 479–489. [Google Scholar] [CrossRef]
- Kaufmann, A.; Köppel, R.; Widmer, M. Determination of microbial transglutaminase in meat and meat products. Food Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess. 2012, 29, 1364–1373. [Google Scholar] [CrossRef]
- Lerner, A.; Aminov, R.; Matthias, T. Transglutaminases in dysbiosis as potential environmental drivers of autoimmunity. Front. Microbiol. 2017, 8, 66. [Google Scholar] [CrossRef]
- Hartung, T. Rebooting the generally recognized as safe (GRAS) approach for food additive safety in the US. ALTEX 2018, 35, 3–25. [Google Scholar] [CrossRef]
- Vandenberg, L.N.; Zoeller, R.T.; Prins, G.S.; Trasande, L. Evaluating adverse effects of environmental agents in food: A brief critique of the US FDA’s criteria. Environ. Health 2023, 22, 38. [Google Scholar] [CrossRef]
- Kolotylo, V.; Piwowarek, K.; Kieliszek, M. Microbiological transglutaminase: Biotechnological application in the food industry. Open Life Sci. 2023, 18, 20220737. [Google Scholar] [CrossRef]
- Lerner, A.; Matthias, T. Microbial transglutaminase should be considered as an environmental inducer of celiac disease. World J. Clin. Cases 2019, 7, 3912–3914. [Google Scholar] [CrossRef]
- Matthias, T.; Lerner, A. Microbial Transglutaminase Is Immunogenic and Potentially Pathogenic in Pediatric Celiac Disease. Front. Pediatr. 2018, 6, 389. [Google Scholar] [CrossRef]
- Lerner, A.; Matthias, T. Microbial transglutaminase: A new potential player in celiac disease. Clin. Immunol. 2019, 199, 37–43. [Google Scholar] [CrossRef]
- Su, T.; Qin, X.Y.; Furutani, Y. Transglutaminase 2 as a marker for inflammation and therapeutic target in sepsis. Int. J. Mol. Sci. 2021, 22, 1897. [Google Scholar] [CrossRef]
- Elli, L.; Bergamini, C.M.; Bardella, M.T.; Schuppan, D. Transglutaminases in inflammation and fibrosis of the gastrointestinal tract and the liver. Dig. Liver Dis. 2009, 41, 541–550. [Google Scholar] [CrossRef]
- Kieliszek, M.; Misiewicz, A. Microbial transglutaminase and its application in the food industry. A review. Folia Microbiol. 2014, 59, 241–250. [Google Scholar] [CrossRef]
- Duarte, L.; Matte, C.R.; Bizarro, C.V.; Ayub, M.A.Z. Review transglutaminases: Part II-industrial applications in food, biotechnology, textiles and leather products. World J. Microbiol. Biotechnol. 2019, 36, 11. [Google Scholar] [CrossRef]
- Paolella, G.; Martucciello, S.; Vasi’cvasi’c, K.; Knez, Ž.; Leitgeb, M. Transglutaminase in Foods and Biotechnology. Int. J. Mol. Sci. 2023, 24, 12402. [Google Scholar] [CrossRef]
- Martins, I.M.; Matos, M.; Costa, R.; Lopes-da-Silva, F.; Pascoal, A.; Estevinho, L.M.; Choupina, A.B. Transglutaminases: Recent achievements and new sources. Appl. Microbiol. Biotechnol. 2014, 98, 6957–6964. [Google Scholar] [CrossRef]
- Fuchsbauer, H.L. Approaching transglutaminase from Streptomyces bacteria over three decades. FEBS J. 2022, 289, 4680–4703. [Google Scholar] [CrossRef]
- Miwa, N. Innovation in the food industry using microbial transglutaminase: Keys to success and future prospects. Anal. Biochem. 2020, 597, 113638. [Google Scholar] [CrossRef]
- Ponting, C.P.; Aravind, L.; Schultz, J.; Bork, P.; Koonin, E.V. Eukaryotic signalling domain homologues in archaea and bacteria. Ancient ancestry and horizontal gene transfer. J. Mol. Biol. 1999, 289, 729–745. [Google Scholar] [CrossRef]
- Krishnan, A.; Burroughs, A.M.; Iyer, L.M.; Aravind, L. Unexpected Evolution of Lesion-Recognition Modules in Eukaryotic NER and Kinetoplast DNA Dynamics Proteins from Bacterial Mobile Elements. iScience 2018, 9, 192–208. [Google Scholar] [CrossRef]
- Horne, T.; Orr, V.T.; Hall, J.P. How do interactions between mobile genetic elements affect horizontal gene transfer? Curr. Opin. Microbiol. 2023, 73, 102282. [Google Scholar] [CrossRef]
- Lerner, A.; Benzvi, C.; Vojdani, A. Cross-reactivity and sequence similarity between microbial transglutaminase and human tissue antigens. Sci. Rep. 2023, 13, 17526. [Google Scholar] [CrossRef]
- Fatima, S.W.; Khare, S.K. Effect of key regulators in augmenting transcriptional expression of Transglutaminase in Streptomyces mobaraensis. Bioresour. Technol. 2021, 340, 125627. [Google Scholar] [CrossRef]
- Lerner, A.; Shoenfeld, Y.; Matthias, T. Probiotics: If it does not help it does not do any harm. really? Microorganisms 2019, 7, 104. [Google Scholar] [CrossRef]
- Lerner, A.; Matthias, T. There Are Many More Cons for Probiotics. ISR Med. Assoc. J. 2020, 22, 131. [Google Scholar]
- Imperial, I.C.V.J.; Ibana, J.A. Addressing the antibiotic resistance problem with probiotics: Reducing the risk of its double-edged sword effect. Front. Microbiol. 2016, 7, 1983. [Google Scholar] [CrossRef]
- Dou, W.; Abdalla, H.B.; Chen, X.; Sun, C.; Chen, X.; Tian, Q.; Wang, J.; Zhou, W.; Chi, W.; Zhou, X.; et al. ProbResist: A database for drug-resistant probiotic bacteria. Database 2022, 2022, baac064. [Google Scholar] [CrossRef]
- Merenstein, D.; Pot, B.; Leyer, G.; Ouwehand, A.C.; Preidis, G.A.; Elkins, C.A.; Hill, C.; Lewis, Z.T.; Shane, A.L.; Zmora, N.; et al. Emerging issues in probiotic safety: 2023 perspectives. Gut Microbes 2023, 15, 2185034. [Google Scholar] [CrossRef]
- Ma, J.; Lyu, Y.; Liu, X.; Jia, X.; Cui, F.; Wu, X.; Deng, S.; Yue, C. Engineered probiotics. Microb. Cell Fact. 2022, 21, 72. [Google Scholar] [CrossRef]
- Wu, T.; Wang, G.; Xiong, Z.; Xia, Y.; Song, X.; Zhang, H.; Wu, Y.; Ai, L. Probiotics Interact with Lipids Metabolism and Affect Gut Health. Front. Nutr. 2022, 9, 917043. [Google Scholar] [CrossRef]
- Philips, J.G.; Martin-Avila, E.; Robold, A.V. Horizontal gene transfer from genetically modified plants—Regulatory considerations. Front. Bioeng. Biotechnol. 2022, 10, 971402. [Google Scholar] [CrossRef]
- Halford, N.G. Legislation governing genetically modified and genome-edited crops in Europe: The need for change. J. Sci. Food Agric. 2019, 99, 8–12. [Google Scholar] [CrossRef]
- Kleter, G.A.; Peijnenburg, A.A.C.M.; Aarts, H.J.M. Health considerations regarding horizontal transfer of microbial transgenes present in genetically modified crops. J. Biomed. Biotechnol. 2005, 2005, 326–352. [Google Scholar] [CrossRef]
- Kamle, S.; Ali, S. Genetically modified crops: Detection strategies and biosafety issues. Gene 2013, 522, 123–132. [Google Scholar] [CrossRef]
- Grover, A.; Ashhar, N.; Patni, P. Why genetically modified food need reconsideration before consumption? J. Fam. Med. Prim. Care 2014, 3, 188–190. [Google Scholar] [CrossRef]
- Mammadov, V.; Mustafayeva, A. The legislation of CIS countries on the issue of genetically modified products. Med. Law 2011, 30, 555–570. [Google Scholar]
- Lee, T.H.; Ho, H.K.; Leung, T.F. Genetically modified foods and allergy. Hong Kong Med. J. 2017, 23, 291–295. [Google Scholar] [CrossRef]
- Boccia, F.; Covino, D.; Sarnacchiaro, P. Genetically modified food versus knowledge and fear: A Noumenic approach for consumer behaviour. Food Res. Int. 2018, 111, 682–688. [Google Scholar] [CrossRef]
- Xu, R.; Wu, Y.; Luan, J. Consumer-perceived risks of genetically modified food in China. Appetite 2020, 147, 104520. [Google Scholar] [CrossRef]
- Lerner, A.; Matthias, T. Food Industrial Microbial Transglutaminase in Celiac Disease: Treat or Trick. Int. J. Celiac Dis. 2015, 3, 1–6. [Google Scholar] [CrossRef][Green Version]
- Lerner, A.; Matthias, T. Don’t forget the exogenous microbial transglutaminases: It is immunogenic and potentially pathogenic. AIMS Biophys. 2016, 3, 546–552. [Google Scholar] [CrossRef]
- Agardh, D.; Matthias, T.; Wusterhausen, P.; Neidhöfer, S.; Heller, A.; Lerner, A. Antibodies against neo-epitope of microbial and human transglutaminase complexes as biomarkers of childhood celiac disease. Clin. Exp. Immunol. 2020, 199, 294–302. [Google Scholar] [CrossRef]
- Matthias, T.; Jeremias, P.; Neidhöfer, S.; Lerner, A. The industrial food additive, microbial transglutaminase, mimics tissue transglutaminase and is immunogenic in celiac disease patients. Autoimmun. Rev. 2016, 15, 1111–1119. [Google Scholar] [CrossRef]
- Lerner, A.; Jeremias, P.; Neidhofer, S.; Matthias, T. Comparison of the Reliability of 17 Celiac Disease Associated Bio-Markers to Reflect Intestinal Damage. J. Clin. Cell. Immunol. 2017, 8, 486. [Google Scholar] [CrossRef]
- Amirdivani, S.; Khorshidian, N.; Fidelis, M.; Granato, D.; Koushki, M.R.; Mohammadi, M.; Khoshtinat, K.; Mortazavian, A.M. Effects of transglutaminase on health properties of food products. Curr. Opin. Food Sci. 2018, 22, 74–80. [Google Scholar] [CrossRef]
- Medina, J.; Perron, H. DNA sequences from mobile genetic elements, a hidden half of the human genome. Medecine/Sciences 2017, 33, 151–158. [Google Scholar] [CrossRef]
- Hall, R.J.; Whelan, F.J.; McInerney, J.O.; Ou, Y.; Domingo-Sananes, M.R. Horizontal Gene Transfer as a Source of Conflict and Cooperation in Prokaryotes. Front. Microbiol. 2020, 11, 1569. [Google Scholar] [CrossRef]
- Hsu, T.Y.; Nzabarushimana, E.; Wong, D.; Luo, C.; Beiko, R.G.; Langille, M.; Huttenhower, C.; Nguyen, L.H.; Franzosa, E.A. Profiling novel lateral gene transfer events in the human microbiome. bioRxiv 2023. [Google Scholar] [CrossRef]
- Liu, Y.; Feng, J.; Pan, H.; Zhang, X.; Zhang, Y. Genetically engineered bacterium: Principles, practices, and prospects. Front. Microbiol. 2022, 13, 997587. [Google Scholar] [CrossRef]
- Lee, J.W.; Chan, C.T.Y.; Slomovic, S.; Collins, J.J. Next-generation biocontainment systems for engineered organisms. Nat. Chem. Biol. 2018, 14, 530–537. [Google Scholar] [CrossRef]
- De Luca, F.; Shoenfeld, Y. The microbiome in autoimmune diseases. Clin. Exp. Immunol. 2019, 195, 74–85. [Google Scholar] [CrossRef]
- Miyauchi, E.; Shimokawa, C.; Steimle, A.; Desai, M.S.; Ohno, H. The impact of the gut microbiome on extra-intestinal autoimmune diseases. Nat. Rev. Immunol. 2023, 23, 9–23. [Google Scholar] [CrossRef]
- English, J.; Patrick, S.; Stewart, L.D. The potential role of molecular mimicry by the anaerobic microbiota in the aetiology of autoimmune disease. Anaerobe 2023, 80, 102721. [Google Scholar] [CrossRef]
- Krishna, V.; Kumar, N.; Banerjee, S. Gut Microbiota and Inflammatory Disorders. Curr. Drug Targets 2021, 23, 156–169. [Google Scholar] [CrossRef]
- Vieira, S.M.; Hiltensperger, M.; Kumar, V.; Zegarra-Ruiz, D.; Dehner, C.; Khan, N.; Costa, F.R.C.; Tiniakou, E.; Greiling, T.; Ruff, W.; et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 2018, 359, 1156–1161. [Google Scholar] [CrossRef]
- Boccuto, L.; Tack, J.; Ianiro, G.; Abenavoli, L.; Scarpellini, E. Human Genes Involved in the Interaction between Host and Gut Microbiome: Regulation and Pathogenic Mechanisms. Genes 2023, 14, 857. [Google Scholar] [CrossRef]
- Lerner, A.; Neidhöfer, S.; Matthias, T. The gut microbiome feelings of the brain: A perspective for non-microbiologists. Microorganisms 2017, 5, 66. [Google Scholar] [CrossRef]
- Wu, N.; Li, X.; Ma, H.; Zhang, X.; Liu, B.; Wang, Y.; Zheng, Q.; Fan, X. The role of the gut microbiota and fecal microbiota transplantation in neuroimmune diseases. Front. Neurol. 2023, 14, 1108738. [Google Scholar] [CrossRef]
- Solanki, R.; Karande, A.; Ranganathan, P. Emerging role of gut microbiota dysbiosis in neuroinflammation and neurodegeneration. Front. Neurol. 2023, 14, 1149618. [Google Scholar] [CrossRef]
- Jönsson, M.E.; Garza, R.; Johansson, P.A.; Jakobsson, J. Transposable Elements: A Common Feature of Neurodevelopmental and Neurodegenerative Disorders. Trends Genet. 2020, 36, 610–623. [Google Scholar] [CrossRef]
- Lerner, A.; Steigerwald, C.; Matthias, T. Feed your microbiome and your heart: The gut-heart axis. Front. Biosci. Landmark 2021, 26, 468–477. [Google Scholar] [CrossRef]
- Lim, Y.Y.; Lee, Y.S.; Ooi, D.S.Q. Engineering the Gut Microbiome for Treatment of Obesity: A Review of Current Understanding and Progress. Biotechnol. J. 2020, 15, e2000013. [Google Scholar] [CrossRef]
- He, Y.J.; You, C.G. The Potential Role of Gut Microbiota in the Prevention and Treatment of Lipid Metabolism Disorders. Int. J. Endocrinol. 2020, 2020, 8601796. [Google Scholar] [CrossRef]
- Sinha, S.; Haque, M. Obesity, Diabetes Mellitus, and Vascular Impediment as Consequences of Excess Processed Food Consumption. Cureus 2022, 14, e28762. [Google Scholar] [CrossRef]
- Vojdani, A.; Lerner, A.; Vojdani, E. Cross-Reactivity and Sequence Homology between Al-Pha-Synuclein and Food Products: A Step Further for Parkinson’s Disease Synucleinopathy. Cells 2021, 10, 1111. [Google Scholar] [CrossRef]
- Zhao, W.; Ho, H.E.; Bunyavanich, S. The gut microbiome in food allergy. Ann. Allergy Asthma Immunol. 2019, 122, 276–282. [Google Scholar] [CrossRef]
- Mullins, E.; Bresson, J.L.; Dalmay, T.; Dewhurst, I.C.; Epstein, M.M.; Firbank, L.G.; Guerche, P.; Hejatko, J.; Naegeli, H.; Nogué, F.; et al. Scientific Opinion on development needs for the allergenicity and protein safety assessment of food and feed products derived from biotechnology. EFSA J. 2022, 20, e07044. [Google Scholar] [CrossRef]
- Sieber, K.B.; Bromley, R.E.; Hotopp, J.C.D. Lateral gene transfer between prokaryotes and eukaryotes. Exp. Cell Res. 2017, 358, 421–426. [Google Scholar] [CrossRef]
- Xue, X.; Li, R.; Chen, Z.; Li, G.; Liu, B.; Guo, S.; Yue, Q.; Yang, S.; Xie, L.; Zhang, Y.; et al. The role of the symbiotic microecosystem in cancer: Gut microbiota, metabolome, and host immunome. Front. Immunol. 2023, 14, 1235827. [Google Scholar] [CrossRef]
- Nougayrède, J.-P.; Chagneau, C.V.; Motta, J.-P.; Bossuet-Greif, N.; Belloy, M.; Taieb, F.; Gratadoux, J.-J.; Thomas, M.; Langella, P.; Oswald, E. A Toxic Friend: Genotoxic and Mutagenic Activity of the Probiotic Strain Escherichia coli Nissle 1917. mSphere 2021, 6, e0062421. [Google Scholar] [CrossRef]
- Sepich-Poore, G.D.; Zitvogel, L.; Straussman, R.; Hasty, J.; Wargo, J.A.; Knight, R. The microbiome and human cancer. Science 2021, 371, 1331. [Google Scholar] [CrossRef]
- Jiang, J.; Mei, J.; Ma, Y.; Jiang, S.; Zhang, J.; Yi, S.; Feng, C.; Liu, Y.; Liu, Y. Tumor hijacks macrophages and microbiota through extracellular vesicles. Exploration 2022, 2, 20210144. [Google Scholar] [CrossRef]
- Riley, D.R.; Sieber, K.B.; Robinson, K.M.; White, J.R.; Ganesan, A.; Nourbakhsh, S.; Hotopp, J.C.D. Bacteria-Human Somatic Cell Lateral Gene Transfer Is Enriched in Cancer Samples. PLoS Comput. Biol. 2013, 9, e1003107. [Google Scholar] [CrossRef]
- Goubet, A.G. Could the tumor-associated microbiota be the new multi-faceted player in the tumor microenvironment? Front. Oncol. 2023, 13, 1185163. [Google Scholar] [CrossRef]
- Yangyanqiu, W.; Shuwen, H. Bacterial DNA involvement in carcinogenesis. Front. Cell. Infect. Microbiol. 2022, 12, 996778. [Google Scholar] [CrossRef]
- Durrant, M.G.; Fanton, A.; Tycko, J.; Hinks, M.; Chandrasekaran, S.S.; Perry, N.T.; Schaepe, J.; Du, P.P.; Lotfy, P.; Bassik, M.C.; et al. Systematic discovery of recombinases for efficient integration of large DNA sequences into the human genome. Nat. Biotechnol. 2023, 41, 488–499. [Google Scholar] [CrossRef]
- Oliero, M.; Calvé, A.; Fragoso, G.; Cuisiniere, T.; Hajjar, R.; Dobrindt, U.; Santos, M.M. Oligosaccharides increase the genotoxic effect of colibactin produced by pks+ Escherichia coli strains. BMC Cancer 2021, 21, 172. [Google Scholar] [CrossRef]
- Pleguezuelos-Manzano, C.; Puschhof, J.; Huber, A.R.; van Hoeck, A.; Wood, H.M.; Nomburg, J.; Gurjao, C.; Manders, F.; Dalmasso, G.; Stege, P.B.; et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 2020, 580, 269–273. [Google Scholar] [CrossRef]
- Li, X.; Fang, C.; Zhao, J.P.; Zhou, X.Y.; Ni, Z.; Niu, D.K. Desiccation does not drastically increase the accessibility of exogenous DNA to nuclear genomes: Evidence from the frequency of endosymbiotic DNA transfer. BMC Genom. 2020, 21, 452. [Google Scholar] [CrossRef]
- Fujihara, H.; Matsunaga, M.; Ueda, E.; Kajiwara, T.; Takeda, A.K.; Watanabe, S.; Baba, K.; Hagihara, K.; Myowa, M. Altered Gut Microbiota Composition Is Associated with Difficulty in Explicit Emotion Regulation in Young Children. Microorganisms 2023, 11, 2245. [Google Scholar] [CrossRef]
- Lerner, A.; Matthias, T. GUT-the Trojan Horse in remote organs’ Autoimmunity. J. Clin. Cell. Immunol. 2016, 7, 401. [Google Scholar] [CrossRef]
- Grau-Del Valle, C.; Fernández, J.; Solá, E.; Montoya-Castilla, I.; Morillas, C.; Bañuls, C. Association between gut microbiota and psychiatric disorders: A systematic review. Front. Psychol. 2023, 14, 1215674. [Google Scholar] [CrossRef]
- Bhatia, N.Y.; Jalgaonkar, M.P.; Hargude, A.B.; Sherje, A.P.; Oza, M.J.; Doshi, G.M. Gut-Brain Axis and Neurological Disorders-How Microbiomes Affect our Mental Health. CNS Neurol. Disord. Drug Targets 2022, 22, 1008–1030. [Google Scholar] [CrossRef]
- Alagiakrishnan, K.; Halverson, T. Microbial Therapeutics in Neurocognitive and Psychiatric Disorders. J. Clin. Med. Res. 2021, 13, 439–459. [Google Scholar] [CrossRef]
- Zang, Y.; Lai, X.; Li, C.; Ding, D.; Wang, Y.; Zhu, Y. The Role of Gut Microbiota in Various Neurological and Psychiatric Disorders—An Evidence Mapping Based on Quantified Evidence. Mediat. Inflamm. 2023, 2023, 5127157. [Google Scholar] [CrossRef]
- Rutsch, A.; Kantsjö, J.B.; Ronchi, F. The Gut-Brain Axis: How Microbiota and Host Inflammasome Influence Brain Physiology and Pathology. Front. Immunol. 2020, 11, 604179. [Google Scholar] [CrossRef]
- Yuan, C.; He, Y.; Xie, K.; Feng, L.; Gao, S.; Cai, L. Review of microbiota gut brain axis and innate immunity in inflammatory and infective diseases. Front. Cell. Infect. Microbiol. 2023, 13, 1282431. [Google Scholar] [CrossRef]
- Zhang, F.; Xiong, Y.; Wu, K.; Wang, L.; Ji, Y.; Zhang, B. Genetic Insights into Intestinal Microbiota and Risk of Infertility: A Mendelian Randomization Study. Microorganisms 2023, 11, 2319. [Google Scholar] [CrossRef]
- Li, T.; Shao, W.; Wang, Y.; Zhou, R.; Yun, Z.; He, Y.; Wu, Y. A two-sample mendelian randomization analysis investigates associations between gut microbiota and infertility. Sci. Rep. 2023, 13, 11426. [Google Scholar] [CrossRef]
- Lerner, A.; Benzvi, C.; Vojdani, A. SARS-CoV-2 Gut-Targeted Epitopes: Sequence Similarity and Cross-Reactivity Join Together for Molecular Mimicry. Biomedicines 2023, 11, 1937. [Google Scholar] [CrossRef]
- Arnold, J.; Glazier, J.; Mimee, M. Genetic Engineering of Resident Bacteria in the Gut Microbiome. J. Bacteriol. 2023, 205, e0012723. [Google Scholar] [CrossRef]
- Jiang, C.; She, Q.; Wang, H. Editorial: Insights in genome editing tools and mechanisms: 2022. Front. Genome Ed. 2023, 5, 1240576. [Google Scholar] [CrossRef]
- Ledford, H. CRISPR tools found in thousands of viruses could boost gene editing. Nature 2022, 612, 21. [Google Scholar] [CrossRef]
- Li, P.; Roos, S.; Luo, H.; Ji, B.; Nielsen, J. Metabolic engineering of human gut microbiome: Recent developments and future perspectives. Metab. Eng. 2023, 79, 1–13. [Google Scholar] [CrossRef]
- Venugopalan, V.; Shriner, K.A.; Wong-Beringer, A. Regulatory oversight and safety of probiotic use. Emerg. Infect. Dis. 2010, 16, 1661–1665. [Google Scholar] [CrossRef]
- Heavey, M.K.; Anselmo, A.C. Modulating Oral Delivery and Gastrointestinal Kinetics of Recombinant Proteins via Engineered Fungi. AAPS J. 2021, 23, 76. [Google Scholar] [CrossRef]
- Amrofell, M.B.; Rottinghaus, A.G.; Moon, T.S. Engineering microbial diagnostics and therapeutics with smart control. Curr. Opin. Biotechnol. 2020, 66, 11–17. [Google Scholar] [CrossRef]
- Airola, C.; Severino, A.; Porcari, S.; Fusco, W.; Mullish, B.H.; Gasbarrini, A.; Cammarota, G.; Ponziani, F.R.; Ianiro, G. Future Modulation of Gut Microbiota: From Eubiotics to FMT, Engineered Bacteria, and Phage Therapy. Antibiotics 2023, 12, 868. [Google Scholar] [CrossRef]
- Voorhees, P.J.; Cruz-Teran, C.; Edelstein, J.; Lai, S.K. Challenges & opportunities for phage-based in situ microbiome engineering in the gut. J. Control. Release 2020, 326, 106–119. [Google Scholar]
- Spees, A.M.; Wangdi, T.; Lopez, C.A.; Kingsbury, D.D.; Xavier, M.N.; Winter, S.E.; Tsolis, R.M.; Bäumler, A.J. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. MBio 2013, 4, 10-1128. [Google Scholar] [CrossRef]
- Plavec, T.V.; Berlec, A. Safety aspects of genetically modified lactic acid bacteria. Microorganisms 2020, 8, 297. [Google Scholar] [CrossRef]
- Tellechea-Luzardo, J.; Stiebritz, M.T.; Carbonell, P. Transcription factor-based biosensors for screening and dynamic regulation. Front. Bioeng. Biotechnol. 2023, 11, 1118702. [Google Scholar] [CrossRef]
- Zhou, G.J.; Zhang, F. Applications and Tuning Strategies for Transcription Factor-Based Metabolite Biosensors. Biosensors 2023, 13, 428. [Google Scholar] [CrossRef]
- Saak, C.C.; Dinh, C.B.; Dutton, R.J. Experimental approaches to tracking mobile genetic elements in microbial communities. FEMS Microbiol. Rev. 2020, 44, 606–630. [Google Scholar] [CrossRef]
- Van Dijk, B.; Buffard, P.; Farr, A.D.; Giersdorf, F.; Meijer, J.; Dutilh, B.E.; Rainey, P.B. Identifying and tracking mobile elements in evolving compost communities yields insights into the nanobiome. ISME Commun. 2023, 3, 90. [Google Scholar] [CrossRef]
- Kang, M.; Choe, D.; Kim, K.; Cho, B.K.; Cho, S. Synthetic biology approaches in the development of engineered therapeutic microbes. Int. J. Mol. Sci. 2020, 21, 8744. [Google Scholar] [CrossRef]
- De Sousa, J.M.; Lourenço, M.; Gordo, I. Horizontal gene transfer among host-associated microbes. Cell Host Microbe 2023, 31, 513–527. [Google Scholar] [CrossRef]
- Wozniak, C.A.; McClung, G.; Gagliardi, J.; Segal, M.; Matthews, K. Chapter 4—Regulation of Genetically Engineered Microorganisms under FIFRA, FFDCA and TSCA. In Regulation of Agricultural Biotechnology: 57 The United States and Canada; Wozniak, C.A., McHughen, A., Eds.; Springer: Dordrecht, The Netherlands, 2012. [Google Scholar] [CrossRef]
- Hanlon, P.; Sewalt, V. GEMs: Genetically engineered microorganisms and the regulatory oversight of their uses in modern food production. Crit. Rev. Food Sci. Nutr. 2021, 61, 959–970. [Google Scholar] [CrossRef]
- Parigi, T.L.; Vieujean, S.; Paridaens, K.; Dalgaard, K.; Peyrin-Biroulet, L.; Danese, S. Efficacy, Safety, and Concerns on Microbiota Modulation, Antibiotics, Probiotics, and Fecal Microbial Transplant for Inflammatory Bowel Disease and Other Gastrointestinal Conditions: Results from an International Survey. Microorganisms 2023, 11, 2806. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).