M-Batches to Simulate Luminal and Mucosal Human Gut Microbial Ecosystems: A Case Study of the Effects of Coffee and Green Tea
Abstract
1. Introduction
2. Materials and Methods
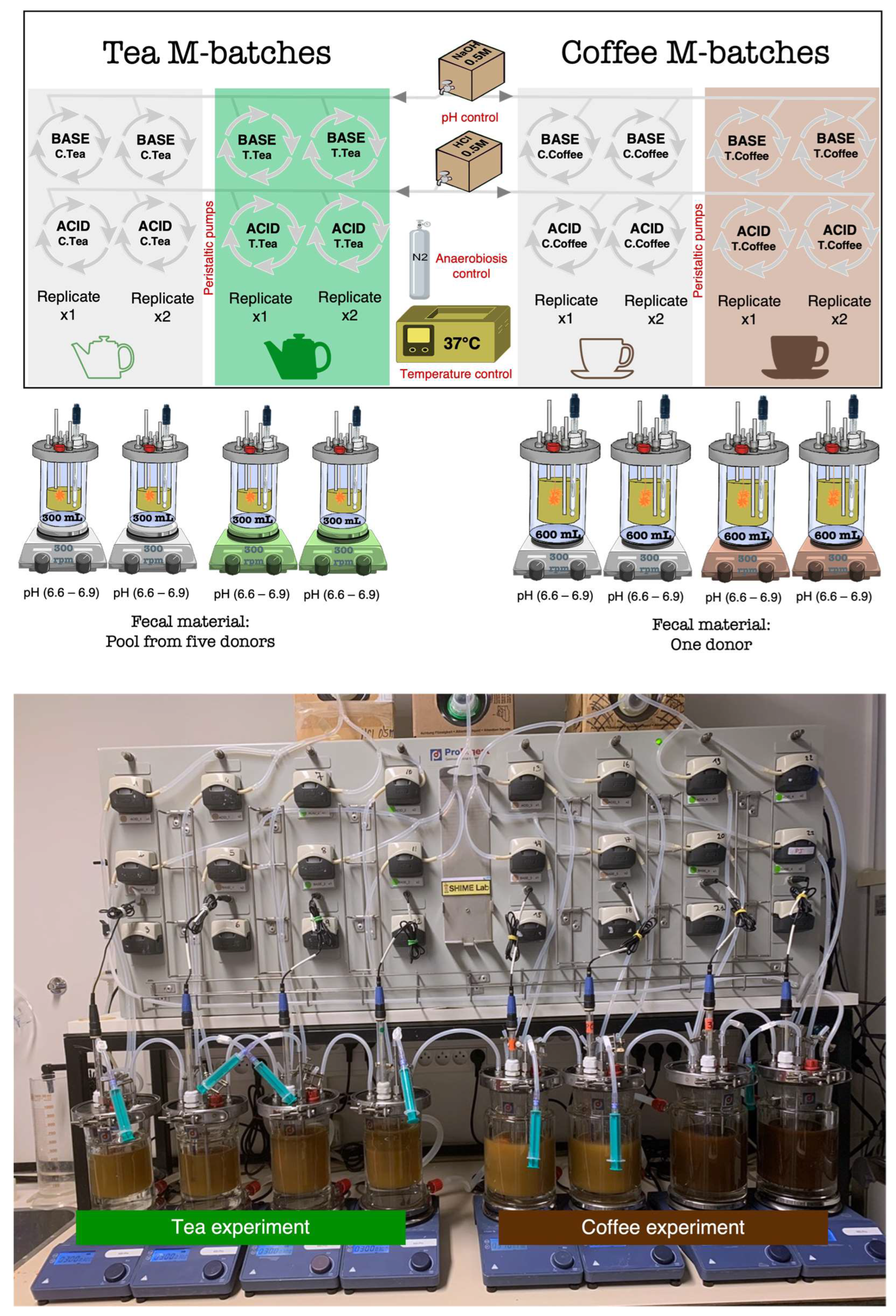
2.1. M-Batches Experiment
2.1.1. Design of the Experiment
2.1.2. Human Stool Samples: Collection, Storage, and Inoculation
2.1.3. Chemicals, Media, and Treatments
- -
- Adult M-SHIME medium: nutritional media for fermentation experiments were obtained from Prodigest (Ghent, Belgium). The composition of the pre-mixed powder (g/L) was: 1.2 g arabinogalactan, 2.0 g pectin, 0.5 g xylan, 0.4 g glucose, 3.0 g yeast extract, 1.0 g special peptone, 2.0 g mucin, 0.5 g L-cysteine-HCl, and 4.0 g starch. It was prepared in demineralized water and autoclaved prior to use.
- -
- Green tea treatment: One dose (~60 mL) taken from a cup of tea (250 mL) prepared using one tea bag from Lipton brand (acquired in the supermarket) was added to vessels with a 300 mL final volume vessel (in duplicate). The same amount of water was added as a blank control (in duplicate).
- -
- Coffee treatment: the equivalent of two ristretto cups (~50 mL) of coffee from the Nespresso brand, the Ispirazione Napoli variety (acquired in the supermarket), was added to vessels with a 600 mL final volume (in duplicate). The same amount of water was added as a blank control (in duplicate).
2.1.4. Mucin-Covered Microcosms
2.2. Analysis of Microbial-Derived Metabolites
2.2.1. SCFA by SPME-GC-MS
2.2.2. Ammonia and Biogenic Amines by UPLC-FLD
2.3. Analysis of Colonic Bacterial Populations
2.4. Cell-Based Analysis of AhR Induction
3. Results
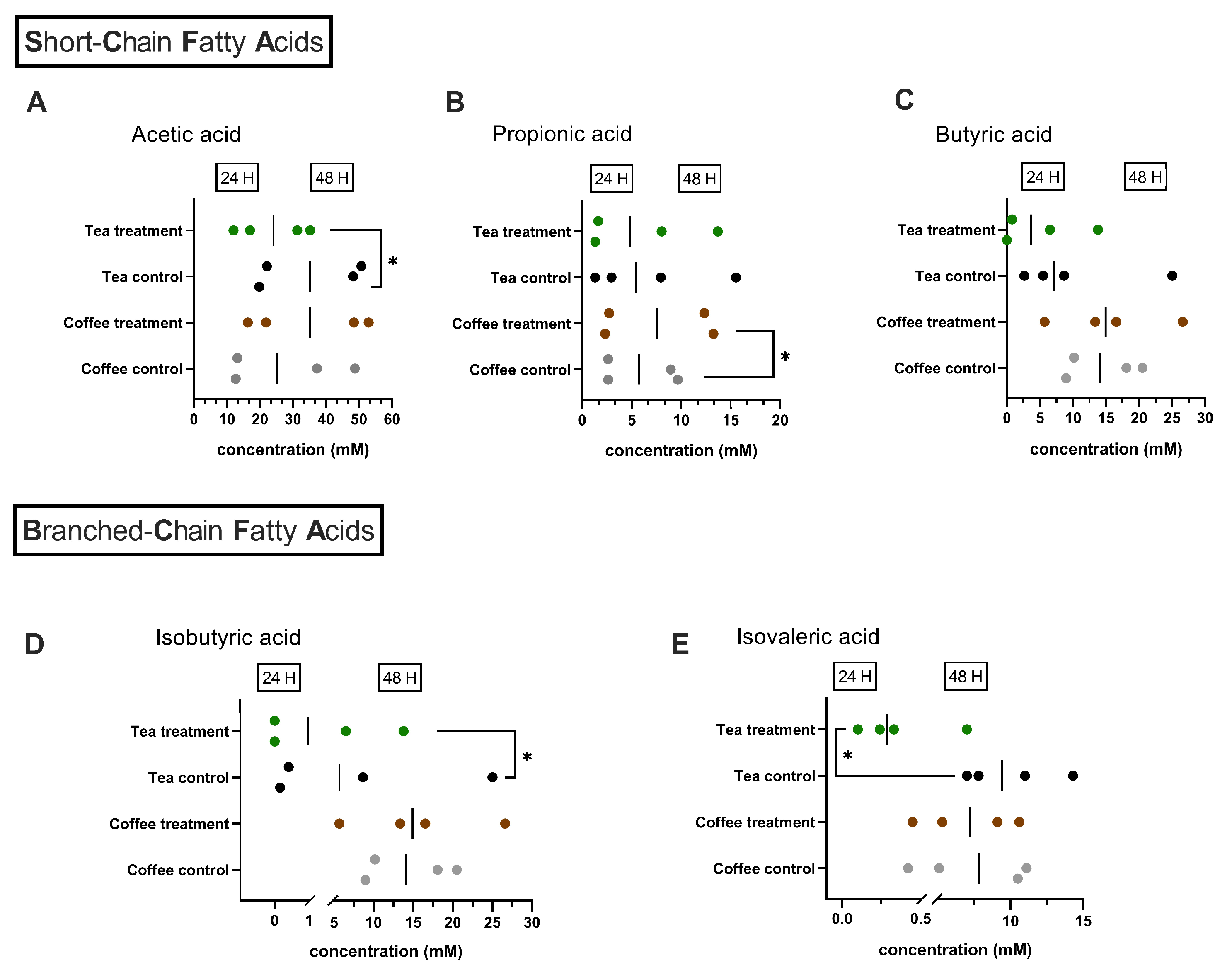
3.1. Chromatographic Quantification of Metabolites
3.2. Evolution of Colon Bacterial Populations
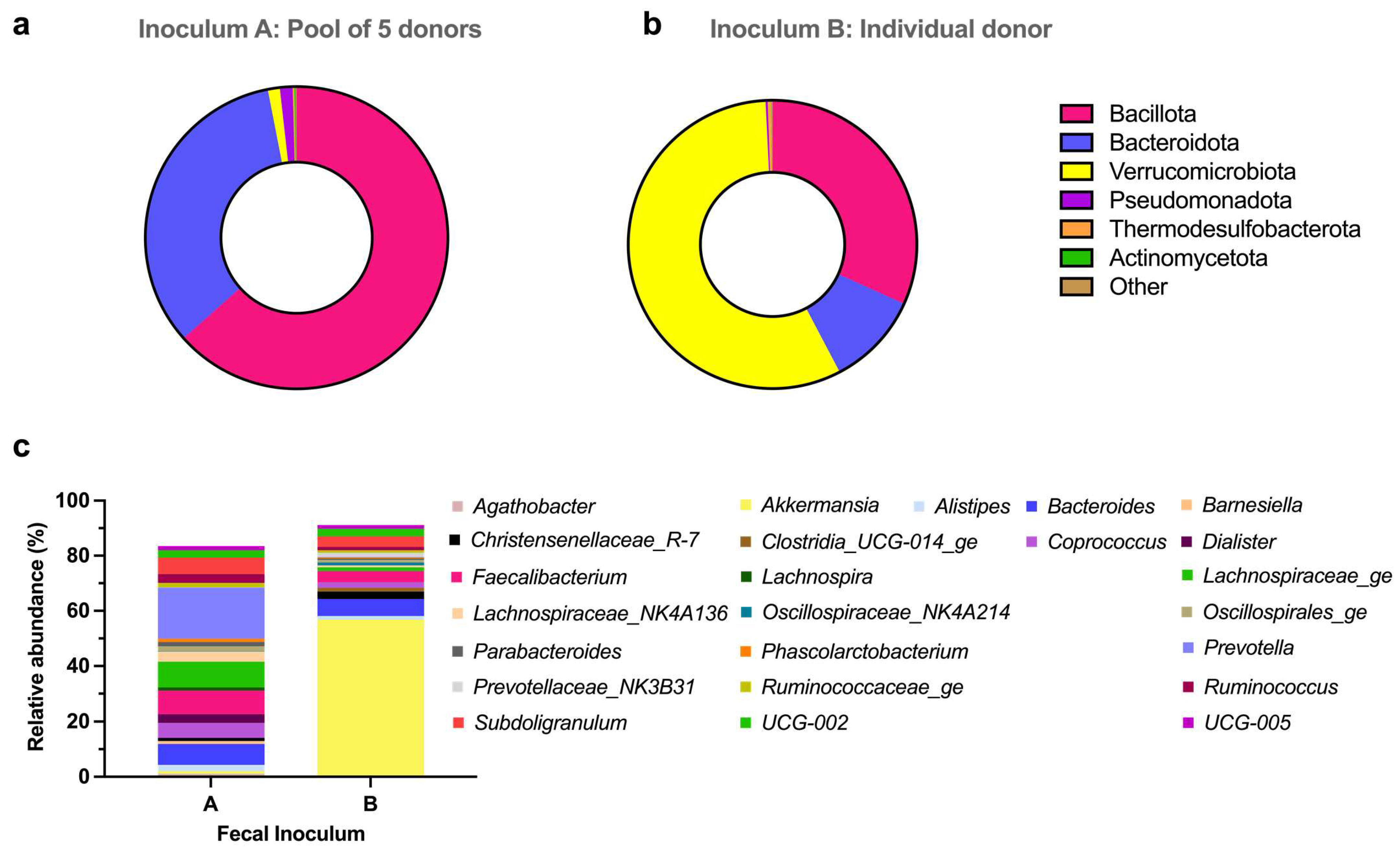
3.2.1. Metataxonomic Analysis of the Fecal Inocula
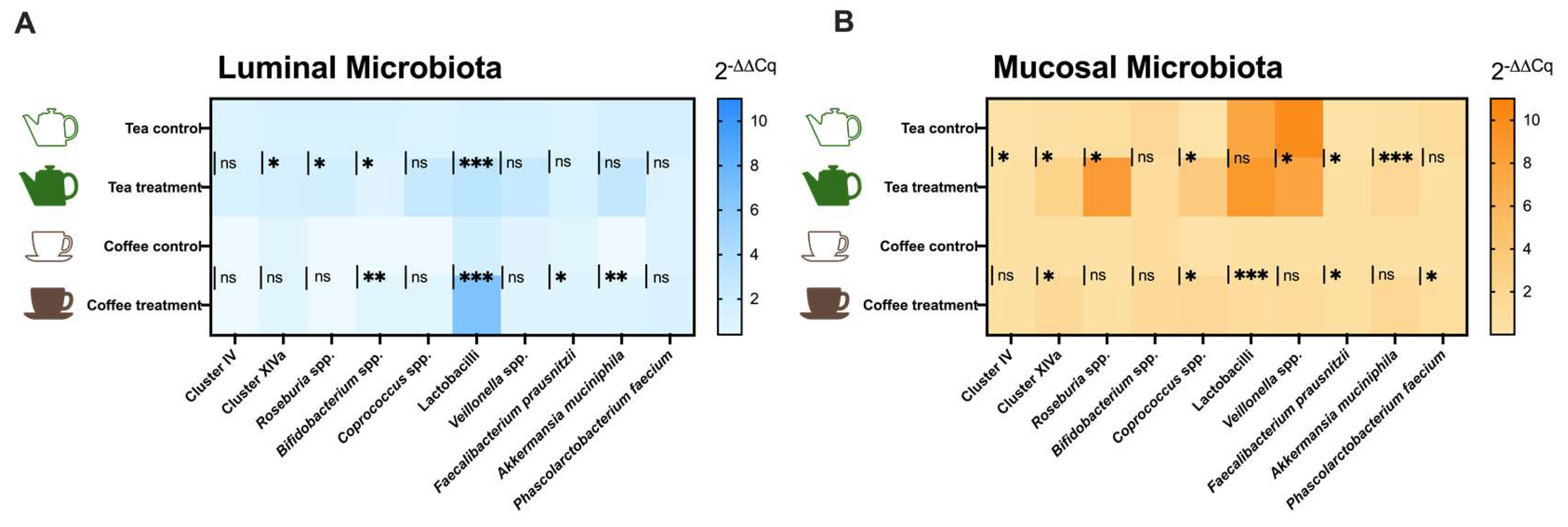
3.2.2. qPCR Results
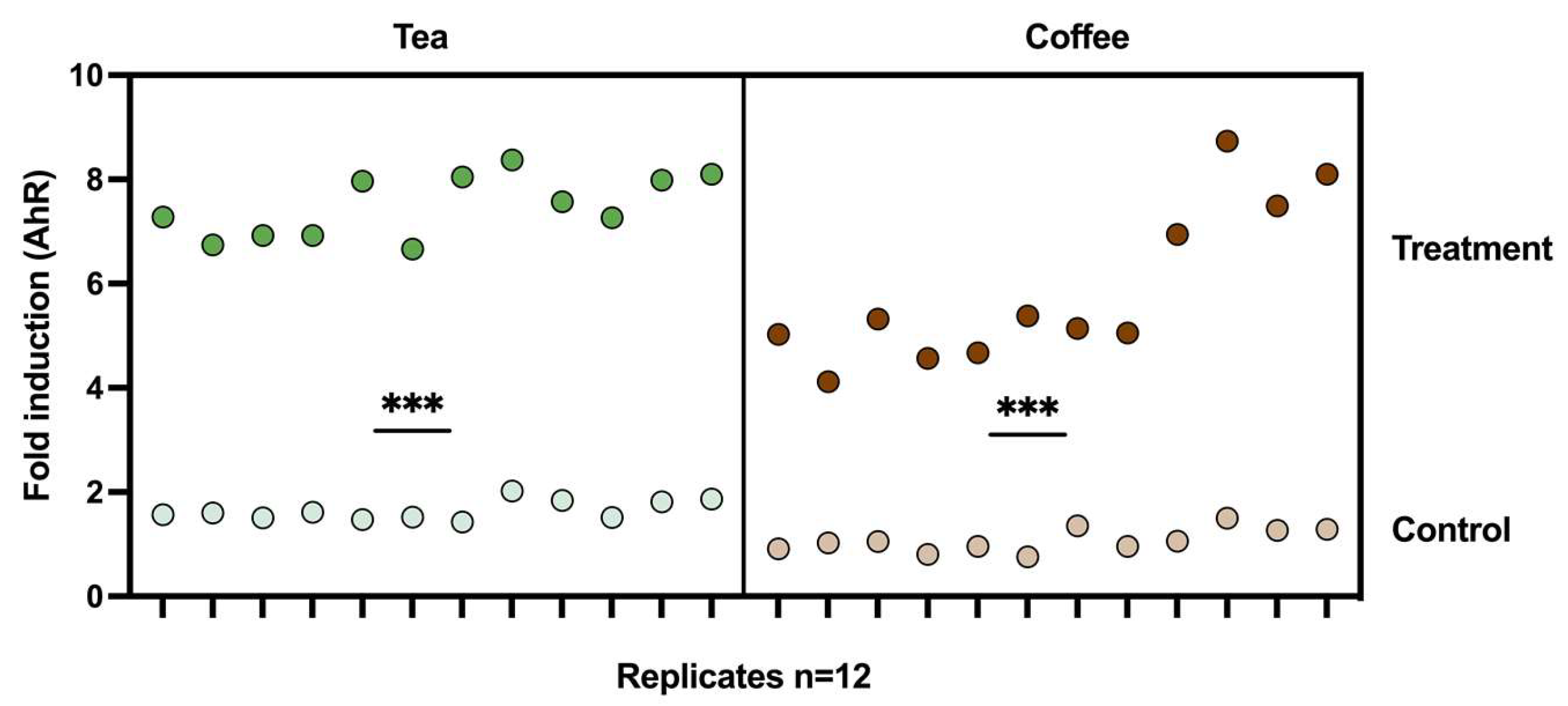
3.3. Induction of AhR Activation Caused by the Metabolic Output of the M-Batches
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roupar, D.; Berni, P.; Martins, J.T.; Caetano, A.C.; Teixeira, J.A.; Nobre, C. Bioengineering Approaches to Simulate Human Colon Microbiome Ecosystem. Trends Food Sci. Technol. 2021, 112, 808–822. [Google Scholar] [CrossRef]
- Veintimilla-Gozalbo, E.; Asensio-Grau, A.; Calvo-Lerma, J.; Heredia, A.; Andrés, A. In Vitro Simulation of Human Colonic Fermentation: A Practical Approach towards Models’ Design and Analytical Tools. Appl. Sci. 2021, 11, 8135. [Google Scholar] [CrossRef]
- Biagini, F.; Daddi, C.; Calvigioni, M.; De Maria, C.; Zhang, Y.S.; Ghelardi, E.; Vozzi, G. Designs and Methodologies to Recreate in Vitro Human Gut Microbiota Models. Biodes Manuf. 2023, 6, 298–318. [Google Scholar] [CrossRef]
- Isenring, J.; Bircher, L.; Geirnaert, A.; Lacroix, C. In Vitro Human Gut Microbiota Fermentation Models: Opportunities, Challenges, and Pitfalls. Microbiome Res. Rep. 2023, 2, 2. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Burillo, S.; Molino, S.; Navajas-Porras, B.; Valverde-Moya, Á.J.; Hinojosa-Nogueira, D.; López-Maldonado, A.; Pastoriza, S.; Rufián-Henares, J.Á. An in Vitro Batch Fermentation Protocol for Studying the Contribution of Food to Gut Microbiota Composition and Functionality. Nat. Protoc. 2021, 16, 3186–3209. [Google Scholar] [CrossRef] [PubMed]
- Goya-Jorge, E.; Bondue, P.; Gonza, I.; Laforêt, F.; Antoine, C.; Boutaleb, S.; Douny, C.; Scippo, M.-L.; de Ribaucourt, J.C.; Crahay, F.; et al. Butyrogenic, Bifidogenic and Slight Anti-Inflammatory Effects of a Green Kiwifruit Powder (Kiwi FFG®) in a Human Gastrointestinal Model Simulating Mild Constipation. Food Res. Int. 2023, 173, 113348. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Son, H.; Lee, G.; Lee, S.; Unno, T.; Shin, J.-H. Role, Relevance, and Possibilities of in Vitro Fermentation Models in Human Dietary, and Gut-Microbial Studies. Biotechnol. Bioeng. 2022, 119, 3044–3061. [Google Scholar] [CrossRef]
- Song, C.; Chai, Z.; Chen, S.; Zhang, H.; Zhang, X.; Zhou, Y. Intestinal Mucus Components and Secretion Mechanisms: What We Do and Do Not Know. Exp. Mol. Med. 2023, 55, 681–691. [Google Scholar] [CrossRef]
- Luis, A.S.; Hansson, G.C. Intestinal Mucus and Their Glycans: A Habitat for Thriving Microbiota. Cell Host Microbe 2023, 31, 1087–1100. [Google Scholar] [CrossRef]
- Tailford, L.E.; Crost, E.H.; Kavanaugh, D.; Juge, N. Mucin Glycan Foraging in the Human Gut Microbiome. Front. Genet. 2015, 6, 81. [Google Scholar] [CrossRef]
- Glover, J.S.; Ticer, T.D.; Engevik, M.A. Characterizing the Mucin-Degrading Capacity of the Human Gut Microbiota. Sci. Rep. 2022, 12, 8456. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Barua, N.; Ip, M. Mucin-Degrading Gut Commensals Isolated from Healthy Faecal Donor Suppress Intestinal Epithelial Inflammation and Regulate Tight Junction Barrier Function. Front. Immunol. 2022, 13, 1021094. [Google Scholar] [CrossRef] [PubMed]
- Van Herreweghen, F.; De Paepe, K.; Roume, H.; Kerckhof, F.-M.; Van de Wiele, T. Mucin Degradation Niche as a Driver of Microbiome Composition and Akkermansia Muciniphila Abundance in a Dynamic Gut Model Is Donor Independent. FEMS Microbiol. Ecol. 2018, 94, fiy186. [Google Scholar] [CrossRef] [PubMed]
- Vaga, S.; Lee, S.; Ji, B.; Andreasson, A.; Talley, N.J.; Agréus, L.; Bidkhori, G.; Kovatcheva-Datchary, P.; Park, J.; Lee, D.; et al. Compositional and Functional Differences of the Mucosal Microbiota along the Intestine of Healthy Individuals. Sci. Rep. 2020, 10, 14977. [Google Scholar] [CrossRef] [PubMed]
- Van den Abbeele, P.; Roos, S.; Eeckhaut, V.; MacKenzie, D.A.; Derde, M.; Verstraete, W.; Marzorati, M.; Possemiers, S.; Vanhoecke, B.; Van Immerseel, F.; et al. Incorporating a Mucosal Environment in a Dynamic Gut Model Results in a More Representative Colonization by Lactobacilli. Microb. Biotechnol. 2012, 5, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.H.T.; Boudry, C.; Everaert, N.; Théwis, A.; Portetelle, D.; Daube, G.; Nezer, C.; Taminiau, B.; Bindelle, J. Adding Mucins to an in Vitro Batch Fermentation Model of the Large Intestine Induces Changes in Microbial Population Isolated from Porcine Feces Depending on the Substrate. FEMS Microbiol. Ecol. 2016, 92, fiv165. [Google Scholar] [CrossRef] [PubMed]
- Douny, C.; Dufourny, S.; Brose, F.; Verachtert, P.; Rondia, P.; Lebrun, S.; Marzorati, M.; Everaert, N.; Delcenserie, V.; Scippo, M.-L. Development of an Analytical Method to Detect Short-Chain Fatty Acids by SPME-GC-MS in Samples Coming from an in Vitro Gastrointestinal Model. J. Chromatogr. B 2019, 1124, 188–196. [Google Scholar] [CrossRef]
- Goya-Jorge, E.; Gonza, I.; Bondue, P.; Douny, C.; Taminiau, B.; Daube, G.; Scippo, M.-L.; Delcenserie, V. Human Adult Microbiota in a Static Colon Model: AhR Transcriptional Activity at the Crossroads of Host-Microbe Interaction. Foods 2022, 11, 1946. [Google Scholar] [CrossRef]
- Douny, C.; Benmedjadi, S.; Brose, F.; Afé, O.H.I.; Igout, A.; Hounhouigan, D.J.; Anihouvi, V.B.; Scippo, M.-L. Development of an Analytical Method for the Simultaneous Measurement of 10 Biogenic Amines in Meat: Application to Beninese Grilled Pork Samples. Food Anal. Methods 2019, 12, 2392–2400. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Belzer, C.; Goossens, M.; Kleerebezem, M.; De Vos, W.M.; Thas, O.; De Weirdt, R.; Kerckhof, F.-M.; Van de Wiele, T. Butyrate-Producing Clostridium Cluster XIVa Species Specifically Colonize Mucins in an in Vitro Gut Model. ISME J. 2013, 7, 949–961. [Google Scholar] [CrossRef] [PubMed]
- Goya-Jorge, E.; Gonza, I.; Bondue, P.; Druart, G.; Al-Chihab, M.; Boutaleb, S.; Douny, C.; Scippo, M.-L.; Thonart, P.; Delcenserie, V. Evaluation of Four Multispecies Probiotic Cocktails in a Human Colonic Fermentation Model. Probiotics Antimicrob. Proteins 2023, 15. [Google Scholar] [CrossRef] [PubMed]
- Firrman, J.; Liu, L.; Mahalak, K.; Tanes, C.; Bittinger, K.; Tu, V.; Bobokalonov, J.; Mattei, L.; Zhang, H.; Van den Abbeele, P. The Impact of Environmental PH on the Gut Microbiota Community Structure and Short Chain Fatty Acid Production. FEMS Microbiol. Ecol. 2022, 98, fiac038. [Google Scholar] [CrossRef] [PubMed]
- Ghyselinck, J.; Verstrepen, L.; Moens, F.; Van den Abbeele, P.; Said, J.; Smith, B.; Bjarnason, I.; Basit, A.W.; Gaisford, S. A 4-Strain Probiotic Supplement Influences Gut Microbiota Composition and Gut Wall Function in Patients with Ulcerative Colitis. Int. J. Pharm. 2020, 587, 119648. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, M.; Eck, A.; Koenen, M.E.; Savelkoul, P.H.M.; Budding, A.E.; Venema, K. Evaluation of an Optimal Preparation of Human Standardized Fecal Inocula for in Vitro Fermentation Studies. J. Microbiol. Methods 2015, 117, 78–84. [Google Scholar] [CrossRef]
- Aguirre, M.; Ramiro-Garcia, J.; Koenen, M.E.; Venema, K. To Pool or Not to Pool? Impact of the Use of Individual and Pooled Fecal Samples for in Vitro Fermentation Studies. J. Microbiol. Methods 2014, 107, 1–7. [Google Scholar] [CrossRef]
- Chen, J.; Tang, H.; Zhang, M.; Sang, S.; Jia, L.; Ou, C. Exploration of the Roles of Microbiota on Biogenic Amines Formation during Traditional Fermentation of Scomber Japonicus. Front. Microbiol. 2022, 13, 1030789. [Google Scholar] [CrossRef]
- Pérez-Burillo, S.; Mehta, T.; Esteban-Muñoz, A.; Pastoriza, S.; Paliy, O.; Ángel Rufián-Henares, J. Effect of in Vitro Digestion-Fermentation on Green and Roasted Coffee Bioactivity: The Role of the Gut Microbiota. Food Chem. 2019, 279, 252–259. [Google Scholar] [CrossRef]
- Preda, M.; Popa, M.I.; Mihai, M.M.; Oţelea, T.C.; Holban, A.M. Effects of Coffee on Intestinal Microbiota, Immunity, and Disease. In Caffeinated and Cocoa Based Beverages; Grumezescu, A.M., Holban, A.M., Eds.; Woodhead Publishing: Sawston, UK, 2019; pp. 391–421. ISBN 978-0-12-815864-7. [Google Scholar]
- González, S.; Salazar, N.; Ruiz-Saavedra, S.; Gómez-Martín, M.; de Los Reyes-Gavilán, C.G.; Gueimonde, M. Long-Term Coffee Consumption Is Associated with Fecal Microbial Composition in Humans. Nutrients 2020, 12, 1287. [Google Scholar] [CrossRef]
- Wu, Z.; Huang, S.; Li, T.; Li, N.; Han, D.; Zhang, B.; Xu, Z.Z.; Zhang, S.; Pang, J.; Wang, S.; et al. Gut Microbiota from Green Tea Polyphenol-Dosed Mice Improves Intestinal Epithelial Homeostasis and Ameliorates Experimental Colitis. Microbiome 2021, 9, 184. [Google Scholar] [CrossRef]
- Pérez-Burillo, S.; Navajas-Porras, B.; López-Maldonado, A.; Hinojosa-Nogueira, D.; Pastoriza, S.; Rufián-Henares, J.Á. Green Tea and Its Relation to Human Gut Microbiome. Molecules 2021, 26, 3907. [Google Scholar] [CrossRef] [PubMed]
- Chapkin, R.S.; Davidson, L.A.; Park, H.; Jin, U.-H.; Fan, Y.-Y.; Cheng, Y.; Hensel, M.E.; Landrock, K.K.; Allred, C.; Menon, R.; et al. Role of the Aryl Hydrocarbon Receptor (AhR) in Mediating the Effects of Coffee in the Colon. Mol. Nutr. Food Res. 2021, 65, e2100539. [Google Scholar] [CrossRef]
- Palermo, C.M.; Hernando, J.I.M.; Dertinger, S.D.; Kende, A.S.; Gasiewicz, T.A. Identification of Potential Aryl Hydrocarbon Receptor Antagonists in Green Tea. Chem. Res. Toxicol. 2003, 16, 865–872. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.N.; Shih, H.; Guenette, D.K.; Brackney, W.; Denison, M.S.; Pickwell, G.V.; Quattrochi, L.C. Comparative Studies on the Effects of Green Tea Extracts and Individual Tea Catechins on Human CYP1A Gene Expression. Chem. Biol. Interact. 2000, 128, 211–229. [Google Scholar] [CrossRef] [PubMed]
- Stockinger, B.; Shah, K.; Wincent, E. AHR in the Intestinal Microenvironment: Safeguarding Barrier Function. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 559–570. [Google Scholar] [CrossRef]
- Lamas, B.; Natividad, J.M.; Sokol, H. Aryl Hydrocarbon Receptor and Intestinal Immunity. Mucosal Immunol. 2018, 11, 1024–1038. [Google Scholar] [CrossRef]
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef]
- Bacchetti De Gregoris, T.; Aldred, N.; Clare, A.S.; Burgess, J.G. Improvement of Phylum- and Class-Specific Primers for Real-Time PCR Quantification of Bacterial Taxa. J. Microbiol. Methods 2011, 86, 351–356. [Google Scholar] [CrossRef]
- Matsuki, T.; Watanabe, K.; Fujimoto, J.; Takada, T.; Tanaka, R. Use of 16S RRNA Gene-Targeted Group-Specific Primers for Real-Time PCR Analysis of Predominant Bacteria in Human Feces. Appl. Environ. Microbiol. 2004, 70, 7220–7228. [Google Scholar] [CrossRef]
- Ramirez-Farias, C.; Slezak, K.; Fuller, Z.; Duncan, A.; Holtrop, G.; Louis, P. Effect of Inulin on the Human Gut Microbiota: Stimulation of Bifidobacterium Adolescentis and Faecalibacterium Prausnitzii. Br. J. Nutr. 2009, 101, 541–550. [Google Scholar] [CrossRef]
- Morel, F.B.; Oozeer, R.; Piloquet, H.; Moyon, T.; Pagniez, A.; Knol, J.; Darmaun, D.; Michel, C. Preweaning Modulation of Intestinal Microbiota by Oligosaccharides or Amoxicillin Can Contribute to Programming of Adult Microbiota in Rats. Nutrition 2015, 31, 515–522. [Google Scholar] [CrossRef]
- Amit-Romach, E.; Sklan, D.; Uni, Z. Microflora Ecology of the Chicken Intestine Using 16S Ribosomal DNA Primers. Poult. Sci. 2004, 83, 1093–1098. [Google Scholar] [CrossRef]
- Rinttilä, T.; Kassinen, A.; Malinen, E.; Krogius, L.; Palva, A. Development of an Extensive Set of 16S RDNA-Targeted Primers for Quantification of Pathogenic and Indigenous Bacteria in Faecal Samples by Real-Time PCR. J. Appl. Microbiol. 2004, 97, 1166–1177. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-Talk between Akkermansia Muciniphila and Intestinal Epithelium Controls Diet-Induced Obesity. Proc. Nat. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef]
- Wang, R.F.; Cao, W.W.; Cerniglia, C.E. PCR Detection and Quantitation of Predominant Anaerobic Bacteria in Human and Animal Fecal Samples. Appl. Environ. Microbiol. 1996, 62, 1242–1247. [Google Scholar] [CrossRef]
- Wu, F.; Guo, X.; Zhang, J.; Zhang, M.; Ou, Z.; Peng, Y. Phascolarctobacterium Faecium Abundant Colonization in Human Gastrointestinal Tract. Exp. Ther. Med. 2017, 14, 3122–3126. [Google Scholar] [CrossRef]
- Lyra, A.; Krogius-Kurikka, L.; Nikkilä, J.; Malinen, E.; Kajander, K.; Kurikka, K.; Korpela, R.; Palva, A. Effect of a Multispecies Probiotic Supplement on Quantity of Irritable Bowel Syndrome-Related Intestinal Microbial Phylotypes. BMC Gastroenterol. 2010, 10, 110. [Google Scholar] [CrossRef]
| Metabolites | Control (Tea M-Batches) | Tea Treatment | Control (Coffee M-Batches) | Coffee Treatment |
|---|---|---|---|---|
| Ammonia (mM) | 7.80 ± 2.10 | 9.89 ± 0.63 | 1.83 ± 0.36 | 2.11 ± 0.12 |
| 2-phenylethylamine(μM) | 36.17 ± 3.74 | 42.29 ± 1.94 | ND | ND |
| Tyramine (μM) | 12.68 ± 2.86 | 18.01 ± 0.42 | ND | ND |
| Putrescine (μM) | 531.99 ± 5.59 | 517.18 ± 0.52 | ND | ND |
| Cadaverine (μM) | 523.5 ± 54.11 | 443.7 ± 5.71 | 15.83 ± 0.36 | 15.74 ± 0.34 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goya-Jorge, E.; Gonza, I.; Douny, C.; Scippo, M.-L.; Delcenserie, V. M-Batches to Simulate Luminal and Mucosal Human Gut Microbial Ecosystems: A Case Study of the Effects of Coffee and Green Tea. Microorganisms 2024, 12, 236. https://doi.org/10.3390/microorganisms12020236
Goya-Jorge E, Gonza I, Douny C, Scippo M-L, Delcenserie V. M-Batches to Simulate Luminal and Mucosal Human Gut Microbial Ecosystems: A Case Study of the Effects of Coffee and Green Tea. Microorganisms. 2024; 12(2):236. https://doi.org/10.3390/microorganisms12020236
Chicago/Turabian StyleGoya-Jorge, Elizabeth, Irma Gonza, Caroline Douny, Marie-Louise Scippo, and Véronique Delcenserie. 2024. "M-Batches to Simulate Luminal and Mucosal Human Gut Microbial Ecosystems: A Case Study of the Effects of Coffee and Green Tea" Microorganisms 12, no. 2: 236. https://doi.org/10.3390/microorganisms12020236
APA StyleGoya-Jorge, E., Gonza, I., Douny, C., Scippo, M.-L., & Delcenserie, V. (2024). M-Batches to Simulate Luminal and Mucosal Human Gut Microbial Ecosystems: A Case Study of the Effects of Coffee and Green Tea. Microorganisms, 12(2), 236. https://doi.org/10.3390/microorganisms12020236






