Novel Wild-Type Pediococcus and Lactiplantibacillus Strains as Probiotic Candidates to Manage Obesity-Associated Insulin Resistance
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.1.1. Isolation of Lactic Acid Bacteria (LAB) Strains and Culture Conditions
2.1.2. Pathogenic Microbial Strains
2.2. Bacterial Identification and Classification
2.2.1. Phenotypic Identification
2.2.2. Genetic Identification of the 16S rRNA Gene
2.2.3. Whole-Genome Sequencing (WGS) and De Novo Assembly of the Isolates
2.2.4. Genome Annotation
2.3. In Vitro Functional Properties Associated with Probiotic Characteristics
2.3.1. Ability to Survive in Simulated Gastrointestinal (GI) Conditions
2.3.2. Bacterial Adhesion to Differentiated Caco-2 Cells
2.3.3. Bile Salt Hydrolase (BSH) Secretion
2.3.4. Cholesterol-Lowering Ability
2.3.5. Evaluation of Cholesterol Binding by Bacterial Strains
2.3.6. Growth Inhibition Activity against Food-Borne Pathogens
2.4. Safety Criteria Assays
2.4.1. Hemolytic Activity
2.4.2. Susceptibility to Antibiotics
Acquired Antimicrobial Resistance Genes
Disk Diffusion Assay
Minimum Inhibitory Concentration Determination
2.5. In Vivo Study Design
2.5.1. Mice
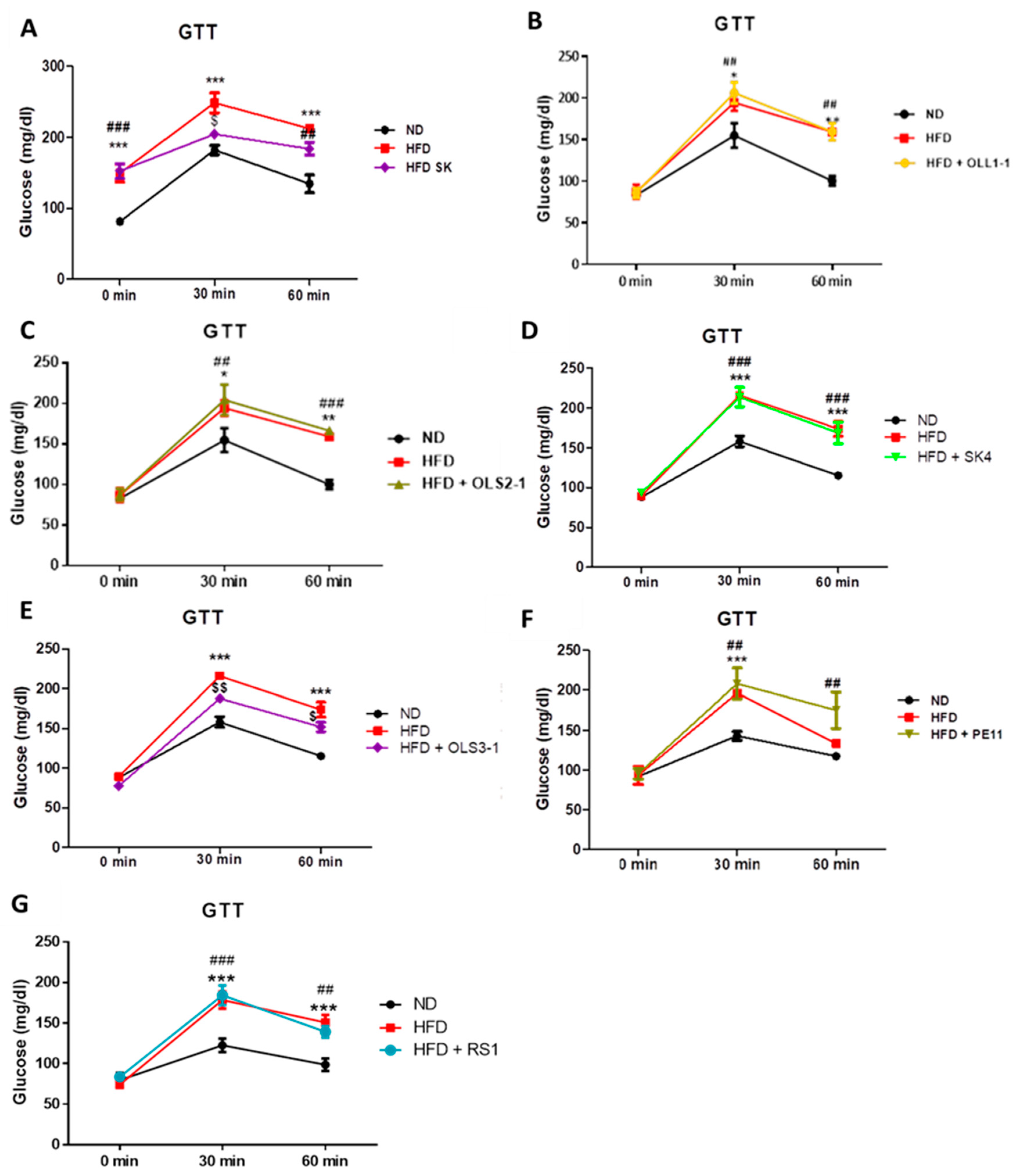
2.5.2. Glucose Tolerance Test (GTT)
2.5.3. Analysis of Fecal Microbiota
2.6. Statistical Analysis
3. Results and Discussion
3.1. Bacterial Isolation and Identification
3.2. Ability to Survive through GI Tract Conditions
3.3. Bacterial Adhesion to Differentiated Caco-2 Cells
3.4. BSH Secretion
3.5. Reduction of Cholesterol Levels
3.5.1. Cholesterol-Lowering Ability
3.5.2. Evaluation of Cholesterol-Binding Ability of Bacterial Strains
3.6. Growth Inhibition Activity against Food-Borne Pathogens
3.7. Safety Criteria
Susceptibility to Antibiotics
3.8. In Vivo Assays
3.8.1. Effect of Probiotic Strains on Glucose Intolerance and Insulin Resistance
3.8.2. Preliminary Evaluation of the Effect of Potential Probiotic Supplementation on Fecal Microbiota
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO/WHO. Joint FAO/WHO Working Group Report on Drafting Guidelines for the Evaluation of Probiotics in Food; FAO/WHO: London, ON, Canada, 2002. [Google Scholar]
- Bouter, K.E.; van Raalte, D.H.; Groen, A.K.; Nieuwdorp, M. Role of the Gut Microbiome in the Pathogenesis of Obesity and Obesity-Related Metabolic Dysfunction. Gastroenterology 2017, 152, 1671–1678. [Google Scholar] [CrossRef] [PubMed]
- Al-Emran, H.M.; Moon, J.F.; Miah, M.L.; Meghla, N.S.; Reuben, R.C.; Uddin, M.J.; Ibnat, H.; Sarkar, S.L.; Roy, P.C.; Rahman, M.S.; et al. Genomic Analysis and in Vivo Efficacy of Pediococcus Acidilactici as a Potential Probiotic to Prevent Hyperglycemia, Hypercholesterolemia and Gastrointestinal Infections. Sci. Rep. 2022, 12, 20429. [Google Scholar] [CrossRef] [PubMed]
- Balakumar, M.; Prabhu, D.; Sathishkumar, C.; Prabu, P.; Rokana, N.; Kumar, R.; Raghavan, S.; Soundarajan, A.; Grover, S.; Batish, V.K.; et al. Improvement in Glucose Tolerance and Insulin Sensitivity by Probiotic Strains of Indian Gut Origin in High-Fat Diet-Fed C57BL/6J Mice. Eur. J. Nutr. 2018, 57, 279–295. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Lee, J.Y.; Kang, C. Hyperglycemia in High-Fat Diet-Induced Mice. Foods 2022, 11, 1–12. [Google Scholar]
- Argyri, A.A.; Zoumpopoulou, G.; Karatzas, K.A.G.; Tsakalidou, E.; Nychas, G.J.E.; Panagou, E.Z.; Tassou, C.C. Selection of Potential Probiotic Lactic Acid Bacteria from Fermented Olives by in Vitro Tests. Food Microbiol. 2013, 33, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Santarmaki, V.; Kourkoutas, Y.; Zoumpopoulou, G.; Mavrogonatou, E.; Kiourtzidis, M.; Chorianopoulos, N.; Tassou, C.; Tsakalidou, E.; Simopoulos, C.; Ypsilantis, P. Survival, Intestinal Mucosa Adhesion, and Immunomodulatory Potential of Lactobacillus Plantarum Strains. Curr. Microbiol. 2017, 74, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Duysburgh, C.; Van den Abbeele, P.; Morera, M.; Marzorati, M. Lacticaseibacillus Rhamnosus GG and Saccharomyces Cerevisiae Boulardii Supplementation Exert Protective Effects on Human Gut Microbiome Following Antibiotic Administration in Vitro. Benef. Microbes 2021, 12, 365–379. [Google Scholar] [CrossRef]
- Fang, Z.; Lu, W.; Zhao, J.; Zhang, H.; Qian, L.; Wang, Q.; Chen, W. Probiotics Modulate the Gut Microbiota Composition and Immune Responses in Patients with Atopic Dermatitis: A Pilot Study. Eur. J. Nutr. 2020, 59, 2119–2130. [Google Scholar] [CrossRef]
- Hor, Y.Y.; Liong, M.T. Use of Extracellular Extracts of Lactic Acid Bacteria and Bifidobacteria for the Inhibition of Dermatological Pathogen Staphylococcus Aureus. Dermatol. Sin. 2014, 32, 141–147. [Google Scholar] [CrossRef]
- Ratsep, M.; Naaber, P.; Kõljalg, S.; Smidt, I.; Shkut, E.; Sepp, E. Effect of Lactobacillus Plantarum Strains on Clinical Isolates of Clostridium Difficile in Vitro. J. Probiotics Health 2014, 2, 1–5. [Google Scholar] [CrossRef]
- Shokryazdan, P.; Sieo, C.C.; Kalavathy, R.; Liang, J.B.; Alitheen, N.B.; Jahromi, M.F.; Ho, Y.W. Probiotic Potential of Lactobacillus Strains with Antimicrobial Activity against Some Human Pathogenic Strains. BioMed Res. Int. 2014, 2014, 927268. [Google Scholar] [CrossRef] [PubMed]
- Chikindas, M.L.; Weeks, R.; Drider, D.; Chistyakov, V.A.; Dicks, L.M. Functions and Emerging Applications of Bacteriocins. Curr. Opin. Biotechnol. 2018, 49, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.A.; Ayivi, R.D.; Zimmerman, T.; Siddiqui, S.A.; Altemimi, A.B.; Fidan, H.; Esatbeyoglu, T.; Bakhshayesh, R.V. Lactic Acid Bacteria as Antimicrobial Agents: Food Safety and Microbial Food Spoilage Prevention. Foods 2021, 10, 3131. [Google Scholar] [CrossRef] [PubMed]
- Şanlıbaba, P.; Güçer, Y. Antimicrobial Activity of Lactic Acid Bacteria. J. Int. Sci. Publ. 2015, 3, 451–457. [Google Scholar]
- Sieuwerts, S.; Bron, P.A.; Smid, E.J. Mutually Stimulating Interactions between Lactic Acid Bacteria and Saccharomyces Cerevisiae in Sourdough Fermentation. LWT 2018, 90, 201–206. [Google Scholar] [CrossRef]
- Porto, M.C.W.; Kuniyoshi, T.M.; Azevedo, P.O.S.; Vitolo, M.; Oliveira, R.P.S. Pediococcus Spp.: An Important Genus of Lactic Acid Bacteria and Pediocin Producers. Biotechnol. Adv. 2017, 35, 361–374. [Google Scholar] [CrossRef]
- Shah, A.A.; Yuan, X.; Khan, R.U.; Shao, T. Effect of Lactic Acid Bacteria-treated King Grass Silage on the Performance Traits and Serum Metabolites in New Zealand White Rabbits (Oryctolagus cuniculus). J. Anim. Physiol. Anim. Nutr. 2018, 102, e902–e908. [Google Scholar] [CrossRef]
- Osman, M.-A.; Neoh, H.; Ab Mutalib, N.-S.; Chin, S.-F.; Jamal, R. 16S rRNA Gene Sequencing for Deciphering the Colorectal Cancer Gut Microbiome: Current Protocols and Workflows. Front. Microbiol. 2018, 9, 767. [Google Scholar] [CrossRef]
- Edwards, U.; Rogall, T.; Blöcker, H.; Emde, M.; Böttger, E.C. Isolation and Direct Complete Nucleotide Determination of Entire Genes. Characterization of a Gene Coding for 16S Ribosomal RNA. Nucleic Acids Res. 1989, 17, 7843–7853. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinforma. 2020, 70, e102. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for Predictions of Phenotypes from Genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A Novel Web Tool for WGS-Based Detection of Antimicrobial Resistance Associated with Chromosomal Point Mutations in Bacterial Pathogens. J. Antimicrob. Chemother. 2017, 72, 2764–2768. [Google Scholar] [CrossRef] [PubMed]
- Munk, P.; Brinch, C.; Møller, F.D.; Petersen, T.N.; Hendriksen, R.S.; Seyfarth, A.M.; Kjeldgaard, J.S.; Svendsen, C.A.; Van Bunnik, B.; Berglund, F.; et al. Genomic Analysis of Sewage from 101 Countries Reveals Global Landscape of Antimicrobial Resistance. Nat. Commun. 2022, 13, 7251. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of Acquired Antimicrobial Resistance Genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Nelios, G.; Santarmaki, V.; Pavlatou, C.; Dimitrellou, D.; Kourkoutas, Y. New Wild-Type Lacticaseibacillus Rhamnosus Strains as Candidates to Manage Type 1 Diabetes. Microorganisms 2022, 10, 272. [Google Scholar] [CrossRef]
- Bosch, M.; Fuentes, M.C.; Audivert, S.; Bonachera, M.A.; Peiró, S.; Cuñé, J. Lactobacillus Plantarum CECT 7527, 7528 and 7529: Probiotic Candidates to Reduce Cholesterol Levels. J. Sci. Food Agric. 2014, 94, 803–809. [Google Scholar] [CrossRef]
- Hernández-Gómez, J.G.; López-Bonilla, A.; Trejo-Tapia, G.; Ávila-Reyes, S.V.; Jiménez-Aparicio, A.R.; Hernández-Sánchez, H. In Vitro Bile Salt Hydrolase (Bsh) Activity Screening of Different Probiotic Microorganisms. Foods 2021, 10, 674. [Google Scholar] [CrossRef]
- Wang, S.C.; Chang, C.K.; Chan, S.C.; Shieh, J.S.; Chiu, C.K.; Duh, P.-D. Effects of Lactic Acid Bacteria Isolated from Fermented Mustard on Lowering Cholesterol. Asian Pac. J. Trop. Biomed. 2014, 4, 523–528. [Google Scholar] [CrossRef]
- Daliri, E.B.M.; Kim, Y.; Do, Y.; Chelliah, R.; Oh, D.H. In Vitro and In Vivo Cholesterol Reducing Ability and Safety of Probiotic Candidates Isolated from Korean Fermented Soya Beans. Probiotics Antimicrob. Proteins 2022, 14, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Mangia, N.P.; Saliba, L.; Deiana, P. Functional and Safety Characterization of Autochthonous Lactobacillus Paracasei FS103 Isolated from Sheep Cheese and Its Survival in Sheep and Cow Fermented Milks during Cold Storage. Ann. Microbiol. 2019, 69, 161–170. [Google Scholar] [CrossRef]
- Papp, M.; Solymosi, N. Review and Comparison of Antimicrobial Resistance Gene Databases. Antibiotics 2022, 11, 339. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E. Comparison of the Web Tools ARG-ANNOT and ResFinder for Detection of Resistance Genes in Bacteria. Antimicrob. Agents Chemother. 2014, 58, 4986. [Google Scholar] [CrossRef]
- Charteris, W.P.; Kelly, P.M.; Morelli, L.; Collins, J.K. Antibiotic Susceptibility of Potentially Probiotic Lactobacillus Species. J. Food Prot. 1998, 61, 1636–1643. [Google Scholar] [CrossRef] [PubMed]
- ISO 10932/IDF 223; Milk and Milk Products. Determination of the Minimal Inhibitory Concentration (MIC) of Antibiotics Applicable to Bifidobacteria and Non-Enterococcal Lactic Acid Bacteria (LAB), 1st ed. International Organization of Standardization/International Dairy Federation: Geneva, Switzerland, 2010.
- Food, E.; Authority, S. Guidance on the Assessment of Bacterial Susceptibility to Antimicrobials of Human and Veterinary Importance 2. EFSA J. 2012, 10, 2740. [Google Scholar] [CrossRef]
- Lee, Y.S.; Li, P.; Huh, J.Y.; Hwang, I.J.; Lu, M.; Kim, J.I.; Ham, M.; Talukdar, S.; Chen, A.; Lu, W.J.; et al. Inflammation Is Necessary for Long-Term but Not Short-Term High-Fat Diet–Induced Insulin Resistance. Diabetes 2011, 60, 2474–2483. [Google Scholar] [CrossRef]
- Wiedemann, M.S.F.; Wueest, S.; Item, F.; Schoenle, E.J.; Konrad, D. Adipose Tissue Inflammation Contributes to Short-Term High-Fat Diet-Induced Hepatic Insulin Resistance. Am. J. Physiol.-Endocrinol. Metab. 2013, 305, E388–E395. [Google Scholar] [CrossRef]
- Lloyd, M.H.; Foden, B.W.; Wolfensohn, S.E. Refinement: Promoting the Three Rs in Practice. Lab. Anim. 2008, 42, 284–293. [Google Scholar] [CrossRef]
- Würbel, H. Ideal Homes? Housing Effects on Rodent Brain and Behaviour. Trends Neurosci. 2001, 24, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Directive 2010/63/EU of The European Parliament and of the Council of 22 September 2010 On the Protection of Animals Used for Scientific Purposes. Off. J. Eur. Union 2010, 276, 33–79.
- Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Hilbert, F.; Lindqvist, R.; et al. Update of the List of QPS-Recommended Biological Agents Intentionally Added to Food or Feed as Notified to EFSA 12: Suitability of Taxonomic Units Notified to EFSA until March 2020. EFSA J. 2020, 18, e06689. [Google Scholar] [CrossRef]
- Minekus, M.; Alminger, M.; Alvito, P.; Ballance, S.; Bohn, T.; Bourlieu, C.; Carrière, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A Standardised Static in Vitro Digestion Method Suitable for Food-an International Consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Zhang, J.; Wang, M.; Du, G.; Chen, J. Lactobacillus Casei Combats Acid Stress by Maintaining Cell Membrane Functionality. J. Ind. Microbiol. Biotechnol. 2012, 39, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wu, C.; Du, G.; Chen, J. Enhanced Acid Tolerance in Lactobacillus Casei by Adaptive Evolution and Compared Stress Response during Acid Stress. Biotechnol. Bioprocess Eng. 2012, 17, 283–289. [Google Scholar] [CrossRef]
- Vasiee, A.; Mortazavi, S.A.; Sankian, M.; Yazdi, F.T.; Mahmoudi, M.; Shahidi, F. Antagonistic Activity of Recombinant Lactococcus Lactis NZ1330 on the Adhesion Properties of Escherichia Coli Causing Urinary Tract Infection. Microb. Pathog. 2019, 133, 103547. [Google Scholar] [CrossRef] [PubMed]
- Vasiee, A.; Falah, F.; Behbahani, B.A.; Tabatabaee-yazdi, F. Probiotic Characterization of Pediococcus Strains Isolated from Iranian Cereal-Dairy Fermented Product: Interaction with Pathogenic Bacteria and the Enteric Cell Line Caco-2. J. Biosci. Bioeng. 2020, 130, 471–479. [Google Scholar] [CrossRef]
- Chen, C.C.; Lai, C.C.; Huang, H.L.; Huang, W.Y.; Toh, H.S.; Weng, T.C.; Chuang, Y.C.; Lu, Y.C.; Tang, H.J. Antimicrobial Activity of Lactobacillus Species against Carbapenem-Resistant Enterobacteriaceae. Front. Microbiol. 2019, 10, 789. [Google Scholar] [CrossRef]
- Krausova, G.; Hyrslova, I.; Hynstova, I. In Vitro Evaluation of Adhesion Capacity, Hydrophobicity, and Auto-Aggregation of Newly Isolated Potential Probiotic Strains. Fermentation 2019, 5, 100. [Google Scholar] [CrossRef]
- Monteagudo-Mera, A.; Rodríguez-Aparicio, L.; Rúa, J.; Martínez-Blanco, H.; Navasa, N.; García-Armesto, M.R.; Ferrero, M. ángel In Vitro Evaluation of Physiological Probiotic Properties of Different Lactic Acid Bacteria Strains of Dairy and Human Origin. J. Funct. Foods 2012, 4, 531–541. [Google Scholar] [CrossRef]
- Xu, H.; Jeong, H.S.; Lee, H.Y.; Ahn, J. Assessment of Cell Surface Properties and Adhesion Potential of Selected Probiotic Strains. Lett. Appl. Microbiol. 2009, 49, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Begley, M.; Hill, C.; Gahan, C.G.M. Bile Salt Hydrolase Activity in Probiotics. Appl. Environ. Microbiol. 2006, 72, 1729–1738. [Google Scholar] [CrossRef]
- Ooi, L.G.; Liong, M.T. Cholesterol-Lowering Effects of Probiotics and Prebiotics: A Review of in Vivo and in Vitro Findings. Int. J. Mol. Sci. 2010, 11, 2499–2522. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y.L.; Ferrell, J.M. Bile Acid Biology, Pathophysiology, and Therapeutics. Clin. Liver Dis. 2020, 15, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Allain, T.; Chaouch, S.; Thomas, M.; Vallée, I.; Buret, A.G.; Langella, P.; Grellier, P.; Polack, B.; Bermúdez-Humarán, L.G.; Florent, I. Bile-Salt-Hydrolases from the Probiotic Strain Lactobacillus Johnsonii La1 Mediate Anti-Giardial Activity in Vitro and in Vivo. Front. Microbiol. 2018, 8, 2707. [Google Scholar] [CrossRef]
- Huang, C.H.; Ho, C.Y.; Chen, C.T.; Hsu, H.F.; Lin, Y.H. Probiotic BSH Activity and Anti-Obesity Potential of Lactobacillus Plantarum Strain TCI378 Isolated from Korean Kimchi. Prev. Nutr. Food Sci. 2019, 24, 434–441. [Google Scholar] [CrossRef]
- Vinderola, C.G.; Reinheimer, J.A. Lactic Acid Starter and Probiotic Bacteria: A Comparative “in Vitro” Study of Probiotic Characteristics and Biological Barrier Resistance. Food Res. Int. 2003, 36, 895–904. [Google Scholar] [CrossRef]
- Prete, R.; Long, S.L.; Gallardo, A.L.; Gahan, C.G.; Corsetti, A.; Joyce, S.A. Beneficial Bile Acid Metabolism from Lactobacillus Plantarum of Food Origin. Sci. Rep. 2020, 10, 1165. [Google Scholar] [CrossRef]
- Tsai, C.C.; Lin, P.P.; Hsieh, Y.M.; Zhang, Z.Y.; Wu, H.C.; Huang, C.C. Cholesterol-Lowering Potentials of Lactic Acid Bacteria Based on Bile-Salt Hydrolase Activity and Effect of Potent Strains on Cholesterol Metabolism In Vitro and In Vivo. Sci. World J. 2014, 2014, 690752. [Google Scholar] [CrossRef]
- Ishimwe, N.; Daliri, E.B.; Lee, B.H.; Fang, F.; Du, G. The Perspective on Cholesterol-Lowering Mechanisms of Probiotics. Mol. Nutr. Food Res. 2015, 59, 94–105. [Google Scholar] [CrossRef]
- Wongrattanapipat, S.; Chiracharoenchitta, A.; Choowongwitthaya, B.; Komsathorn, P.; La-ongkham, O.; Nitisinprasert, S.; Tunsagool, P.; Nakphaichit, M. Selection of Potential Probiotics with Cholesterol-Lowering Properties for Probiotic Yoghurt Production. Food Sci. Technol. Int. 2022, 28, 353–365. [Google Scholar] [CrossRef]
- Mandal, V.; Sen, S.K.; Mandal, N.C. Effect of Prebiotics on Bacteriocin Production and Cholesterol Lowering Activity of Pediococcus Acidilactici LAB 5. World J. Microbiol. Biotechnol. 2009, 25, 1837–1847. [Google Scholar] [CrossRef]
- Albano, C.; Morandi, S.; Silvetti, T.; Casiraghi, M.C.; Manini, F.; Brasca, M. Lactic Acid Bacteria with Cholesterol-Lowering Properties for Dairy Applications: In Vitro and in Situ Activity. J. Dairy Sci. 2018, 101, 10807–10818. [Google Scholar] [CrossRef]
- Gylling, H.; Hallikainen, M.; Pihlajamäki, J.; Simonen, P.; Kuusisto, J.; Laakso, M.; Miettinen, T.A. Insulin Sensitivity Regulates Cholesterol Metabolism to a Greater Extent than Obesity: Lessons from the METSIM Study. J. Lipid Res. 2010, 51, 2422–2427. [Google Scholar] [CrossRef]
- Hoenig, M.R.; Sellke, F.W. Insulin Resistance Is Associated with Increased Cholesterol Synthesis, Decreased Cholesterol Absorption and Enhanced Lipid Response to Statin Therapy. Atherosclerosis 2010, 211, 260–265. [Google Scholar] [CrossRef]
- Simonen, P.P.; Gylling, H.K.; Miettinen, T.A. Diabetes Contributes to Cholesterol Metabolism Regardless of Obesity. Diabetes Care 2002, 25, 1511–1515. [Google Scholar] [CrossRef]
- Ambalam, P.; Kondepudi, K.K.; Balusupati, P.; Nilsson, I.; Wadström, T. Ljungh Prebiotic Preferences of Human Lactobacilli Strains in Co-Culture with Bifidobacteria and Antimicrobial Activity against Clostridium Difficile. J. Appl. Microbiol. 2015, 119, 1672–1682. [Google Scholar] [CrossRef]
- Lee, J.S.; Chung, M.J.; Seo, J.G. In Vitro Evaluation of Antimicrobial Activity of Lactic Acid Bacteria against Clostridium Difficile. Toxicol. Res. 2013, 29, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Alkalbani, N.S.; Turner, M.S.; Ayyash, M.M. Isolation, Identification, and Potential Probiotic Characterization of Isolated Lactic Acid Bacteria and in Vitro Investigation of the Cytotoxicity, Antioxidant, and Antidiabetic Activities in Fermented Sausage. Microb. Cell Factories 2019, 18, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Arena, M.P.; Silvain, A.; Normanno, G.; Grieco, F.; Drider, D.; Spano, G.; Fiocco, D. Use of Lactobacillus Plantarum Strains as a Bio-Control Strategy against Food-Borne Pathogenic Microorganisms. Front. Microbiol. 2016, 7, 1–10. [Google Scholar] [CrossRef]
- Muñoz-Atienza, E.; Gómez-Sala, B.; Araújo, C.; Campanero, C.; Del Campo, R.; Hernández, P.E.; Herranz, C.; Cintas, L.M. Antimicrobial Activity, Antibiotic Susceptibility and Virulence Factors of Lactic Acid Bacteria of Aquatic Origin Intended for Use as Probiotics in Aquaculture. BMC Microbiol. 2013, 13, 15. [Google Scholar] [CrossRef]
- de Melo Pereira, G.V.; de Oliveira Coelho, B.; Magalhães Júnior, A.I.; Thomaz-Soccol, V.; Soccol, C.R. How to Select a Probiotic? A Review and Update of Methods and Criteria. Biotechnol. Adv. 2018, 36, 2060–2076. [Google Scholar] [CrossRef]
- Suokko, A.; Savijoki, K.; Malinen, E.; Palva, A.; Varmanen, P. Characterization of a Mobile clpL Gene from Lactobacillus Rhamnosus. Appl. Environ. Microbiol. 2005, 71, 2061–2069. [Google Scholar] [CrossRef]
- Tao, L.; Biswas, I. ClpL Is Required for Folding of CtsR in Streptococcus Mutans. J. Bacteriol. 2013, 195, 576–584. [Google Scholar] [CrossRef]
- Jana, B.; Biswas, I. Significance of Individual Domains of ClpL: A Novel Chaperone from Streptococcus mutans. Biochemistry 2020, 59, 3368–3379. [Google Scholar] [CrossRef]
- Tran, T.D.-H.; Kwon, H.-Y.; Kim, E.-H.; Kim, K.-W.; Briles, D.E.; Pyo, S.; Rhee, D.-K. Heat-Shock Protein ClpL/HSP100 Increases Penicillin Tolerance in Streptococcus pneumoniae. In Advances in Oto-Rhino-Laryngology; Harabuchi, Y., Hayashi, T., Katada, A., Eds.; S. Karger AG: Basel, Switzerland, 2011; Volume 72, pp. 126–128. ISBN 978-3-8055-9722-7. [Google Scholar]
- Cao, C.; Wang, J.; Liu, Y.; Kwok, L.-Y.; Zhang, H.; Zhang, W. Adaptation of Lactobacillus Plantarum to Ampicillin Involves Mechanisms That Maintain Protein Homeostasis. mSystems 2020, 5, e00853-19. [Google Scholar] [CrossRef]
- Campedelli, I.; Mathur, H.; Salvetti, E.; Rea, M.C.; Torriani, S.; Ross, R.P.; Hill, C. Crossm Genus-Wide Assessment of Antibiotic Resistance in Lactobacillus spp. Appl. Environ. Microbiol. 2019, 85, e01738-18. [Google Scholar] [CrossRef]
- Rojo-Bezares, B.; Sáenz, Y.; Poeta, P.; Zarazaga, M.; Ruiz-Larrea, F.; Torres, C. Assessment of Antibiotic Susceptibility within Lactic Acid Bacteria Strains Isolated from Wine. Int. J. Food Microbiol. 2006, 111, 234–240. [Google Scholar] [CrossRef]
- Klare, I.; Konstabel, C.; Werner, G.; Huys, G.; Vankerckhoven, V.; Kahlmeter, G.; Hildebrandt, B.; Müller-Bertling, S.; Witte, W.; Goossens, H. Antimicrobial Susceptibilities of Lactobacillus, Pediococcus and Lactococcus Human Isolates and Cultures Intended for Probiotic or Nutritional Use. J. Antimicrob. Chemother. 2007, 59, 900–912. [Google Scholar] [CrossRef] [PubMed]
- Lüdin, P.; Roetschi, A.; Wüthrich, D.; Bruggmann, R.; Berthoud, H.; Shani, N. Update on Tetracycline Susceptibility of Pediococcus Acidilactici Based on Strains Isolated from Swiss Cheese and Whey. J. Food Prot. 2018, 81, 1582–1589. [Google Scholar] [CrossRef]
- Oh, Y.J.; Jung, D.S. Evaluation of Probiotic Properties of Lactobacillus and Pediococcus Strains Isolated from Omegisool, a Traditionally Fermented Milletalcoholic Beverage in Korea. LWT 2015, 63, 437–444. [Google Scholar] [CrossRef]
- Salas-Jara, M.J.; Ilabaca, A.; Vega, M.; García, A. Biofilm Forming Lactobacillus: New Challenges for the Development of Probiotics. Microorganisms 2016, 4, 35. [Google Scholar] [CrossRef]
- Grounta, A.; Doulgeraki, A.I.; Nychas, G.J.E.; Panagou, E.Z. Biofilm Formation on Conservolea Natural Black Olives during Single and Combined Inoculation with a Functional Lactobacillus Pentosus Starter Culture. Food Microbiol. 2016, 56, 35–44. [Google Scholar] [CrossRef]
- Montoro, B.P.; Benomar, N.; Lerma, L.L.; Gutiérrez, S.C.; Gálvez, A.; Abriouel, H. Fermented Aloreña Table Olives as a Source of Potential Probiotic Lactobacillus Pentosus Strains. Front. Microbiol. 2016, 7, 1583. [Google Scholar] [CrossRef]
- Perpetuini, G.; Pham-Hoang, B.N.; Scornec, H.; Tofalo, R.; Schirone, M.; Suzzi, G.; Cavin, J.F.; Waché, Y.; Corsetti, A.; Licandro-Seraut, H. In Lactobacillus Pentosus, the Olive Brine Adaptation Genes Are Required for Biofilm Formation. Int. J. Food Microbiol. 2016, 216, 104–109. [Google Scholar] [CrossRef]
- Merenstein, D.; Pot, B.; Leyer, G.; Ouwehand, A.C.; Preidis, G.A.; Elkins, C.A.; Hill, C.; Lewis, Z.T.; Shane, A.L.; Zmora, N.; et al. Emerging Issues in Probiotic Safety: 2023 Perspectives. Gut Microbes 2023, 15, 2185034. [Google Scholar] [CrossRef]
- Updated List of QPS-Recommended Microorganisms for Safety Risk Assessments Carried out by EFSA. Available online: https://zenodo.org/records/8124409 (accessed on 24 November 2023).
- Kim, M.H.; Yun, K.E.; Kim, J.; Park, E.; Chang, Y.; Ryu, S.; Kim, H.L.; Kim, H.N. Gut Microbiota and Metabolic Health among Overweight and Obese Individuals. Sci. Rep. 2020, 10, 19417. [Google Scholar] [CrossRef]
- Yanni, A.E.; Mitropoulou, G.; Prapa, I.; Agrogiannis, G.; Kostomitsopoulos, N.; Bezirtzoglou, E.; Kourkoutas, Y.; Karathanos, V.T. Functional Modulation of Gut Microbiota in Diabetic Rats Following Dietary Intervention with Pistachio Nuts (Pistacia vera L.). Metab. Open 2020, 7, 100040. [Google Scholar] [CrossRef] [PubMed]
- Muscogiuri, G.; Cantone, E.; Cassarano, S.; Tuccinardi, D.; Barrea, L.; Savastano, S.; Colao, A. Gut Microbiota: A New Path to Treat Obesity. Int. J. Obes. Suppl. 2019, 9, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.; Ni, Q.; Sun, W.; Li, L.; Feng, X. The Links between Gut Microbiota and Obesity and Obesity Related Diseases. Biomed. Pharmacother. 2022, 147, 112678. [Google Scholar] [CrossRef]
- Singh, R.K.; Chang, H.W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of Diet on the Gut Microbiome and Implications for Human Health. J. Transl. Med. 2017, 15, 1–17. [Google Scholar] [CrossRef]


| Isolate Code | SK | SK4 | OLL1-1 | OLS2-1 | OLS3-1 | PE11 | RS1 |
|---|---|---|---|---|---|---|---|
| Source of isolation | Human stool samples | Human stool samples | Olive brine | Olive fruit | Olive fruit | Olive fruit | Raisins |
| Classification | Pediococcus acidilactici | Lactiplantibacillus plantarum subsp. plantarum | Pediococcus acidilactici | Pediococcus acidilactici | Pediococcus acidilactici | Lactiplantibacillus pentosus | Lactiplantibacillus plantarum subsp. plantarum |
| Classification method | Illumina/ Nanopore | Illumina | Illumina/ Nanopore | Illumina | Illumina | Illumina | Illumina |
| Number of contigs | 11 | 45 | 610 | 327 | 390 | 150 | 411 |
| Genome length (bp) | 2,044,391 | 3,226,900 | 5,199,416 | 2,171,522 | 2,209,033 | 3,897,459 | 3,421,158 |
| N50 | 1,082,018 | 481,816 | 52,231 | 394,208 | 393,990 | 166,230 | 156,534 |
| Completeness | 99.38 | 99.07 | 98.27 | 99.38 | 99.38 | 99.38 | 99.07 |
| GC content (%) | 42 | 44 | 43 | 41 | 42 | 46 | 44 |
| Predicted genes | 2030 | 3044 | 5875 | 2070 | 2124 | 3574 | 3439 |
| CDSs | 1933 | 3029 | 5485 | 1994 | 1961 | 3508 | 3207 |
| rRNAs | 14 | 8 | 7 | 9 | 41 | 26 | 3 |
| tRNAs | 56 | 65 | 58 | 63 | 54 | 73 | 57 |
| tmRNAs | 1 | 1 | 2 | 1 | 1 | 1 | 1 |
| misc_RNAs | 26 | 44 | 0 | 40 | 0 | 52 | 0 |
| Antibiotic resistance genes | No resistance | Resistance gene ClpL | No resistance | No resistance | No resistance | No resistance | Resistance gene ClpL |
| Isolate Code | Classification | Survival Rate (%) during In Vitro Gastrointestinal Tract Conditions | Percentage (%) Adherence to Caco-2 Cells | Bile Acid Precipitation Zone (mm) | Percentage (%) Reduction of Cholesterol Levels | Cellular Cholesterol Binding (%) | ||
|---|---|---|---|---|---|---|---|---|
| Simulated Oral Phase | Simulated Gastric Phase | Simulated Intestinal Phase | ||||||
| SK | Pediococcus acidilactici | 99.69 ± 0.06 | 71.73 ± 1.12 c | 71.83 ± 0.24 d | 11.75 ± 2.47 ab | 17.0 | 29.24 ± 3.57 cd | 0.39 ± 0.03 a |
| SK4 | Lactiplantibacillus plantarum subsp. plantarum | 99.36 ± 0.37 | 63.97 ± 0.37 ab | 62.92 ± 1.34 b | 8.59 ± 0.31 a | 16.5 | 21.22 ± 3.22 ab | 3.42 ± 0.15 ab |
| OLL1-1 | Pediococcu sacidilactici | 99.27 ± 0.14 | 63.93 ± 0.12 ab | 52.51 ± 0.54 a | 16.75 ± 4.26 bc | 14.0 | 15.88 ± 2.15 b | 3.61 ± 2.36 b |
| OLS2-1 | Pediococcus acidilactici | 99.68 ± 0.08 | 76.11 ± 0.41 d | 70.19 ± 0.30 c | 8.65 ± 0.87 a | 16.0 | 22.36 ± 3.45 a | n.d. |
| OLS3-1 | Pediococcus acidilactici | 99.58 ± 0.24 | 71.89 ± 0.97 c | 64.16 ± 0.21 b | 21.42 ± 3.25 c | 18.0 | 32.96 ± 1.84 d | 1.58 ± 0.27 ab |
| PE11 | Lactiplantibacillus pentosus | 99.78 ± 0.41 | 65.41 ± 0.57 b | 76.91 ± 0.13 e | 16.80 ± 3.31 bc | 15.0 | 43.57 ± 1.55 e | 2.46 ± 1.75 ab |
| RS1 | Lactiplantibacillus plantarum subsp. plantarum | 99.65 ± 0.18 | 63.28 ± 0.06 a | 52.14 ± 0.91 a | 6.83 ± 1.48 a | 9.0 | 25.24 ± 0.47 ac | 0.89 ± 0.49 ab |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Somalou, P.; Ieronymaki, E.; Feidaki, K.; Prapa, I.; Stylianopoulou, E.; Spyridopoulou, K.; Skavdis, G.; Grigoriou, M.E.; Panas, P.; Argiriou, A.; et al. Novel Wild-Type Pediococcus and Lactiplantibacillus Strains as Probiotic Candidates to Manage Obesity-Associated Insulin Resistance. Microorganisms 2024, 12, 231. https://doi.org/10.3390/microorganisms12020231
Somalou P, Ieronymaki E, Feidaki K, Prapa I, Stylianopoulou E, Spyridopoulou K, Skavdis G, Grigoriou ME, Panas P, Argiriou A, et al. Novel Wild-Type Pediococcus and Lactiplantibacillus Strains as Probiotic Candidates to Manage Obesity-Associated Insulin Resistance. Microorganisms. 2024; 12(2):231. https://doi.org/10.3390/microorganisms12020231
Chicago/Turabian StyleSomalou, Paraskevi, Eleftheria Ieronymaki, Kyriaki Feidaki, Ioanna Prapa, Electra Stylianopoulou, Katerina Spyridopoulou, George Skavdis, Maria E. Grigoriou, Panayiotis Panas, Anagnostis Argiriou, and et al. 2024. "Novel Wild-Type Pediococcus and Lactiplantibacillus Strains as Probiotic Candidates to Manage Obesity-Associated Insulin Resistance" Microorganisms 12, no. 2: 231. https://doi.org/10.3390/microorganisms12020231
APA StyleSomalou, P., Ieronymaki, E., Feidaki, K., Prapa, I., Stylianopoulou, E., Spyridopoulou, K., Skavdis, G., Grigoriou, M. E., Panas, P., Argiriou, A., Tsatsanis, C., & Kourkoutas, Y. (2024). Novel Wild-Type Pediococcus and Lactiplantibacillus Strains as Probiotic Candidates to Manage Obesity-Associated Insulin Resistance. Microorganisms, 12(2), 231. https://doi.org/10.3390/microorganisms12020231








