Comparative Genomics and Biosynthetic Cluster Analysis of Antifungal Secondary Metabolites of Three Strains of Streptomyces albidoflavus Isolated from Rhizospheric Soils
Abstract
1. Introduction
2. Materials and Methods
2.1. Streptomyces Isolation, Culture, and Molecular Identification
2.2. Candida spp. Cultures
2.3. Fungal Susceptibility of Candida spp. Strains
2.4. Bioassay for Candida spp. Growth Inhibitory Activity of Streptomyces Strains
2.5. Obtention and Lyophilisation of Streptomyces Supernatants
2.6. Confirmation of Antifungal Activity of Supernatants
2.7. Statistical Analysis
2.8. Library Preparation and Genome Sequencing of Streptomyces A1, J25, and J29 ori2
2.9. De Novo Assembly and Genome Annotation
2.10. Comparative Genomics
2.11. Bioassay for the Production of Extracellular Hydrolytic Enzymes and Siderophore of Streptomyces
2.12. Prediction and Genomic Context of BGCs of Antifungal Metabolites
3. Results
3.1. Colonial Morphology of Streptomyces Strains
3.2. Fungal Susceptibility of Candida spp. Strains to Determine Fluconazole Phenotype
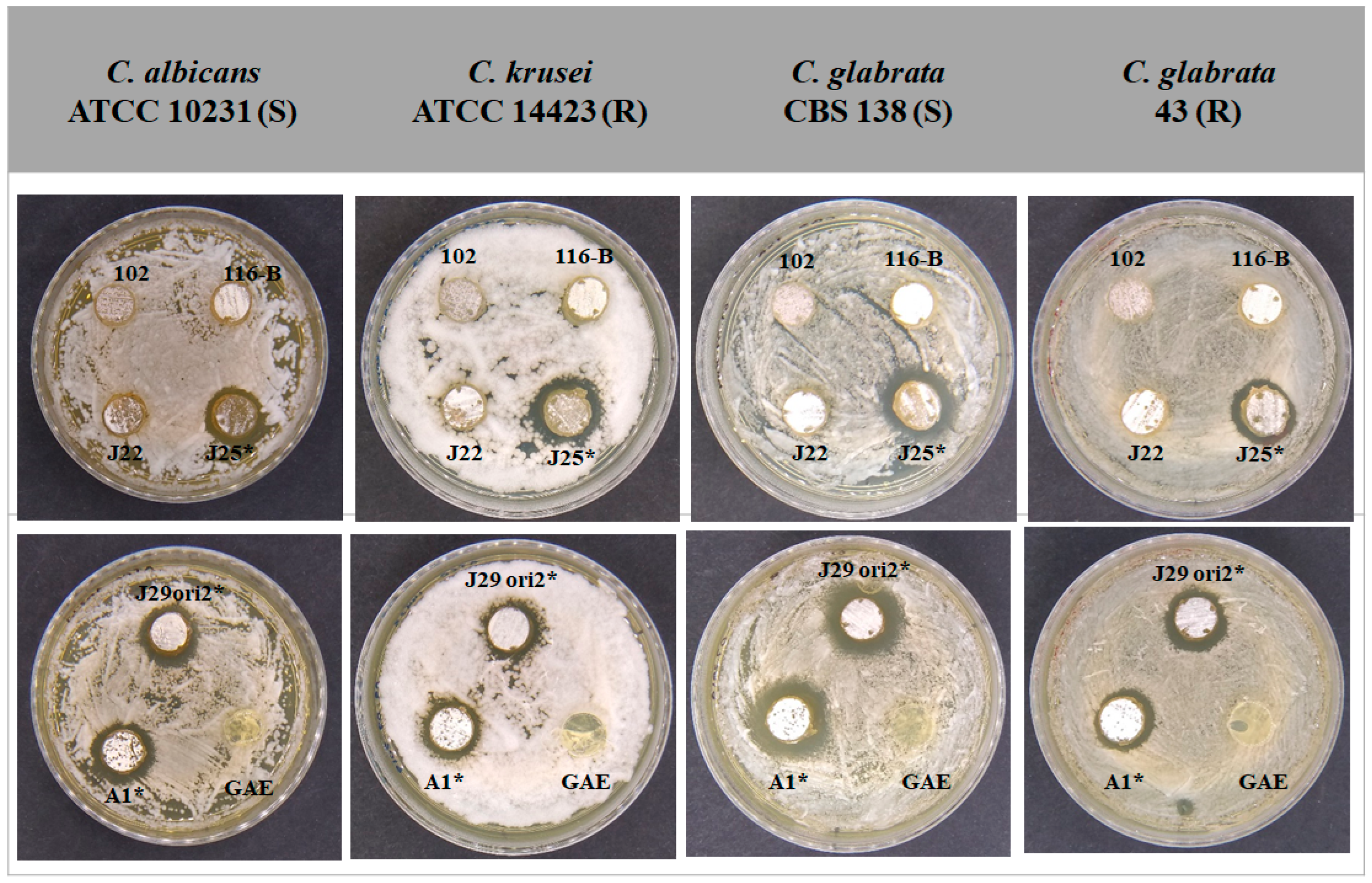
3.3. Bioassay for Selection of Streptomyces spp. with Inhibitory Capacity for the Growth of Candida spp. Strains
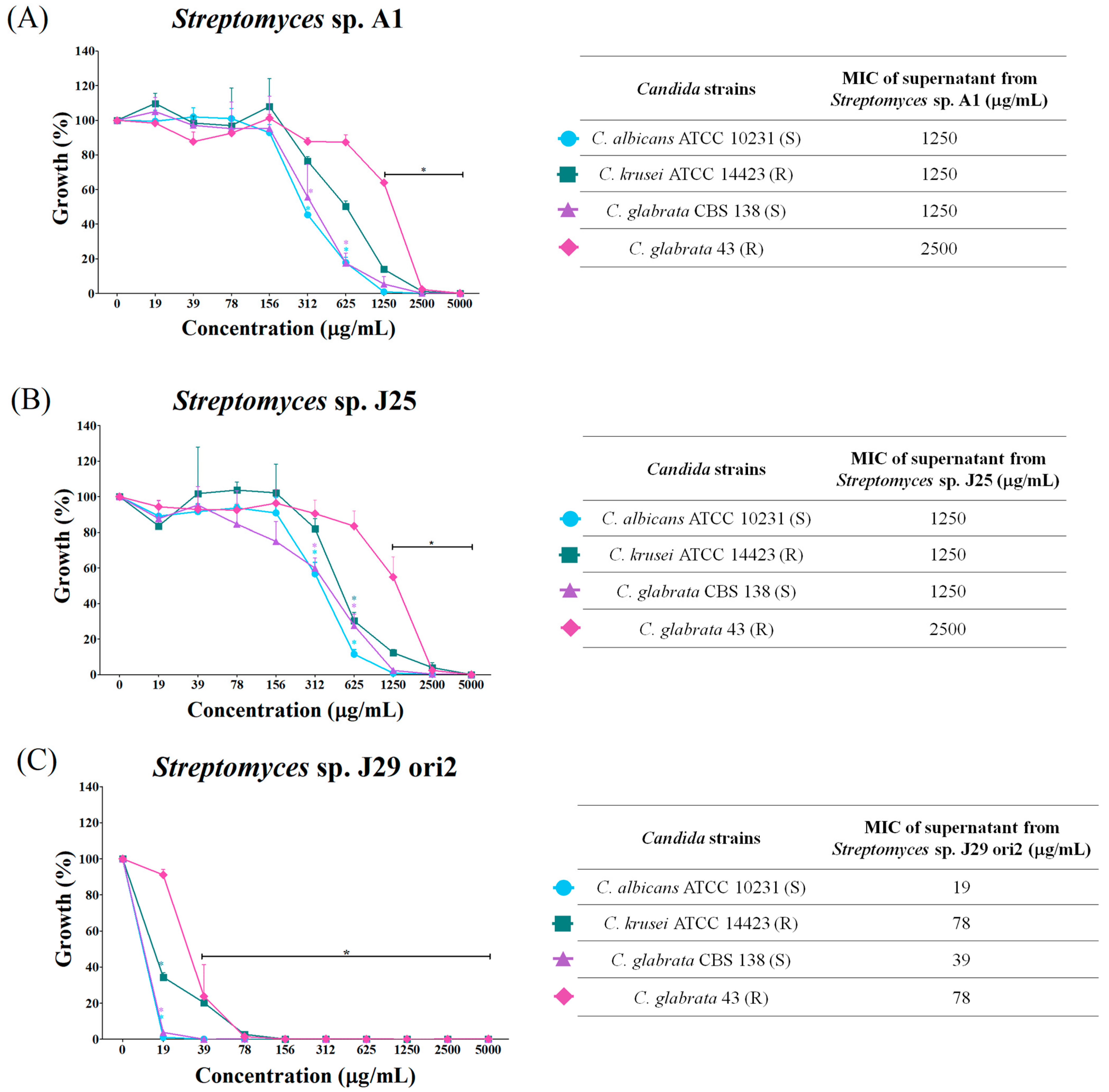
3.4. Determination of the Minimum Inhibitory Concentration of Supernatants
3.5. Genome Sequencing, Assembly, and Annotation of S. albidoflavus Strains
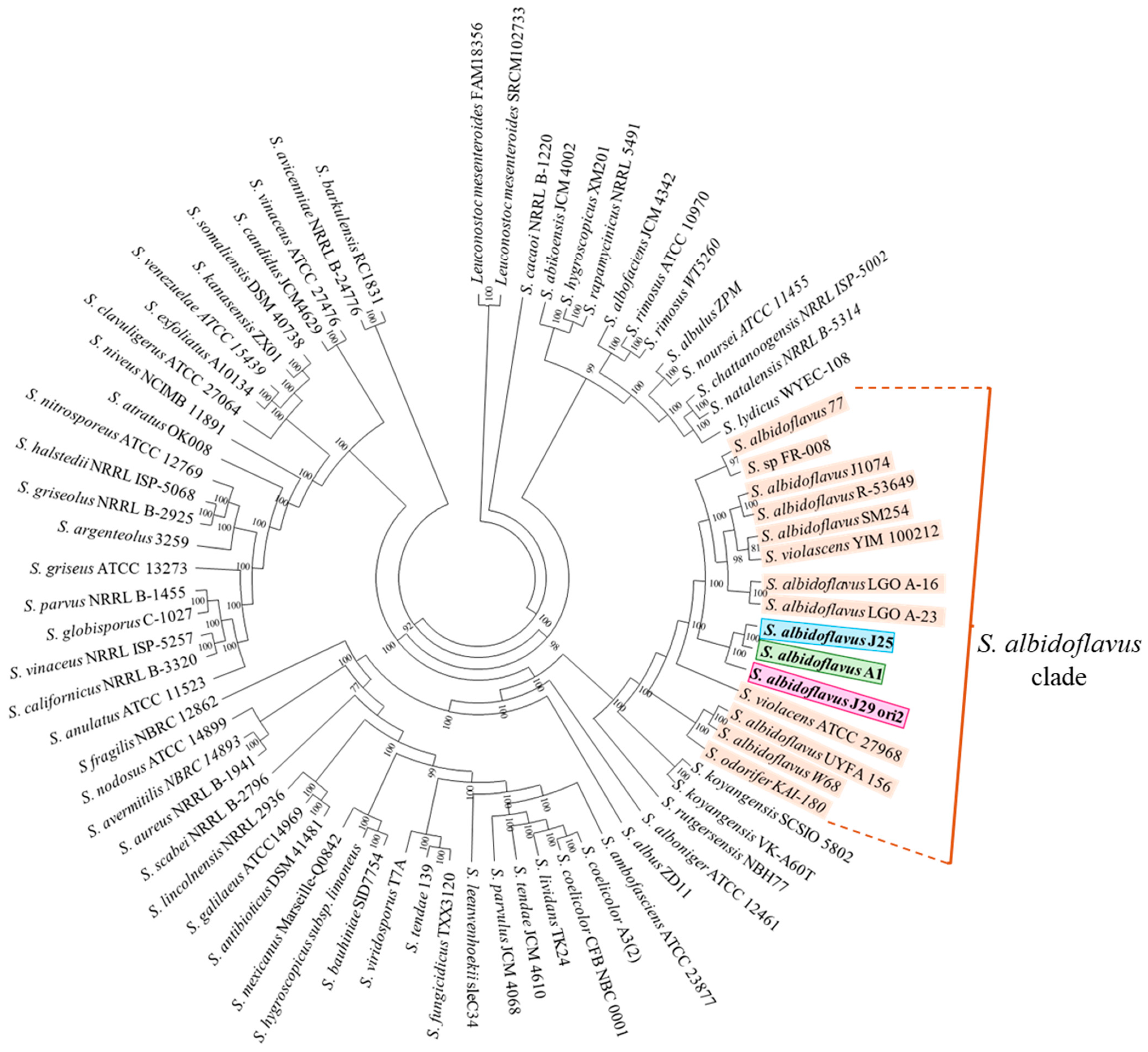
3.6. Genome-Based Taxonomic Identification of the Streptomyces Species
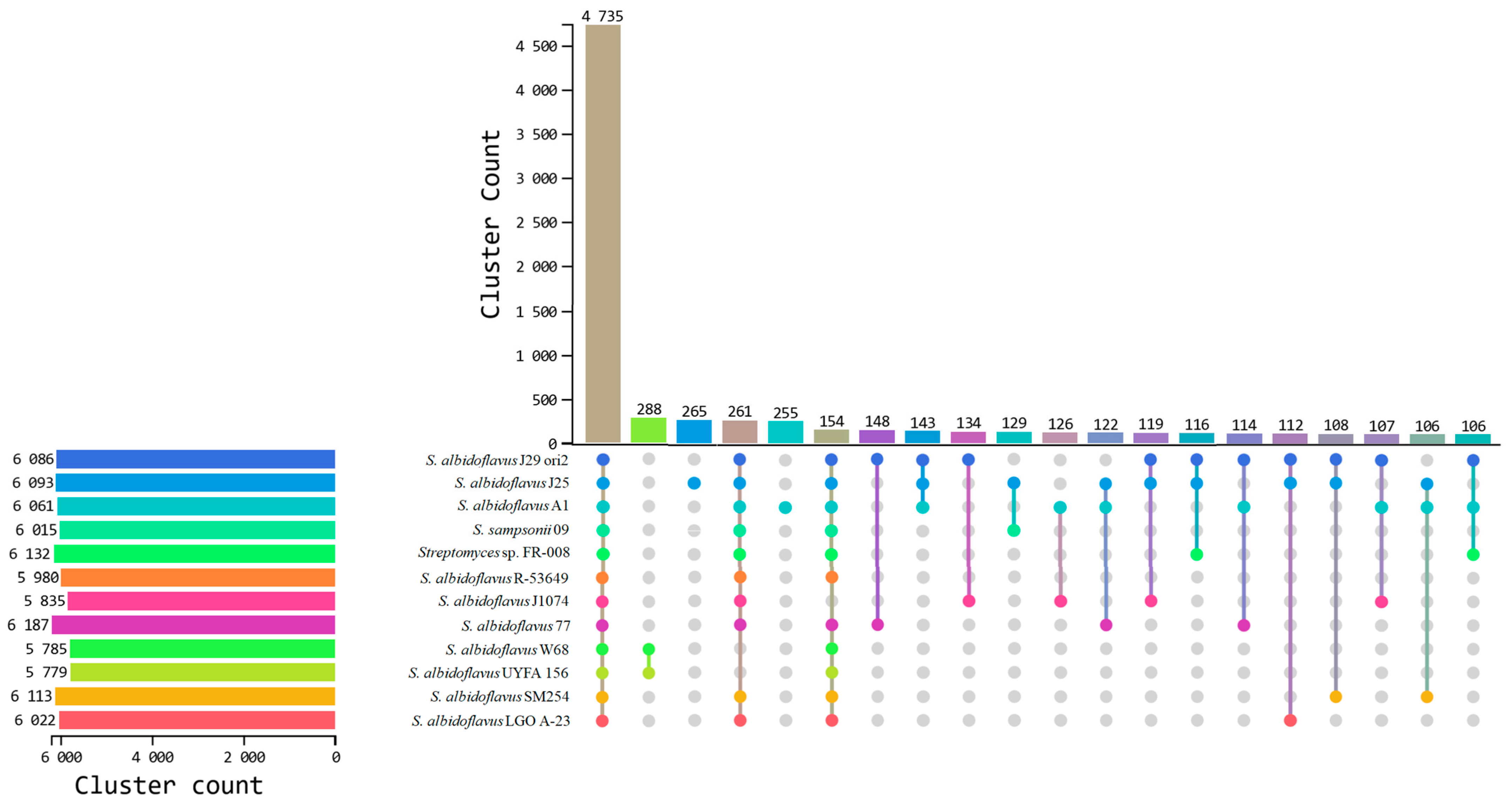
3.7. Comparative Genomics Among S. albidoflavus Strains
3.8. Analysis of Extracellular Hydrolytic Enzymes and Siderophore Production of S. albidoflavus Strains
3.9. Analysis of BGCs of Metabolites with Antifungal Activity from S. albidoflavus Strains
3.10. Synteny Analysis of the BGC of Candicidin and Surugamide A/D of Streptomyces spp.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Youseif, S.H.; Abd El-Megeed, F.H.; Humm, E.A.; Maymon, M.; Mohamed, A.H.; Saleh, S.A.; Hirsch, A.M. Comparative Analysis of the Cultured and Total Bacterial Community in the Wheat Rhizosphere Microbiome Using Culture-Dependent and Culture-Independent Approaches. Microbiol. Spectr. 2021, 9, e0067821. [Google Scholar] [CrossRef]
- Osburn, E.D.; Yang, G.; Rillig, M.C.; Strickland, M.S. Evaluating the role of bacterial diversity in supporting soil ecosystem functions under anthropogenic stress. ISME Commun. 2023, 3, 66. [Google Scholar] [CrossRef]
- Nonthakaew, N.; Panbangred, W.; Songnuan, W.; Intra, B. Plant growth-promoting properties of Streptomyces spp. isolates and their impact on mung bean plantlets’ rhizosphere microbiome. Front. Microbiol. 2022, 13, 967415. [Google Scholar] [CrossRef]
- Park, H.S.; Nah, H.J.; Kang, S.H.; Choi, S.S.; Kim, E.S. Screening and Isolation of a Novel Polyene-Producing Streptomyces Strain Inhibiting Phytopathogenic Fungi in the Soil Environment. Front. Bioeng. Biotechnol. 2021, 9, 692340. [Google Scholar] [CrossRef]
- Bekker, V.; Dodd, A.; Brady, D.; Rumbold, K. Tools for metabolic engineering in Streptomyces. Bioengineered 2014, 5, 293–299. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hwang, K.S.; Kim, H.U.; Charusanti, P.; Palsson, B.Ø.; Lee, S.Y. Systems biology and biotechnology of Streptomyces species for the production of secondary metabolites. Biotechnol. Adv. 2014, 32, 255–268. [Google Scholar] [CrossRef]
- Li, L.; Liu, X.; Jiang, W.; Lu, Y. Recent advances in synthetic biology approaches to optimize production of bioactive natural products in Actinobacteria. Front. Microbiol. 2019, 10, 2467. [Google Scholar] [CrossRef]
- Alam, K.; Hao, J.; Zhong, L.; Fan, G.; Ouyang, Q.; Islam, M.M.; Saiful, I.; Sun, H.; Zhang, Y.; Li, R.; et al. Complete genome sequencing and in silico genome mining reveal the promising metabolic potential in Streptomyces strain CS-7. Front. Microbiol. 2022, 13, 939919. [Google Scholar] [CrossRef] [PubMed]
- García-Gutiérrez, C.; Pérez-Victoria, I.; Montero, I.; Fernández-De la Hoz, J.; Malmierca, M.G.; Martín, J.; Salas, J.A.; Olano, C.; Reyes, F.; Méndez, C. Unearthing a cryptic biosynthetic gene cluster for the piperazic acid-bearing depsipeptide diperamycin in the ant-dweller Streptomyces sp. CS113. Int. J. Mol. Sci. 2024, 25, 2347. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Muñoz, J.C.; Selem-Mojica, N.; Mullowney, M.W.; Kautsar, S.A.; Tryon, J.H.; Parkinson, E.I.; De Los Santos, E.L.C.; Yeong, M.; Cruz-Morales, P.; Abubucker, S.; et al. A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 2020, 16, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Manteca, Á.; Yagüe, P. Streptomyces differentiation in liquid cultures as a trigger of secondary metabolism. Antibiotics 2018, 7, 41. [Google Scholar] [CrossRef]
- Sarmiento-Tovar, A.A.; Silva, L.; Sánchez-Suárez, J.; Diaz, L. Streptomyces-Derived Bioactive Pigments: Ecofriendly Source of Bioactive Compounds. Coatings 2022, 12, 1858. [Google Scholar] [CrossRef]
- Sadeghi, A.; Soltani, B.M.; Nekouei, M.K.; Jouzani, G.S.; Mirzaei, H.H.; Sadeghizadeh, M. Diversity of the ectoines biosynthesis genes in the salt tolerant Streptomyces and evidence for inductive effect of ectoines on their accumulation. Microbiol. Res. 2014, 169, 699–708. [Google Scholar] [CrossRef] [PubMed]
- van Bergeijk, D.A.; Terlouw, B.R.; Medema, M.H.; van Wezel, G.P. Ecology and genomics of Actinobacteria: New concepts for natural product discovery. Nat. Rev. Microbiol. 2020, 18, 546–558. [Google Scholar] [CrossRef]
- Zhang, M.; Kong, L.; Gong, R.; Iorio, M.; Donadio, S.; Deng, Z.; Sosio, M.; Chen, W. Biosynthesis of C-nucleoside antibiotics in actinobacteria: Recent advances and future developments. Microb. Cell Factories 2022, 21, 2. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, M.; Yang, Y.; Ding, X.; Yang, P.; Huang, K.; Hu, X.; Zhang, M.; Liu, X.; Yu, H. Structures and mechanism of chitin synthase and its inhibition by antifungal drug Nikkomycin Z. Cell Discov. 2022, 8, 129. [Google Scholar] [CrossRef] [PubMed]
- Osada, H. Discovery and applications of nucleoside antibiotics beyond polyoxin. J. Antibiot. 2019, 72, 855–864. [Google Scholar] [CrossRef]
- Xu, F.; Nazari, B.; Moon, K.; Bushin, L.B.; Seyedsayamdost, M.R. Discovery of a cryptic antifungal compound from Streptomycess albus J1074 using high-throughput elicitor screens. J. Am. Chem. Soc. 2017, 139, 9203–9212. [Google Scholar] [CrossRef]
- Caffrey, P.; De Poire, E.; Sheehan, J.; Sweeney, P. Polyene macrolide biosynthesis in streptomycetes and related bacteria: Recent advances from genome sequencing and experimental studies. Appl. Microbiol. Biotechnol. 2016, 100, 3893–3908. [Google Scholar] [CrossRef]
- Krespach, M.K.C.; Stroe, M.C.; Netzker, T.; Rosin, M.; Zehner, L.M.; Komor, A.J.; Beilmann, J.M.; Krüger, T.; Scherlach, K.; Kniemeyer, O.; et al. Streptomyces polyketides mediate bacteria-fungi interactions across soil environments. Nat. Microbiol. 2023, 8, 1348–1361. [Google Scholar] [CrossRef] [PubMed]
- Simpson, A.J.; Simpson, M.J.; Smith, E.; Kelleher, B.P. Microbially derived inputs to soil organic matter: Are cur-rent estimates too low? Environ. Sci. Technol. 2007, 41, 8070–8076. [Google Scholar] [CrossRef] [PubMed]
- Schulten, H.R.; Schnitzer, M. The chemistry of soil organic nitrogen: A review. Biol. Fertil Soils. 1997, 26, 1–15. [Google Scholar] [CrossRef]
- Hu, Y.; Zheng, Q.; Noll, L.; Zhang, S.; Wanek, W. Direct measurement of the in-situ decomposition of microbial-derived soil organic matter. Soil Biol. Biochem. 2020, 141, 107660. [Google Scholar] [CrossRef]
- Chater, K.F.; Biró, S.; Lee, K.J.; Palmer, T.; Schrempf, H. The complex extracellular biology of Streptomyces. FEMS Microbiol. Rev. 2010, 34, 171–198. [Google Scholar] [CrossRef] [PubMed]
- Book, A.J.; Lewin, G.R.; McDonald, B.R.; Takasuka, T.E.; Wendt-Pienkowski, E.; Doering, D.T.; Suh, S.; Raffa, K.F.; Fox, B.G.; Currie, C.R. Evolution of high cellulolytic activity in symbiotic Streptomyces through selection of expanded gene content and coordinated gene expression. PLoS Biol. 2016, 14, e1002475. [Google Scholar] [CrossRef]
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef]
- Kumar, M.; Kumar, P.; Das, P.; Solanki, R.; Kapur, M.K. Potential applications of extracellular enzymes from Streptomyces spp. in various industries. Arch. Microbiol. 2020, 202, 1597–1615. [Google Scholar] [CrossRef] [PubMed]
- Méndez, C.; Braña, A.F.; Manzanal, M.B.; Hardisson, C. Role of substrate mycelium in colony development in Streptomyces. Can. J. Microbiol. 1985, 31, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Stubner, S. Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreen™ detection. J. Microbiol. Methods 2002, 50, 155–164. [Google Scholar] [CrossRef] [PubMed]
- CLSI Standard M27; Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. 4th ed, Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2017.
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 1 August 2024).
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Sun, J.; Lu, F.; Luo, Y.; Bie, L.; Xu, L.; Wang, Y. OrthoVenn3: An integrated platform for exploring and visualizing orthologous data across genomes. Nucleic Acids Res. 2023, 51, W397–W403. [Google Scholar] [CrossRef] [PubMed]
- Suriyachadkun, C.; Chunhachart, O.; Srithaworn, M.; Tangchitcharoenkhul, R.; Tangjitjareonkun, J. Zinc-solubilizing Streptomyces spp. as bioinoculants for promoting the growth of soybean (Glycine max (L.) Merrill). J. Microbiol. Biotechnol. 2022, 32, 1435–1446. [Google Scholar] [CrossRef] [PubMed]
- Narayana, K.J.; Vijayalakshmi, M. Chitinase production by Streptomyces sp. ANU 6277. Braz. J. Microbiol. 2009, 40, 725–733. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.B.; Chou, K.C. Gpos-mPLoc: A top-down approach to improve the quality of predicting subcellular localization of Gram-positive bacterial proteins. Protein Pept. Lett. 2009, 16, 1478–1484. [Google Scholar] [CrossRef]
- Rehan, M.; Barakat, H.; Almami, I.S.; Qureshi, K.A.; Alsohim, A.S. Production and Potential Genetic Pathways of Three Different Siderophore Types in Streptomyces tricolor Strain HM10. Fermentation 2022, 8, 346. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed]
- Terlouw, B.R.; Blin, K.; Navarro-Munoz, J.C.; Avalon, N.E.; Chevrette, M.G.; Egbert, S.; Lee, S.; Meijer, D.; Recchia, M.J.J.; Reitz, Z.L.; et al. MIBiG 3.0: A community-driven effort to annotate experimentally validated biosynthetic gene clusters. Nucleic Acids Res. 2023, 51, D603–D610. [Google Scholar] [CrossRef]
- Nofiani, R.; Rudiyansyah; Ardiningsih, P.; Rizky; Zahra, S.T.A.; Sukito, A.; Weisberg, A.J.; Chang, J.H.; Mahmud, T. Genome features and secondary metabolite potential of the marine symbiont Streptomyces sp. RS2. Arch. Microbiol. 2023, 205, 244. [Google Scholar] [CrossRef]
- Bautista-Crescencio, C.; Casimiro-Ramos, A.; Fragoso-Vázquez, M.J.; Correa-Basurto, J.; Olano, C.; Hernández-Rodríguez, C.; Villa-Tanaca, L. Streptomyces albidoflavus Q antifungal metabolites inhibit the ergosterol biosynthesis pathway and yeast growth in fluconazole-resistant Candida glabrata: Phylogenomic and metabolomic analyses. Microbiol. Spectr. 2023, 11, e0127123. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Zhang, Z.; Huang, J.; Wei, S.; Sun, X.; Chen, Y.; Liu, H.; Li, S. Candicidin isomer production is assential for biocontrol of cucumber Rhizoctonia rot by Streptomyces albidoflavus W68. Appl. Environ. Microbiol. 2021, 87, e03078-20. [Google Scholar] [CrossRef]
- Martins, N.D.R.C.; Rodrigues da Silva, A.; Ratcliffe, N.; Evangelho, V.G.O.; Castro, H.C.; Quinn, G.A. Streptomyces: A natural source of anti-Candida agents. J. Med. Microbiol. 2023, 72, 001777. [Google Scholar] [CrossRef]
- Lai, J.; Liu, B.; Xiong, G.; Luo, Q.; Song, S.; Jiang, J.; Wei, H.; Wang, J. Inhibitory mechanism of 4-ethyl-1,2-dimethoxybenzene produced by Streptomyces albidoflavus strain ML27 against Colletotrichum gloeosporioides. Pestic. Biochem. Physiol. 2024, 204, 106086. [Google Scholar] [CrossRef] [PubMed]
- Sottorff, I.; Wiese, J.; Lipfert, M.; Preußke, N.; Sönnichsen, F.D.; Imhoff, J.F. Different secondary metabolite profiles of phylogenetically almost identical Streptomyces griseus strains originating from geographically remote locations. Microorganisms 2019, 7, 166. [Google Scholar] [CrossRef]
- Wen, Y.; Zhang, G.; Bahadur, A.; Liu, Y.; Zhang, Z.; Chen, T.; Liu, G.; Zhang, W. Streptomyces gobiensis sp. nov., an antimicrobial producing actinobacterium isolated from soil under black Gobi rocks. Int. J. Syst. Evol. Microbiol. 2022, 72, 005318. [Google Scholar] [CrossRef] [PubMed]
- Feng, N.X.; Li, D.W.; Zhang, F.; Bin, H.; Huang, Y.T.; Xiang, L.; Liu, B.L.; Cai, Q.Y.; Li, Y.W.; Xu, D.L.; et al. Biodegradation of phthalate acid esters and whole-genome analysis of a novel Streptomyces sp. FZ201 isolated from natural habitats. J. Hazard. Mater. 2024, 469, 133972. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.; Rong, X.; Pinto-Tomás, A.A.; Fernández-Villalobos, M.; Murillo-Cruz, C.; Huang, Y. Population genetic analysis of Streptomyces albidoflavus reveals habitat barriers to homologous recombination in the diversification of streptomycetes. Appl. Environ. Microbiol. 2015, 81, 966–975. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Deng, M.R.; Guo, J.; Li, X.; Zhu, C.H.; Zhu, H.H. Granaticins and their biosynthetic gene cluster from Streptomyces vietnamensis: Evidence of horizontal gene transfer. Antonie Van Leeuwenhoek 2011, 100, 607–617. [Google Scholar] [CrossRef]
- Li, J.; Zhao, Z.; Zhong, W.; Zhong, C.; Zong, G.; Fu, J.; Cao, G. Impacts of horizontal gene transfer on the compact genome of the clavulanic acid-producing Streptomyces strain F613-1. 3 Biotech 2018, 8, 472. [Google Scholar] [CrossRef] [PubMed]
- Ayala-Ruano, S.; Santander-Gordón, D.; Tejera, E.; Perez-Castillo, Y.; Armijos-Jaramillo, V. A putative antimicrobial peptide from Hymenoptera in the megaplasmid pSCL4 of Streptomyces clavuligerus ATCC 27064 reveals a singular case of horizontal gene transfer with potential applications. Ecol. Evol. 2019, 9, 2602–2614. [Google Scholar] [CrossRef]
- Tidjani, A.R.; Lorenzi, J.N.; Toussaint, M.; van Dijk, E.; Naquin, D.; Lespinet, O.; Bontemps, C.; Leblond, P. Massive gene flux drives genome diversity between sympatric Streptomyces conspecifics. MBio 2019, 10, e01533-19. [Google Scholar] [CrossRef]
- Choufa, C.; Tidjani, A.R.; Gauthier, A.; Harb, M.; Lao, J.; Leblond-Bourget, N.; Vos, M.; Leblond, P.; Bontemps, C. Prevalence and mobility of integrative and conjugative elements within a Streptomyces natural population. Front. Microbiol. 2022, 13, 970179. [Google Scholar] [CrossRef]
- Badalamenti, J.P.; Erickson, J.D.; Salomon, C.E. Complete genome sequence of Streptomyces albus SM254, a potent antagonist of bat white-nose syndrome pathogen Pseudogymnoascus destructans. Genome Announc. 2016, 4, e00290-16. [Google Scholar] [CrossRef]
- de los Santos, M.C.; Taulé, C.; Mareque, C.; Beracochea, M.; Battistoni, F. Identification and characterization of the part of the bacterial community associated with field-grown tall fescue (Festuca arundinacea) cv. SFRO Don Tomás in Uruguay. Ann. Microbiol. 2016, 66, 329–342. [Google Scholar] [CrossRef]
- Peng, F.; Zhang, M.Y.; Hou, S.Y.; Chen, J.; Wu, Y.Y.; Zhang, Y.X. Insights into Streptomyces spp. isolated from the rhizospheric soil of Panax notoginseng: Isolation, antimicrobial activity and biosynthetic potential for polyketides and non-ribosomal peptides. BMC Microbiol. 2020, 20, 143. [Google Scholar] [CrossRef]
- Du, Y.; Wang, T.; Jiang, J.; Wang, Y.; Lv, C.; Sun, K.; Sun, J.; Yan, B.; Kang, C.; Guo, L.; et al. Biological control and plant growth promotion properties of Streptomyces albidoflavus St-220 isolated from Salvia miltiorrhiza rhizosphere. Front. Plant Sci. 2022, 13, 976813. [Google Scholar] [CrossRef]
- Li, J.; Zhang, L.; Yao, G.; Zhu, L.; Lin, J.; Wang, C.; Du, B.; Ding, Y.; Mei, X. Synergistic effect of co-culture rhizosphere Streptomyces: A promising strategy to enhance antimicrobial activity and plant growth-promoting function. Front. Microbiol. 2022, 13, 976484. [Google Scholar] [CrossRef]
- Amaresan, N.; Kumar, K.; Naik, J.H.; Bapatla, K.G.; Mishra, R.K. Streptomyces in plant growth promotion: Mechanisms and role. In New and Future Developments in Microbial Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2018; pp. 125–135. [Google Scholar] [CrossRef]
- Vijayalakshmi, M.; Narayana, K.J.P. Production of extracellular a-amylase by Streptomyces albidoflavus. Asian J. Biochem. 2008, 3, 194–197. [Google Scholar] [CrossRef]
- Vijayalakshmi, M.; Narayana, K.J.P. Production of extracellular protease by Streptomyces albidoflavus. Asian J. Biochem. 2008, 3, 198–202. [Google Scholar] [CrossRef]
- Sugimori, D.; Kano, K.; Matsumoto, Y. Purification, characterization, molecular cloning and extracellular production of a phospholipase A1 from Streptomyces albidoflavus NA297. FEBS Open Bio 2012, 2, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Umar, A.A.; Hussaini, A.B.; Yahayya, J.; Sani, I.; Aminu, H. Chitinolytic and antagonistic activity of Streptomyces isolated from fadama soil against phytopathogenic fungi. Trop. Life Sci. Res. 2021, 32, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Kotb, E.; Alabdalall, A.H.; Alghamdi, A.I.; Ababutain, I.M.; Aldakeel, S.A.; Al-Zuwaid, S.K.; Algarudi, B.M.; Algarudi, S.M.; Ahmed, A.A.; Albarrag, A.M. Screening for chitin degrading bacteria in the environment of Saudi Arabia and characterization of the most potent chitinase from Streptomyces variabilis Am1. Sci. Rep. 2023, 13, 11723. [Google Scholar] [CrossRef] [PubMed]
- Rillig, M.C.; Caldwell, B.A.; Wösten, H.A.B.; Sollins, P. Role of proteins in soil carbon and nitrogen storage: Controls on persistence. Biogeochemistry 2007, 85, 25–44. [Google Scholar] [CrossRef]
- Greenfield, L.M.; Hill, P.W.; Seaton, F.M.; Paterson, E.; Baggs, E.M.; Jones, D.L. Is soluble protein mineralisation and protease activity in soil regulated by supply or demand? Soil Biol. Biochem. 2020, 150, 108007. [Google Scholar] [CrossRef]
- Kim, J.; Copley, S.D. Why metabolic enzymes are essential or nonessential for growth of Escherichia coli K12 on glucose. Biochemistry 2007, 46, 12501–12511. [Google Scholar] [CrossRef]
- Schniete, J.K.; Reumerman, R.; Kerr, L.; Tucker, N.P.; Hunter, I.S.; Herron, P.R.; Hoskisson, P.A. Differential transcription of expanded gene families in central carbon metabolism of Streptomyces coelicolor A3(2). Access Microbiol. 2020, 2, acmi000122. [Google Scholar] [CrossRef]
- Belknap, K.C.; Park, C.J.; Barth, B.M.; Andam, C.P. Genome mining of biosynthetic and chemotherapeutic gene clusters in Streptomyces bacteria. Sci. Rep. 2020, 10, 2003. [Google Scholar] [CrossRef] [PubMed]
- Diabankana, R.G.C.; Frolov, M.; Keremli, S.; Validov, S.Z.; Afordoanyi, D.M. Genomic insights into the microbial agent Streptomyces albidoflavus MGMM6 for various biotechnology applications. Microorganisms 2023, 11, 2872. [Google Scholar] [CrossRef] [PubMed]
- Thekkangil, A.; George, B.; Prakash, S.M.U.; Suchithra, T.V. Mechanism of Streptomyces albidoflavus STV1572a derived 1-heneicosanol as an inhibitor against squalene epoxidase of Trichophyton mentagrophytes. Microb. Pathog. 2021, 154, 104853. [Google Scholar] [CrossRef]
- Ye, S.; Magadán-Corpas, P.; Pérez-Valero, Á.; Villar, C.J.; Lombó, F. Metabolic engineering strategies for naringenin production enhancement in Streptomyces albidoflavus J1074. Microb. Cell Factories 2023, 22, 167. [Google Scholar] [CrossRef]
- Abdelshafy Mohamad, O.A.; Liu, Y.H.; Huang, Y.; Kuchkarova, N.; Dong, L.; Jiao, J.Y.; Fang, B.Z.; Ma, J.B.; Hatab, S.; Li, W.J. Metabonomic analysis to identify exometabolome changes underlying antifungal and growth promotion mechanisms of endophytic actinobacterium Streptomyces albidoflavus for sustainable agriculture practice. Front. Microbiol. 2024, 15, 1439798. [Google Scholar] [CrossRef]
- Jørgensen, H.; Fjaervik, E.; Hakvåg, S.; Bruheim, P.; Bredholt, H.; Klinkenberg, G.; Ellingsen, T.E.; Zotchev, S.B. Candicidin biosynthesis gene cluster is widely distributed among Streptomyces spp. isolated from the sediments and the neuston layer of the Trondheim fjord, Norway. Appl. Environ. Microbiol. 2009, 75, 3296–3303. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, J.F.; Caffrey, P.; Gil, J.A.; Zotchev, S.B. Polyene antibiotic biosynthesis gene clusters. Appl. Microbiol. Biotechnol. 2003, 61, 179–188. [Google Scholar] [CrossRef]
- Chen, S.; Huang, X.; Zhou, X.; Bai, L.; He, J.; Jeong, K.J.; Lee, S.Y.; Deng, Z. organizational and mutational analysis of a complete FR-008/candicidin gene cluster encoding a structural related polyene complex. Chem. Biol. 2003, 10, 1065–1076. [Google Scholar] [CrossRef] [PubMed]
- Gil, J.; Campelo-Diez, A. Candicidin biosynthesis in Streptomyces griseus. Appl. Microbiol. Biotechnol. 2003, 60, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Olano, C.; García, I.; González, A.; Rodriguez, M.; Rozas, D.; Rubio, J.; Sánchez-Hidalgo, M.; Braña, A.F.; Méndez, C.; Salas, J.A. Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb. Biotechnol. 2014, 7, 242–256. [Google Scholar] [CrossRef]
- Liu, X.; Sun, X.; Wang, T.; Zhang, X.; Tian, X.; Zhuang, Y.; Chu, J. Enhancing candicidin biosynthesis by medium optimization and pH stepwise control strategy with process metabolomics analysis of Streptomyces ZYJ-6. Bioprocess Biosyst. Eng. 2018, 41, 1743–1755. [Google Scholar] [CrossRef]
- Szwarc, K.; Szczeblewski, P.; Sowiński, P.; Borowski, E.; Pawlak, J. The stereostructure of candicidin D. J. Antibiot. 2015, 68, 504–510. [Google Scholar] [CrossRef]
- Haeder, S.; Wirth, R.; Herz, H.; Spiteller, D. Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 4742–4746. [Google Scholar] [CrossRef] [PubMed]
- Ridley, C.P.; Lee, H.Y.; Khosla, C. Evolution of polyketide synthases in bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 4595–4600. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, K.; Kuranaga, T.; Sano, A.; Ninomiya, A.; Takada, K.; Wakimoto, T. The revised structure of the cyclic octapeptide surugamide A. Chem. Pharm. Bull. 2019, 67, 476–480. [Google Scholar] [CrossRef]
- Takeuchi, A.; Hirata, A.; Teshima, A.; Ueki, M.; Satoh, T.; Matsuda, K.; Wakimoto, T.; Arakawa, K.; Ishikawa, M.; Suzuki, T. Characterization of the surugamide biosynthetic gene cluster of TUA-NKU25, a Streptomyces diastaticus strain isolated from Kusaya, and its effects on salt-dependent growth. Biosci. Biotechnol. Biochem. 2023, 87, 320–329. [Google Scholar] [CrossRef]
- Attia, M.A.; Thibodeaux, C.J. Cycling back to biocatalysis. Nat. Catal. 2020, 3, 476–477. [Google Scholar] [CrossRef]
- Fazal, A.; Webb, M.E.; Seipke, R.F. The desotamide family of antibiotics. Antibiotics 2020, 9, 452. [Google Scholar] [CrossRef]
- Almeida, E.L.; Kaur, N.; Jennings, L.K.; Carrillo Rincón, A.F.; Jackson, S.A.; Thomas, O.P.; Dobson, A.D.W. Genome mining coupled with OSMAC-based cultivation reveal differential production of surugamide A by the marine sponge isolate Streptomyces sp. SM17 when compared to its terrestrial relative S. albidoflavus J1074. Microorganisms 2019, 7, 394. [Google Scholar] [CrossRef]
- Maw, Z.A.; Haltli, B.; Guo, J.J.; Baldisseri, D.M.; Cartmell, C.; Kerr, R.G. Discovery of acyl-surugamide A2 from marine Streptomyces albidoflavus RKJM-0023-a new cyclic nonribosomal peptide containing an N-E-acetyl-L-lysine residue. Molecules 2024, 29, 1482. [Google Scholar] [CrossRef] [PubMed]
- Schroeckh, V.; Scherlach, K.; Nützmann, H.W.; Shelest, E.; Schmidt-Heck, W.; Schuemann, J.; Martin, K.; Hertweck, C.; Brakhage, A.A. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 2009, 106, 14558–14563. [Google Scholar] [CrossRef]
- Onaka, H.; Mori, Y.; Igarashi, Y.; Furumai, T. Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl. Environ. Microbiol. 2011, 77, 400–406. [Google Scholar] [CrossRef]
- Wu, C.; Zacchetti, B.; Ram, A.F.; van Wezel, G.P.; Claessen, D.; Hae Choi, Y. Expanding the chemical space for natural products by Aspergillus-Streptomyces co-cultivation and biotransformation. Sci. Rep. 2015, 5, 10868. [Google Scholar] [CrossRef] [PubMed]

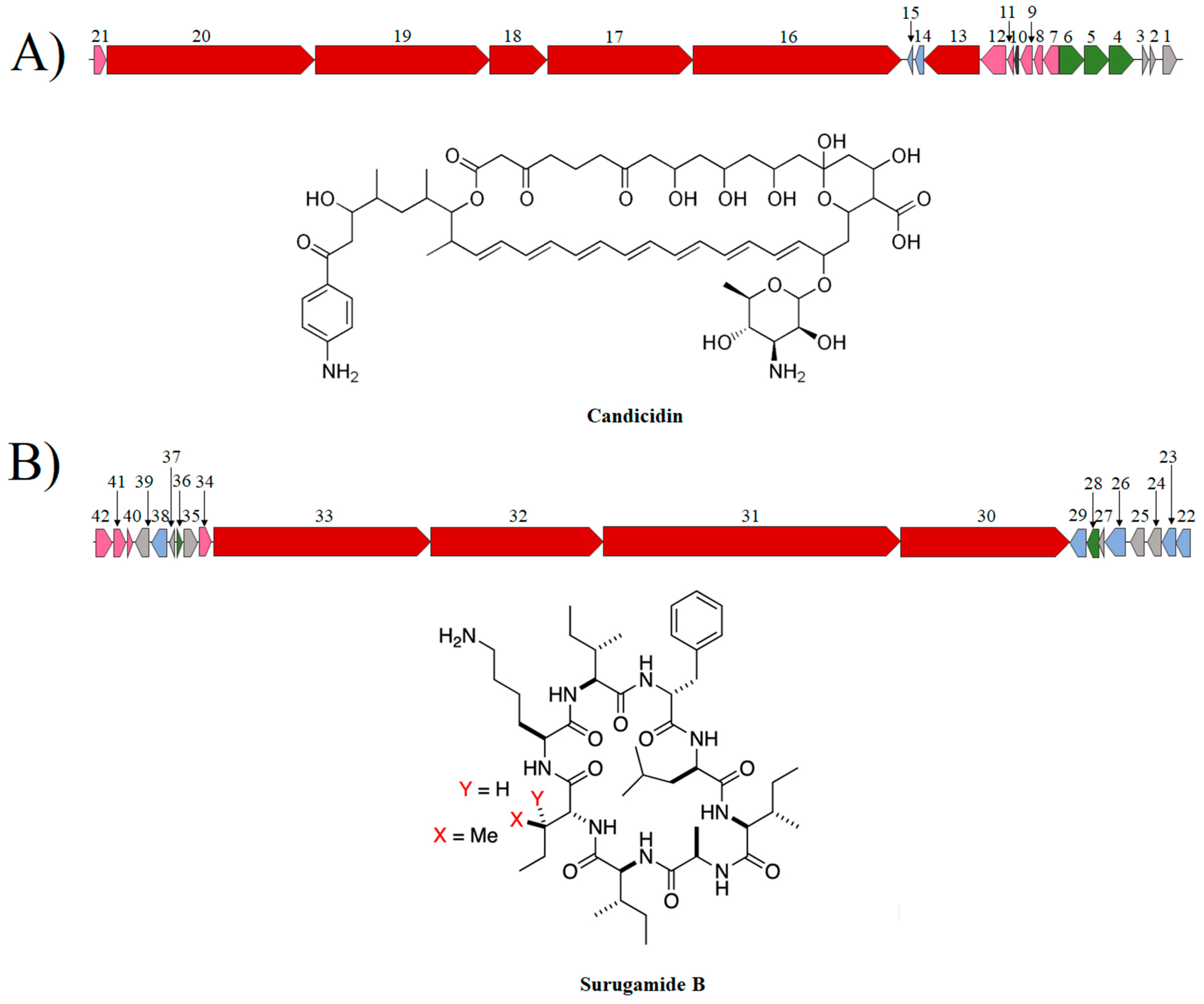
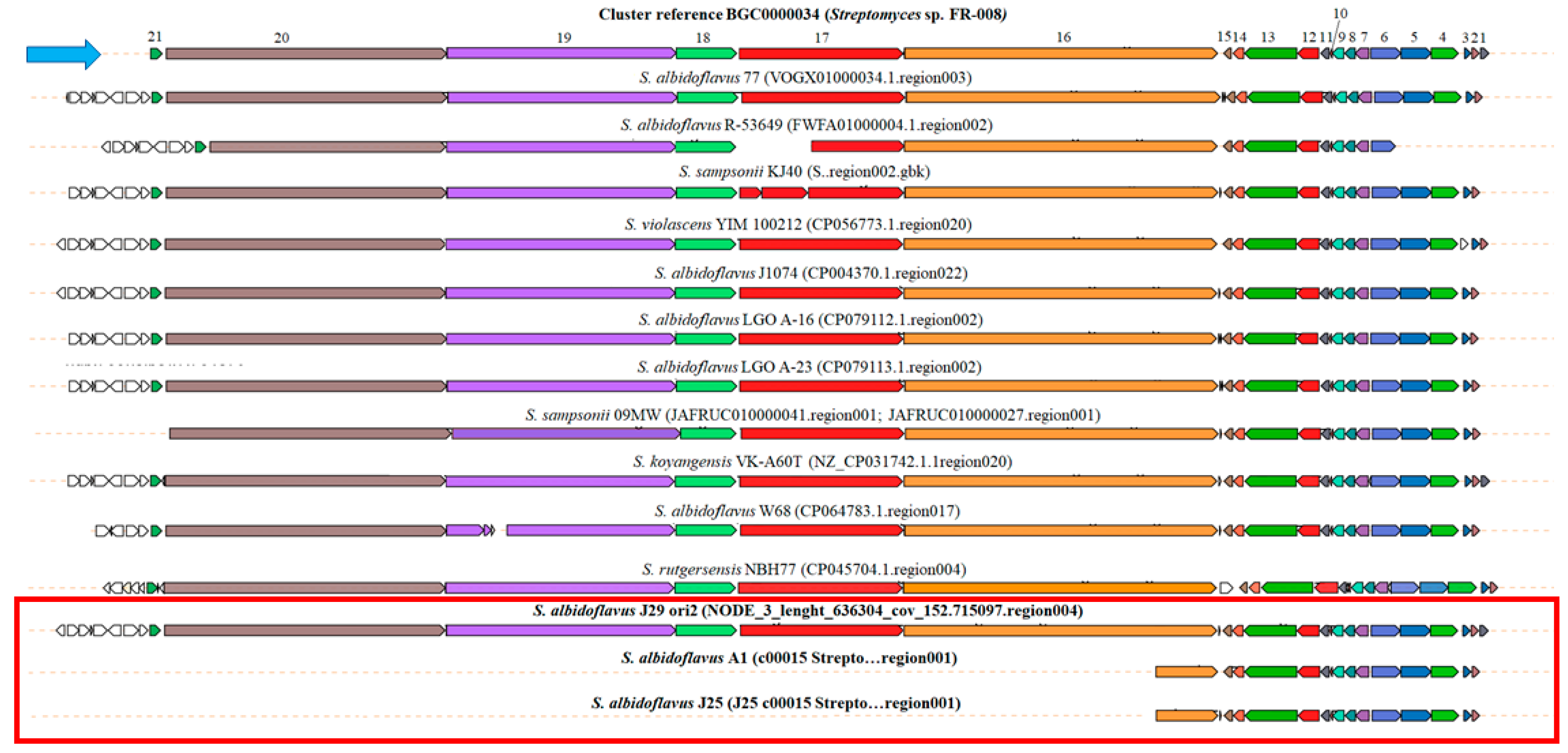

| Reference Genome (GeneBank Access Number) | Streptomyces sp. A1 | Streptomyces sp. J25 | Streptomyces sp. J29 ori2 |
|---|---|---|---|
| Streptomyces albidoflavus 77 (GCA_007896905.1) | 98.62 | 98.62 | 98.60 |
| Streptomyces albidoflavus J1074 (GCA_000359525.1) | 98.62 | 98.61 | 98.65 |
| Streptomyces albidoflavus LGO A-23 (GCA_019286295.1) | 98.67 | 98.64 | 98.63 |
| Streptomyces albidoflavus SM254 (GCA_001577385.1) | 98.71 | 98.67 | 98.65 |
| Streptomyces albidoflavus R-53649 (GCA_900171555.1) | 98.68 | 98.64 | 98.66 |
| Streptomyces sp. FR-008 (GCA_001431765.1) | 98.61 | 98.59 | 98.60 |
| Streptomyces koyangensis VK-A60T (GCA_003428925) | 95.79 | 95.78 | 95.75 |
| Streptomyces odorifer KAI-180 (GCA_013363465.1) | 95.98 | 95.95 | 96.05 |
| Streptomyces vinaceus ATCC 27476 (GCA_008704935.1) | 78.25 | 78.26 | 78.13 |
| Streptomyces violascens ATCC 27968 (GCA_009429105.1) | 96.00 | 95.99 | 95.98 |
| S. albidoflavus Strain | Phenotypic Extracellular Enzyme | Number of Encoding Genes | Solubilization Index (SI) |
|---|---|---|---|
| A1 | Amylase | 1 | 4.93 |
| Cellulase | 3 | 2.28 | |
| Chitinase | 11 | 2.16 | |
| Protease | 22 | 3.26 | |
| J25 | Amylase | 1 | 5.23 |
| Cellulase | 3 | 2.13 | |
| Chitinase | 11 | 2.17 | |
| Protease | 25 | 3.16 | |
| J29 ori2 | Amylase | 3 | 6.17 |
| Cellulase | 2 | 2.22 | |
| Chitinase | 10 | 2.10 | |
| Protease | 28 | 4.24 |
| Type | Activity | Most Similar Known Cluster | Similitude Percentage (%) |
|---|---|---|---|
| S. albidoflavus A1 | |||
| T1PKS, NRPS | AF, AB | SGR PTMs | 100 |
| T1PKS, NRPS-like | AF | Candicidin | 90 |
| T2PKS | AF, ATM, AB | Fredericamycin A | 60 |
| NRPS, RRE-containing | AF | Surugamide A/Surugamide D | 90 |
| Siderophore | Q, AF | Desferrioxamin B | 100 |
| Siderophore | AF, AB | Ficellomycin | 5 |
| LAP, Thiopeptide | AF | Fluostatins M-Q | 4 |
| S. albidoflavus J25 | |||
| T1PKS, NRPS | AF, AB | SGR PTMs | 100 |
| T1PKS, NRPS-like, NRPS, lanthipeptide Class II | AF | Candicidin | 95 |
| T1PKS | AF | Mediomycin A | 28 |
| T2PKS | AF, ATM, AB | Fredericamycin A | 60 |
| NRPS, RRE-containing | AF | Surugamide A/Surugamide D | 100 |
| Siderophore | AF, AB | Ficellomycin | 5 |
| Siderophore | Q, AF | Desferrioxamin B | 100 |
| LAP, Thiopeptide | AF | Fluostatins M-Q | 4 |
| S. albidoflavus J29 ori2 | |||
| T1PKS, NRPS-like, NRPS, lanthipeptide Class II | AF | Candicidin | 100 |
| T1PKS, NRPS | AF, AB | SGR PTMs/SGR PTM Compound b-d | 100 |
| T2PKS, NRPS | AF, ATM, AB | Fredericamycin A | 96 |
| NRPS | AF, AB | Cyclofaulknamycin | 75 |
| NRPS, LAP | AF | Surugamide A/Surugamide D | 100 |
| NI-Siderophore | Q, AF | Desferrioxamin B | 100 |
| LAP, thiopeptide, RRE-containing | AF | Fluostatins M-Q | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez-Silva, A.; San Juan-Mendo, M.; Delgado-Prudencio, G.; Hernández-García, J.A.; Larios-Serrato, V.; Aguilar, C.; Villa-Tanaca, L.; Hernández-Rodríguez, C. Comparative Genomics and Biosynthetic Cluster Analysis of Antifungal Secondary Metabolites of Three Strains of Streptomyces albidoflavus Isolated from Rhizospheric Soils. Microorganisms 2024, 12, 2637. https://doi.org/10.3390/microorganisms12122637
Gonzalez-Silva A, San Juan-Mendo M, Delgado-Prudencio G, Hernández-García JA, Larios-Serrato V, Aguilar C, Villa-Tanaca L, Hernández-Rodríguez C. Comparative Genomics and Biosynthetic Cluster Analysis of Antifungal Secondary Metabolites of Three Strains of Streptomyces albidoflavus Isolated from Rhizospheric Soils. Microorganisms. 2024; 12(12):2637. https://doi.org/10.3390/microorganisms12122637
Chicago/Turabian StyleGonzalez-Silva, Adilene, Magali San Juan-Mendo, Gustavo Delgado-Prudencio, Juan Alfredo Hernández-García, Violeta Larios-Serrato, César Aguilar, Lourdes Villa-Tanaca, and César Hernández-Rodríguez. 2024. "Comparative Genomics and Biosynthetic Cluster Analysis of Antifungal Secondary Metabolites of Three Strains of Streptomyces albidoflavus Isolated from Rhizospheric Soils" Microorganisms 12, no. 12: 2637. https://doi.org/10.3390/microorganisms12122637
APA StyleGonzalez-Silva, A., San Juan-Mendo, M., Delgado-Prudencio, G., Hernández-García, J. A., Larios-Serrato, V., Aguilar, C., Villa-Tanaca, L., & Hernández-Rodríguez, C. (2024). Comparative Genomics and Biosynthetic Cluster Analysis of Antifungal Secondary Metabolites of Three Strains of Streptomyces albidoflavus Isolated from Rhizospheric Soils. Microorganisms, 12(12), 2637. https://doi.org/10.3390/microorganisms12122637






