A Tachyplesin Antimicrobial Peptide from Theraphosidae Spiders with Potent Antifungal Activity Against Cryptococcus neoformans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Sample Preparation
2.3. Transcriptome Sequencing
2.4. Sequence Screening and Characterization of Antimicrobial Peptides
2.5. Peptide Synthesis and Post-Synthesis Modification
2.6. Structural Analysis
2.7. Antifungal Susceptibility Testing
2.8. Stability and Resistance to Proteolytic Degradation
2.9. In Vitro and In Vivo Toxicity Test
2.10. Time-Killing Kinetics Assay
2.11. Electron Microscopy
2.12. Membrane Permeability Test
2.13. Biofilm Inhibition and Eradication Assays
2.14. Two-Photon Laser Scanning Microscope (TPLSM)
2.15. Evaluation of the Therapeutic Potential of QS18 In Vivo
2.15.1. Mice Infection and Treatment
2.15.2. Histopathological Examination
2.15.3. Quantification of Cytokine Levels
2.16. Statistical Analysis
3. Results
3.1. Identification of AMPs from the Venom Gland Transcriptome of C. liboensis
3.2. Physicochemical Parameters of AMPs in C. liboensis Venom Gland Transcriptome
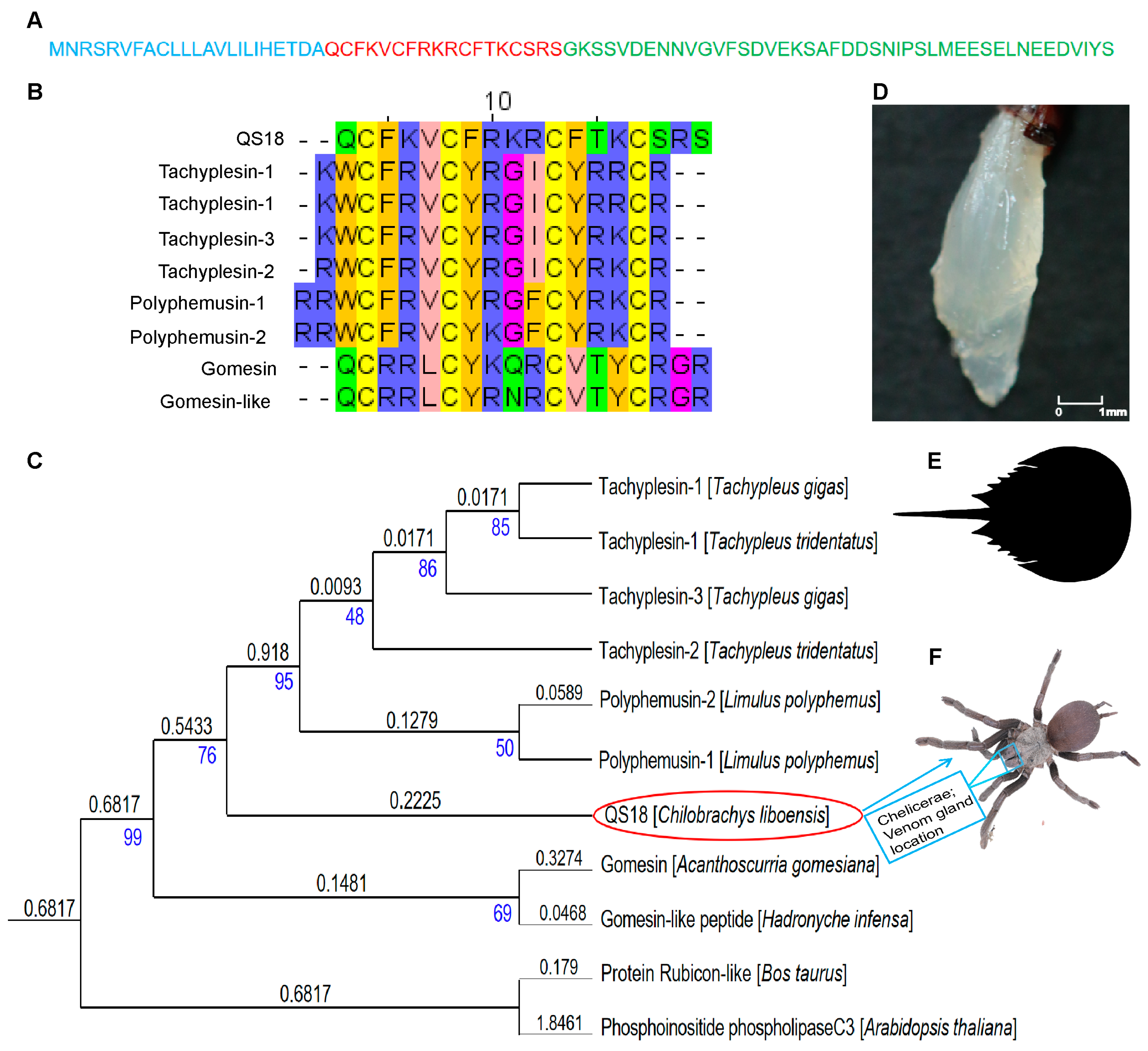
3.3. Sequence Alignment and Phylogenetic Analysis of QS18
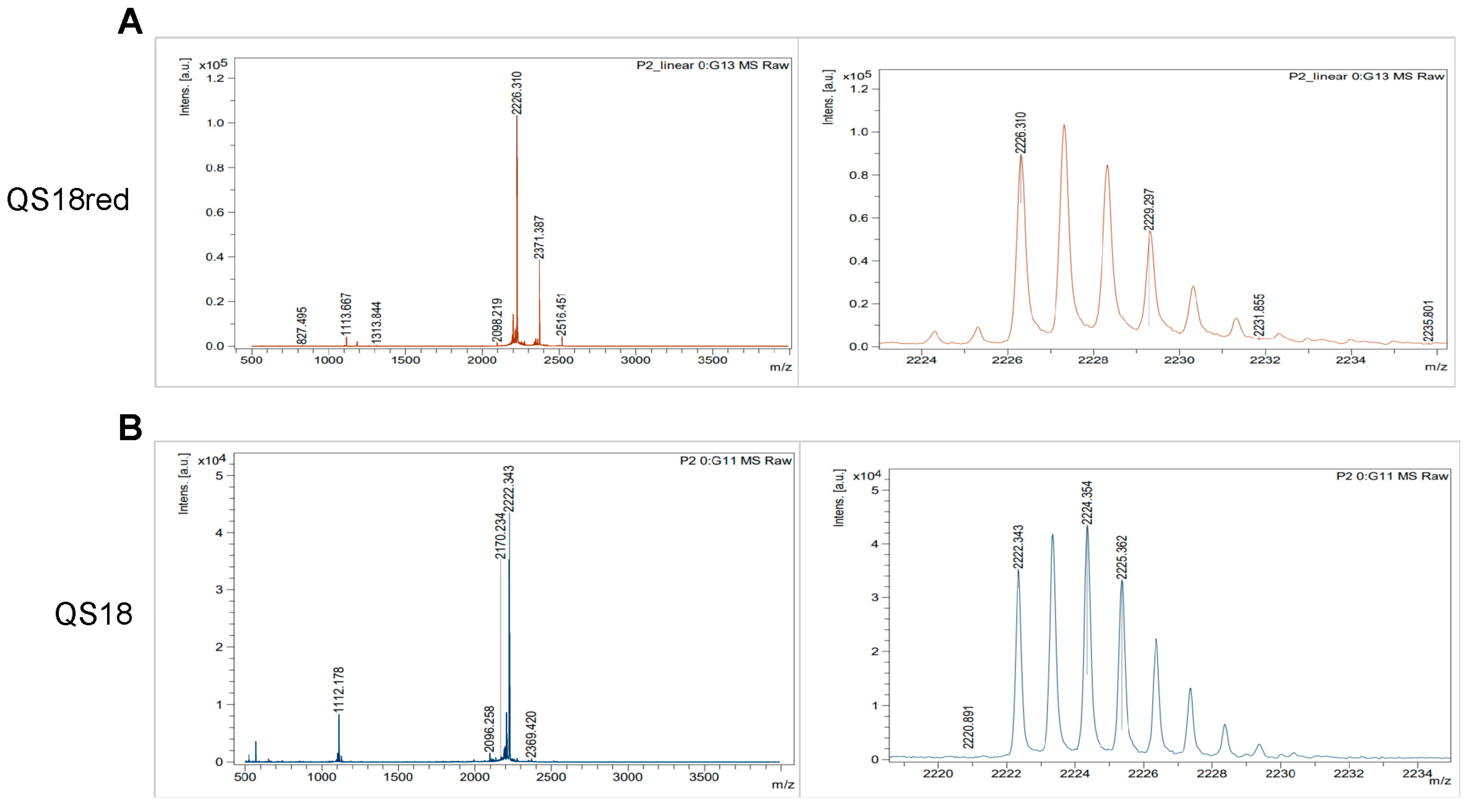
3.4. Synthesis and Purification of Novel Antimicrobial Peptide QS18
3.5. Structural Analysis of QS18
3.6. Evaluation of Toxicity of QS18 to Mammalian Cells
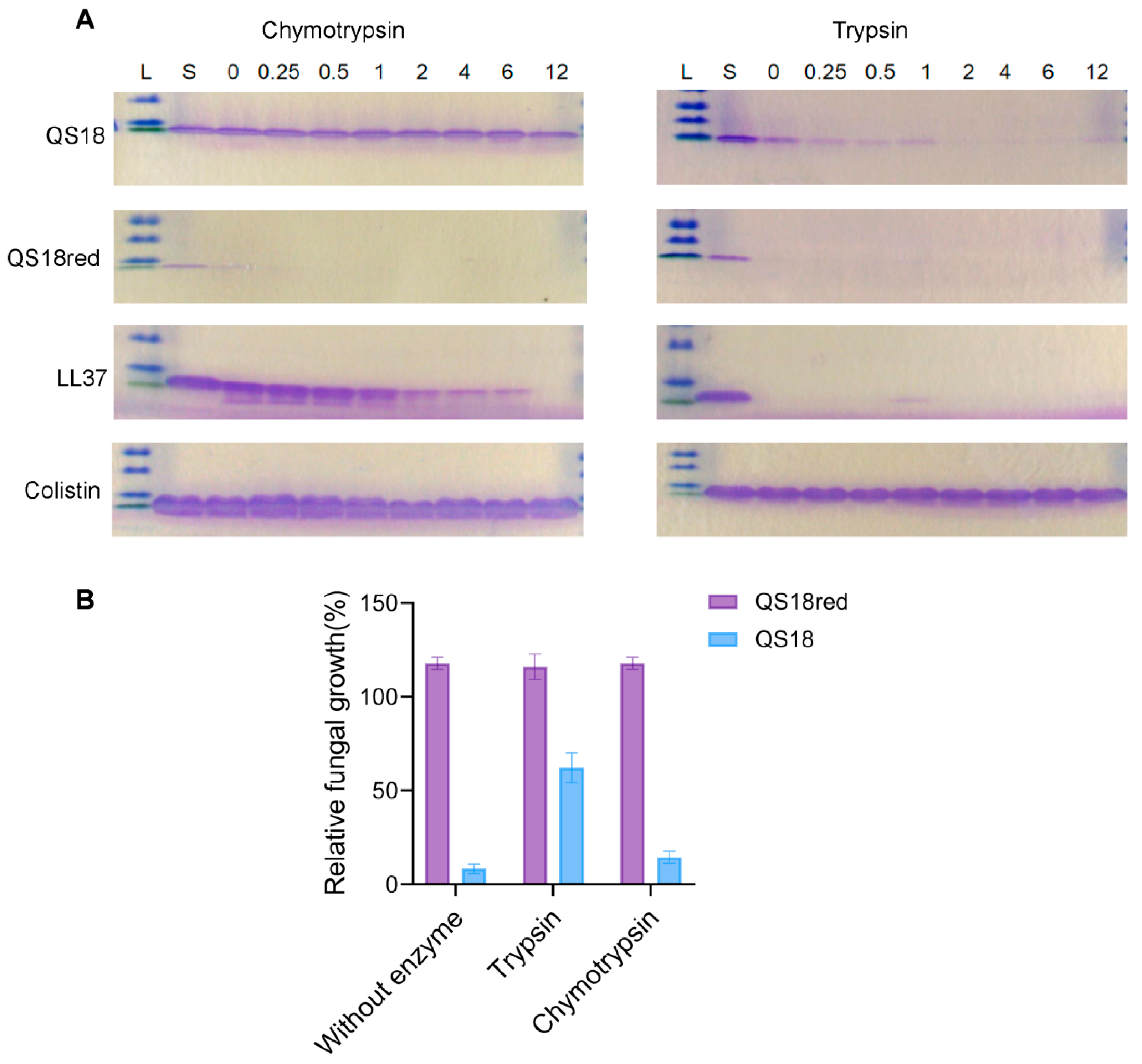
3.7. Protease Resistance
3.8. Antifungal Activity of QS18
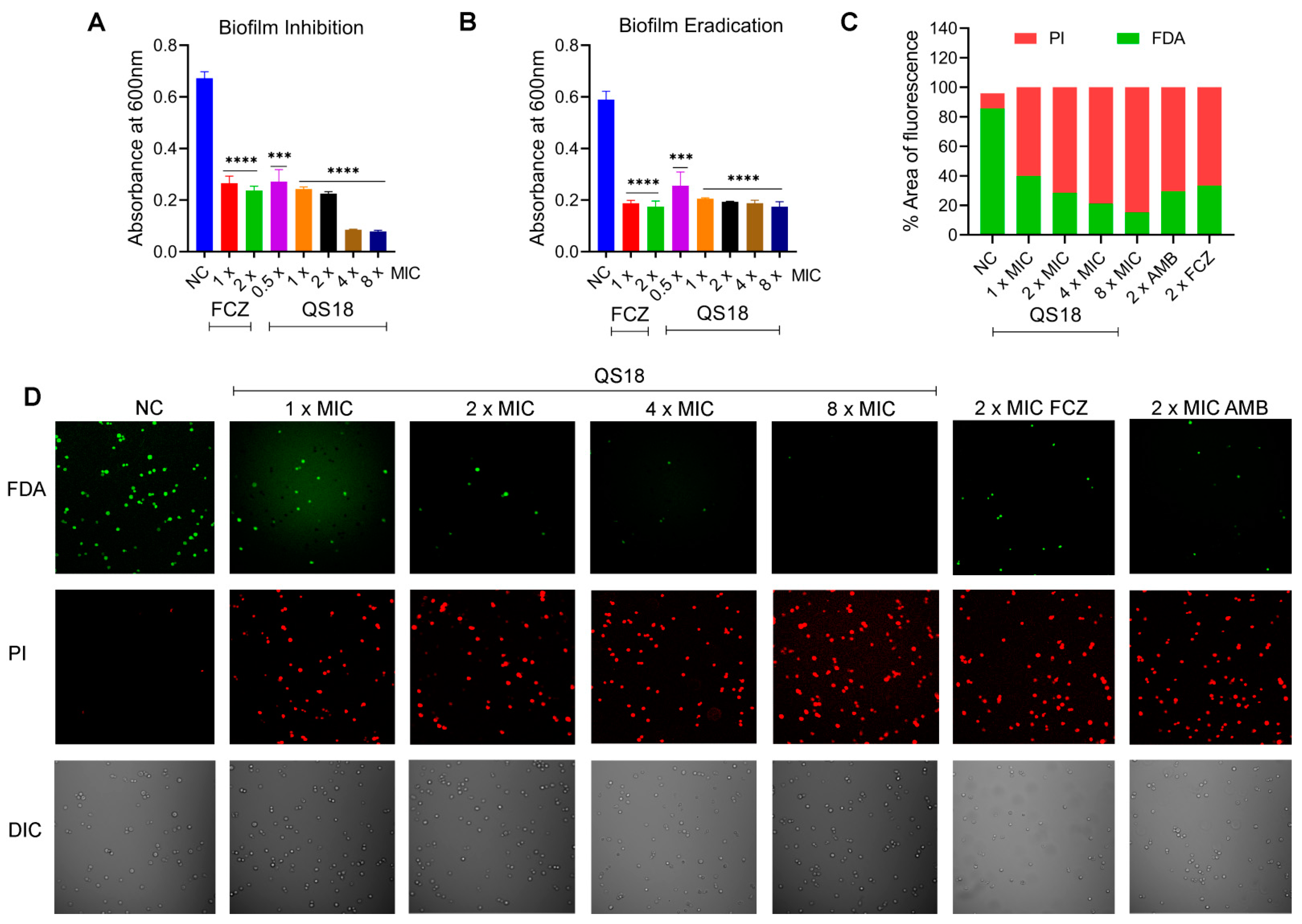
3.9. QS18 Displays Effective Antibiofilm Activity
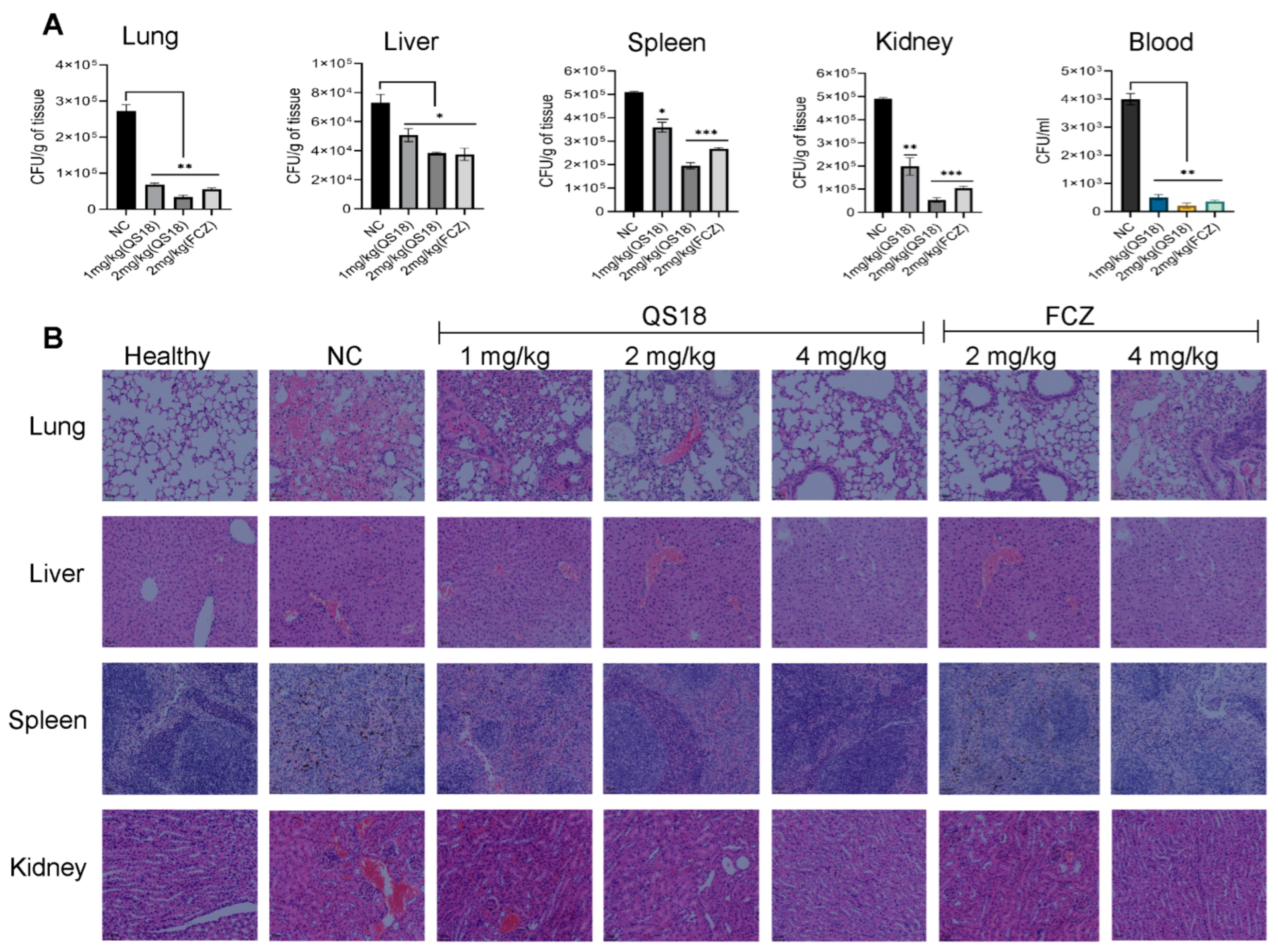
3.10. QS18 Suppresses the Dissemination of Fungi to Target Organs
4. Discussion
5. Conclusions
6. Ethics Approval
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rathore, S.S.; Sathiyamoorthy, J.; Lalitha, C.; Ramakrishnan, J. A holistic review on Cryptococcus neoformans. Microb. Pathog. 2022, 166, 105521. [Google Scholar] [CrossRef] [PubMed]
- Cafarchia, C.; Romito, D.; Iatta, R.; Camarda, A.; Montagna, M.T.; Otranto, D. Role of birds of prey as carriers and spreaders of Cryptococcus neoformans and other zoonotic yeasts. Med. Mycol. 2006, 44, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Rajasingham, R.; Smith, R.M.; Park, B.J.; Jarvis, J.N.; Govender, N.P.; Chiller, T.M.; Denning, D.W.; Loyse, A.; Boulware, D.R. Global burden of disease of HIV-associated cryptococcal meningitis: An updated analysis. Lancet Infect. Dis. 2017, 17, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Decoté-Ricardo, D.; LaRocque-de-Freitas, I.F.; Rocha, J.D.B.; Nascimento, D.d.O.; Nunes, M.P.; Morrot, A.; Freire-de-Lima, L.; Previato, J.O.; Mendonça-Previato, L.; Freire-de-Lima, C.G. Immunomodulatory Role of Capsular Polysaccharides Constituents of Cryptococcus neoformans. Front. Med. 2019, 6, 129. [Google Scholar] [CrossRef] [PubMed]
- Diniz-Lima, I.; Fonseca, L.M.d.; Silva-Junior, E.B.d.; Guimarães-de-Oliveira, J.C.; Freire-de-Lima, L.; Nascimento, D.O.; Morrot, A.; Previato, J.O.; Mendonça-Previato, L.; Decote-Ricardo, D.; et al. Cryptococcus: History, Epidemiology and Immune Evasion. Appl. Sci. 2022, 12, 7086. [Google Scholar] [CrossRef]
- Doering, T.L.; Nosanchuk, J.D.; Roberts, W.K.; Casadevall, A. Melanin as a potential cryptococcal defence against microbicidal proteins. Med. Mycol. 1999, 37, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Kwon-Chung, K.J.; Fraser, J.A.; Doering, T.L.; Wang, Z.; Janbon, G.; Idnurm, A.; Bahn, Y.-S. Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb. Perspect. Med. 2014, 4, a019760. [Google Scholar] [CrossRef] [PubMed]
- Ngan, N.T.T.; Flower, B.; Day, J.N. Treatment of Cryptococcal meningitis: How have we got here and where are we going? Drugs 2022, 82, 1237–1249. [Google Scholar] [CrossRef]
- Muhaj, F.F.; George, S.J.; Nguyen, C.D.; Tyring, S.K. Antimicrobials and resistance part II: Antifungals, antivirals, and antiparasitics. J. Am. Acad. Dermatol. 2022, 86, 1207–1226. [Google Scholar] [CrossRef]
- Kuhn-Nentwig, L. Antimicrobial and cytolytic peptides of venomous arthropods. Cell. Mol. Life Sci. CMLS 2003, 60, 2651–2668. [Google Scholar] [CrossRef]
- Yacoub, T.; Rima, M.; Karam, M.; Sabatier, J.-M.; Fajloun, Z. Antimicrobials from venomous animals: An overview. Molecules 2020, 25, 2402. [Google Scholar] [CrossRef] [PubMed]
- Akef, H.M. Anticancer, antimicrobial, and analgesic activities of spider venoms. Toxicol. Res. 2018, 7, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Adams, M.E. Lycotoxins, antimicrobial peptides from venom of the wolf spider Lycosa carolinensis. J. Biol. Chem. 1998, 273, 2059–2066. [Google Scholar] [CrossRef] [PubMed]
- Opatova, V.; Hamilton, C.A.; Hedin, M.; De Oca, L.M.; Král, J.; Bond, J.E. Phylogenetic systematics and evolution of the spider infraorder Mygalomorphae using genomic scale data. Syst. Biol. 2020, 69, 671–707. [Google Scholar] [CrossRef]
- World Spider Catalog. World Spider Catalog Version 25.5. 2024. Available online: http://wsc.nmbe.ch (accessed on 20 September 2024).
- Viquez, C.; Rojas-Gätjens, D.; Mesén-Porras, E.; Avendaño, R.; Sasa, M.; Lomonte, B.; Chavarría, M. Venom-microbiomics of eight species of Neotropical spiders from the Theraphosidae family. J. Appl. Microbiol. 2024, 135, lxae113. [Google Scholar] [CrossRef]
- Pineda, S.S.; Chin, Y.K.-Y.; Undheim, E.A.; Senff, S.; Mobli, M.; Dauly, C.; Escoubas, P.; Nicholson, G.M.; Kaas, Q.; Guo, S. Structural venomics reveals evolution of a complex venom by duplication and diversification of an ancient peptide-encoding gene. Proc. Natl. Acad. Sci. USA 2020, 117, 11399–11408. [Google Scholar] [CrossRef] [PubMed]
- Herzig, V.; King, G.F. The neurotoxic mode of action of venoms from the spider family Theraphosidae. In Spider Ecophysiology; Springer: Berlin/Heidelberg, Germany, 2012; pp. 203–215. [Google Scholar]
- Estrada-Gómez, S.; Vargas-Muñoz, L.J.; Saldarriaga-Córdoba, M.; Cifuentes, Y.; Perafan, C. Identifying different transcribed proteins in the newly described Theraphosidae Pamphobeteus verdolaga. Toxicon 2017, 129, 81–88. [Google Scholar] [CrossRef]
- Silva, P.I.; Daffre, S.; Bulet, P. Isolation and characterization of gomesin, an 18-residue cysteine-rich defense peptide from the spider Acanthoscurria gomesiana hemocytes with sequence similarities to horseshoe crab antimicrobial peptides of the tachyplesin family. J. Biol. Chem. 2000, 275, 33464–33470. [Google Scholar] [CrossRef] [PubMed]
- Ayroza, G.; Ferreira, I.L.; Sayegh, R.S.; Tashima, A.K.; da Silva, P.I., Jr. Juruin: An antifungal peptide from the venom of the Amazonian Pink Toe spider, Avicularia juruensis, which contains the inhibitory cystine knot motif. Front. Microbiol. 2012, 3, 324. [Google Scholar] [CrossRef] [PubMed]
- Muniz Seif, E.J.; Icimoto, M.Y.; Silva Júnior, P.I. In silico bioprospecting of receptors associated with the mechanism of action of Rondonin, an antifungal peptide from spider Acanthoscurria rondoniae haemolymph. Silico Pharmacol. 2024, 12, 55. [Google Scholar] [CrossRef]
- Shin, M.K.; Hwang, I.-W.; Jang, B.-Y.; Bu, K.-B.; Yoo, J.S.; Sung, J.-S. In silico identification of novel antimicrobial peptides from the venom gland transcriptome of the spider Argiope bruennichi (Scopoli, 1772). Front. Microbiol. 2023, 14, 1249175. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Gao, J.; Zhang, S.; Wu, S.; Xie, Z.; Ling, G.; Kuang, Y.-Q.; Yang, Y.; Yu, H.; Wang, Y. Identification and characterization of the first cathelicidin from sea snakes with potent antimicrobial and anti-inflammatory activity and special mechanism. J. Biol. Chem. 2015, 290, 16633–16652. [Google Scholar] [CrossRef] [PubMed]
- Gerdol, M.; Puillandre, N.; Moro, G.D.; Guarnaccia, C.; Lucafò, M.; Benincasa, M.; Zlatev, V.; Manfrin, C.; Torboli, V.; Giulianini, P.G. Identification and characterization of a novel family of cysteine-rich peptides (MgCRP-I) from Mytilus galloprovincialis. Genome Biol. Evol. 2015, 7, 2203–2219. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.K.; Hwang, I.-W.; Kim, Y.; Kim, S.T.; Jang, W.; Lee, S.; Bang, W.Y.; Bae, C.-H.; Sung, J.-S. Antibacterial and anti-inflammatory effects of novel peptide toxin from the spider Pardosa astrigera. Antibiotics 2020, 9, 422. [Google Scholar] [CrossRef]
- Ajish, C.; Kumar, S.D.; Kim, E.Y.; Yang, S.; Shin, S.Y. A short novel antimicrobial peptide BP100-W with antimicrobial, antibiofilm and anti-inflammatory activities designed by replacement with tryptophan. J. Anal. Sci. Technol. 2022, 13, 46. [Google Scholar] [CrossRef]
- Rajasekaran, G.; Kumar, S.D.; Yang, S.; Shin, S.Y. The design of a cell-selective fowlicidin-1-derived peptide with both antimicrobial and anti-inflammatory activities. Eur. J. Med. Chem. 2019, 182, 111623. [Google Scholar] [CrossRef]
- Mohammadi, M.; Taheri, B.; Momenzadeh, N.; Salarinia, R.; Nabipour, I.; Farshadzadeh, Z.; Bargahi, A. Identification and characterization of novel antimicrobial peptide from hippocampus comes by In Silico and experimental studies. Mar. Biotechnol. 2018, 20, 718–728. [Google Scholar] [CrossRef]
- Kim, H.; Jang, J.H.; Kim, S.C.; Cho, J.H. De novo generation of short antimicrobial peptides with enhanced stability and cell specificity. J. Antimicrob. Chemother. 2014, 69, 121–132. [Google Scholar] [CrossRef]
- Brunetti, J.; Falciani, C.; Roscia, G.; Pollini, S.; Bindi, S.; Scali, S.; Arrieta, U.C.; Gómez-Vallejo, V.; Quercini, L.; Ibba, E. In vitro and in vivo efficacy, toxicity, bio-distribution and resistance selection of a novel antibacterial drug candidate. Sci. Rep. 2016, 6, 26077. [Google Scholar] [CrossRef]
- Te Winkel, J.D.; Gray, D.A.; Seistrup, K.H.; Hamoen, L.W.; Strahl, H. Analysis of antimicrobial-triggered membrane depolarization using voltage sensitive dyes. Front. Cell Dev. Biol. 2016, 4, 29. [Google Scholar] [CrossRef]
- Yang, L.; Tian, Z.; Zhou, L.; Zhu, L.; Sun, C.; Huang, M.; Peng, J.; Guo, G. In vitro antifungal activity of a novel antimicrobial peptide AMP-17 against planktonic cells and biofilms of Cryptococcus neoformans. Infect. Drug Resist. 2022, 15, 233–248. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-M.; Zhou, S.-W.; Huang, X.-S.; Chen, Y.-F.; Mwangi, J.; Fang, Y.-Q.; Du, T.; Zhao, M.; Shi, L.; Lu, Q.-M. Blap-6, a Novel Antifungal Peptide from the Chinese Medicinal Beetle Blaps rhynchopetera against Cryptococcus neoformans. Int. J. Mol. Sci. 2024, 25, 5336. [Google Scholar] [CrossRef]
- Rossi, D.C.; Muñoz, J.E.; Carvalho, D.D.; Belmonte, R.; Faintuch, B.; Borelli, P.; Miranda, A.; Taborda, C.P.; Daffre, S. Therapeutic use of a cationic antimicrobial peptide from the spider Acanthoscurria gomesiana in the control of experimental candidiasis. BMC Microbiol. 2012, 12, 28. [Google Scholar] [CrossRef] [PubMed]
- Jindal, M.; Le, C.; Mohd Yusof, M.; Sekaran, S. Net charge, hydrophobicity and specific amino acids contribute to the activity of antimicrobial peptides. J. Health Transl. Med. 2014, 17, 1–7. [Google Scholar]
- Kumar, V.; Chugh, A. Peptide-mediated leishmaniasis management strategy: Tachyplesin emerges as an effective anti-leishmanial peptide against Leishmania donovani. Biochim. Biophys. Acta (BBA)-Biomembr. 2021, 1863, 183629. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Yadav, B.K.; Chugh, A. Marine antimicrobial peptide tachyplesin as an efficient nanocarrier for macromolecule delivery in plant and mammalian cells. FEBS J. 2015, 282, 732–745. [Google Scholar] [CrossRef] [PubMed]
- Shigenaga, T.; Muta, T.; Toh, Y.; Tokunaga, F.; Iwanaga, S. Antimicrobial tachyplesin peptide precursor. cDNA cloning and cellular localization in the horseshoe crab (Tachypleus tridentatus). J. Biol. Chem. 1990, 265, 21350–21354. [Google Scholar] [CrossRef]
- Kawano, K.; Yoneya, T.; Miyata, T.; Yoshikawa, K.; Tokunaga, F.; Terada, Y.; Iwanaga, S. Antimicrobial peptide, tachyplesin I, isolated from hemocytes of the horseshoe crab (Tachypleus tridentatus). NMR determination of the beta-sheet structure. J. Biol. Chem. 1990, 265, 15365–15367. [Google Scholar] [CrossRef]
- Muta, T.; Fujimoto, T.; Nakajima, H.; Iwanaga, S. Tachyplesins isolated from hemocytes of Southeast Asian horseshoe crabs (Carcinoscorpius rotundicauda and Tachypleus gigas) identification of a new tachyplesin, tachyplesin III, and a processing intermediate of its precursor. J. Biochem. 1990, 108, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Powers, J.-P.S.; Rozek, A.; Hancock, R.E. Structure–activity relationships for the β-hairpin cationic antimicrobial peptide polyphemusin I. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2004, 1698, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Wang, R.; Mai, S. Advances in delivery systems for the therapeutic application of LL37. J. Drug Deliv. Sci. Technol. 2020, 60, 102016. [Google Scholar] [CrossRef]
- Jabra-Rizk, M.A.; Falkler, W.A.; Meiller, T.F. Fungal biofilms and drug resistance. Emerg. Infect. Dis. 2004, 10, 14. [Google Scholar] [CrossRef]
- Macedo, K.W.R.; Costa, L.J.d.L.; Souza, J.O.d.; Vasconcelos, I.A.d.; Castro, J.S.d.; Santana, C.J.C.d.; Magalhães, A.C.M.; Castro, M.d.S.; Pires, O.R. Brazilian Theraphosidae: A toxicological point of view. J. Venom. Anim. Toxins Incl. Trop. Dis. 2021, 27, e20210004. [Google Scholar] [CrossRef]
- Xiao, Y.; Luo, X.; Kuang, F.; Deng, M.; Wang, M.; Zeng, X.; Liang, S. Synthesis and characterization of huwentoxin-IV, a neurotoxin inhibiting central neuronal sodium channels. Toxicon 2008, 51, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Cai, T.; Zhu, Q.; Deng, M.; Li, J.; Zhou, X.; Zhang, F.; Li, D.; Li, J.; Liu, Y. Structure and function of hainantoxin-III, a selective antagonist of neuronal tetrodotoxin-sensitive voltage-gated sodium channels isolated from the Chinese bird spider Ornithoctonus hainana. J. Biol. Chem. 2013, 288, 20392–20403. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Zhou, X.; Huang, Y.; Zhang, Y.; Hu, Z.; Wang, M.; Chen, P.; Liu, Z.; Liang, S. The tarantula toxin jingzhaotoxin-XI (κ-theraphotoxin-Cj1a) regulates the activation and inactivation of the voltage-gated sodium channel Nav1. 5. Toxicon 2014, 92, 6–13. [Google Scholar] [CrossRef]
- Zhu, M.-S.; Zhang, R. Revision of the theraphosid spiders from China (Araneae: Mygalomorphae). J. Arachnol. 2008, 36, 425–447. [Google Scholar] [CrossRef]
- Kushibiki, T.; Kamiya, M.; Aizawa, T.; Kumaki, Y.; Kikukawa, T.; Mizuguchi, M.; Demura, M.; Kawabata, S.-I.; Kawano, K. Interaction between tachyplesin I, an antimicrobial peptide derived from horseshoe crab, and lipopolysaccharide. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2014, 1844, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. The antimicrobial peptide database is 20 years old: Recent developments and future directions. Protein Sci. 2023, 32, e4778. [Google Scholar] [CrossRef] [PubMed]
- Carroll, L.; Jiang, S.; Rogowska-Wrzesinska, A.; Davies, M. Oxidation of disulfide bonds: A novel pathway to protein glutathionylation. Free Radic. Biol. Med. 2018, 128, S50. [Google Scholar] [CrossRef]
- Lee, W.; Lee, D.G. A novel mechanism of fluconazole: Fungicidal activity through dose-dependent apoptotic responses in Candida albicans. Microbiology 2018, 164, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Kullberg, B.J.; Netea, M.G.; Vonk, A.G.; van der Meer, J.W. Modulation of neutrophil function in host defense against disseminated Candida albicans infection in mice. FEMS Immunol. Med. Microbiol. 1999, 26, 299–307. [Google Scholar] [CrossRef] [PubMed]



| Transcript ID | Mature Sequence | Length | MM | TpI | Nc | GRAVY |
|---|---|---|---|---|---|---|
| >GC-BN-1-1:11507.p1 | RCRSYCFGKRCLTYCLS | 17 | 2059.47 | 9.25 | 4 | −0.135 |
| >GC-BN-1-1:19939.p3 | YCRSVCGRKRCFTYCKEK | 18 | 2230.67 | 9.44 | 5 | −0.900 |
| >GC-BN-1-1:25378.p1 | QCRSVCISWRCYTYCASS | 18 | 2116.43 | 8.53 | 2 | 0.033 |
| >GC-BN-1-1:36375.p2 | QRPDFCKSMRFLKSLKGR | 18 | 2197.65 | 11.01 | 5 | −1.011 |
| >GC-BN-1-1:5740.p2 | QCRSVCISWRCYTYCASS | 18 | 2116.43 | 8.53 | 2 | 0.033 |
| >GC-BN-1-1:78484.p1 | QCRSVCFRSRCITYCSS | 17 | 1999.33 | 8.98 | 3 | −0.041 |
| >GC-BN-1-1:8433.p1 | QCRSYCFGKLCLTYCGK | 17 | 1973.37 | 8.89 | 3 | −0.018 |
| >GC-BN-1-1:979.p1 | QCFKVCFRKRCFTKCSRS | 18 | 2227.71 | 9.94 | 6 | −0.467 |
| >GC-BN-1-1:PDBI077_L03_78483.p4 | QCRSVCFRSRCITYCSS | 17 | 1999.33 | 8.98 | 3 | −0.041 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michira, B.B.; Wang, Y.; Mwangi, J.; Wang, K.; Asmamaw, D.; Tadese, D.A.; Gao, J.; Khalid, M.; Lu, Q.-M.; Lai, R.; et al. A Tachyplesin Antimicrobial Peptide from Theraphosidae Spiders with Potent Antifungal Activity Against Cryptococcus neoformans. Microorganisms 2024, 12, 2648. https://doi.org/10.3390/microorganisms12122648
Michira BB, Wang Y, Mwangi J, Wang K, Asmamaw D, Tadese DA, Gao J, Khalid M, Lu Q-M, Lai R, et al. A Tachyplesin Antimicrobial Peptide from Theraphosidae Spiders with Potent Antifungal Activity Against Cryptococcus neoformans. Microorganisms. 2024; 12(12):2648. https://doi.org/10.3390/microorganisms12122648
Chicago/Turabian StyleMichira, Brenda B., Yi Wang, James Mwangi, Kexin Wang, Demeke Asmamaw, Dawit Adisu Tadese, Jinai Gao, Mehwish Khalid, Qiu-Min Lu, Ren Lai, and et al. 2024. "A Tachyplesin Antimicrobial Peptide from Theraphosidae Spiders with Potent Antifungal Activity Against Cryptococcus neoformans" Microorganisms 12, no. 12: 2648. https://doi.org/10.3390/microorganisms12122648
APA StyleMichira, B. B., Wang, Y., Mwangi, J., Wang, K., Asmamaw, D., Tadese, D. A., Gao, J., Khalid, M., Lu, Q.-M., Lai, R., & Li, J. (2024). A Tachyplesin Antimicrobial Peptide from Theraphosidae Spiders with Potent Antifungal Activity Against Cryptococcus neoformans. Microorganisms, 12(12), 2648. https://doi.org/10.3390/microorganisms12122648










