Microbiome Structures and Beneficial Bacteria in Soybean Roots Under Field Conditions of Prolonged High Temperatures and Drought Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Sample Collection and Preservation
2.2. Field Management
2.2.1. Insecticide and Herbicide Applications
2.2.2. Fungicide and Fertilizer Applications
2.3. DNA Sample Preparation
2.4. Library Construction and High-Throughput DNA Sequencing of 16S rDNA
2.5. Raw Data Import, Quality Checking, and ASV Feature Table Construction
2.6. Taxonomy Assignment
2.7. Diversity Analysis
2.8. Co-Occurrence Microbial Network Analysis
2.9. Isolation and Characterization of Beneficial Bacteria from Soybean Plants Under Drought Stress Conditions
2.10. Identification of Soybean-Associated Candidate Beneficial Bacteria Through 16S Ribosomal RNA Gene Amplification and Sequencing
2.11. Evaluating Bacterial Isolates as Biostimulants to Enhance Soybean Growth Under Drought Stress
2.11.1. Experiment Setup, Bacterial Preparation, and Design for Drought Stress
2.11.2. Determination of Growth-Related Parameters
(SFW = Shoot Fresh Weight, SDW = Shoot Dry Weight),
(RFW = Root Fresh Weight, RDW = Root Dry Weight).
2.11.3. Determination of Drought Stress Index (DSI) and Chlorophyll Content
3. Results
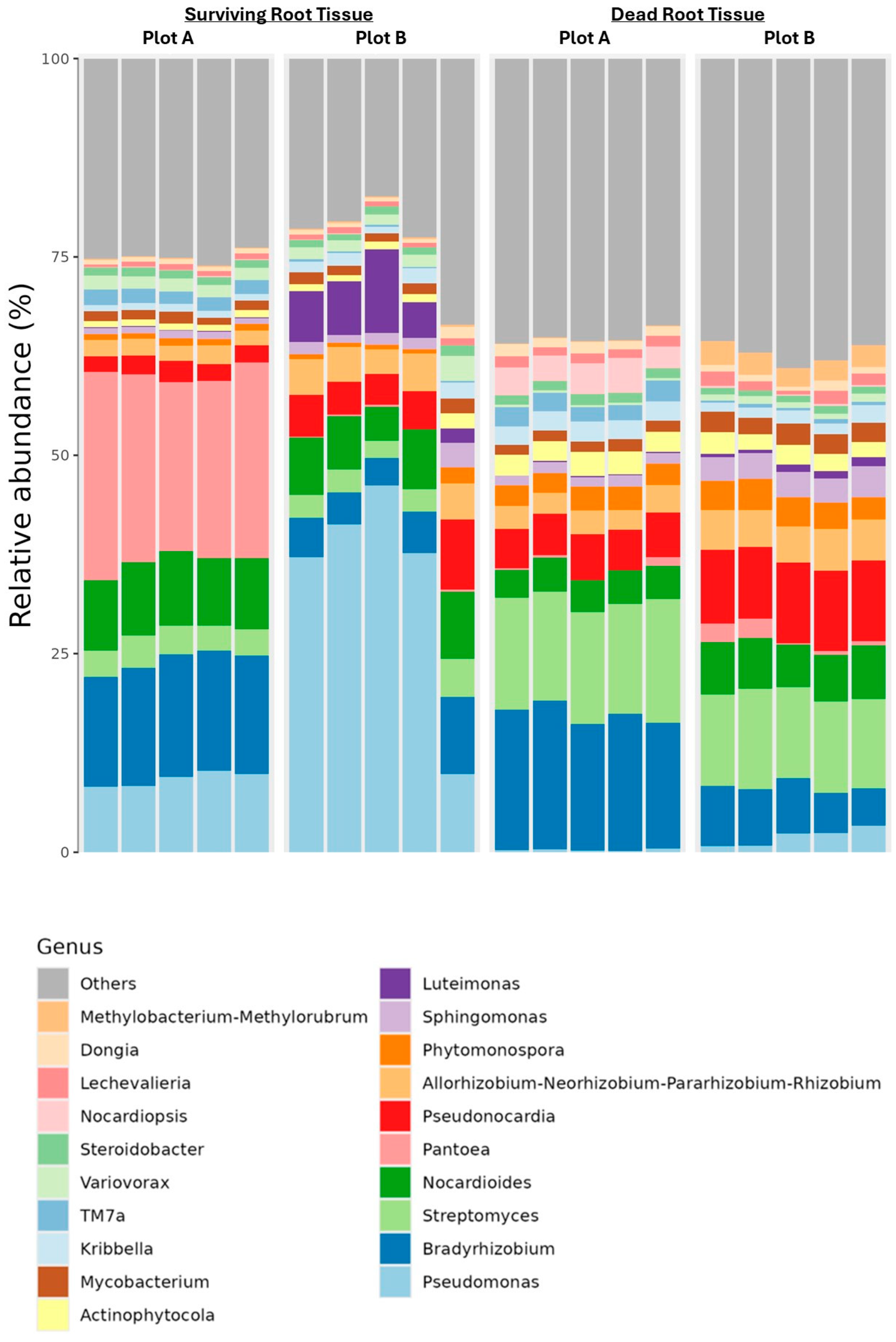
3.1. Alpha-Beta Diversities and Taxa Bar Plots
3.2. Co-Occurrence Networks Analyses
3.3. Enhancement of Soybean Drought Tolerance Through Seed Treatment of Soybean-Associated Bacteria Isolated from a Drought-Conditioned Field
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | abscisic acid |
| ACC | 1aminocyclopropane-1-carboxylate |
| ANOVA | analysis of variance |
| ASV | amplicon sequence variants |
| BLAST | basic local alignment search tool |
| CAS | chrome azurol s |
| CMC | carboxymethyl cellulose |
| C: N | carbon-to-nitrogen |
| ddH2O | double distilled water |
| DNA | deoxyribonucleic acid |
| DSI | drought stress index |
| EPS | exopolysaccharide |
| ET | ethylene |
| HDTMA | hexadecyltrimethylammonium bromide |
| IST | induced systemic tolerance |
| LBA | luria-bertani agar |
| LSU | Louisiana State University |
| mM | millimolar |
| MM9 | minimal media 9 |
| OD | optical density |
| OTUs | operational taxonomic units |
| PCoA | principal coordinates analysis |
| PC | polymerase chain reaction |
| PGP | plant growth-promoting |
| PIPES | piperazine-N,N′-bis(2-ethanesulfonic acid) |
| QIIME | quantitative insights into microbial ecology |
| RAS | root-adhering soil |
| rRNA | ribosomal ribonucleic acid |
| RT | root tissue |
| SPAD | soil plant analysis development |
| TBE | Tris-Borate-EDTA |
| VOCs | volatile organic compounds |
References
- Poudel, S.; Vennam, R.R.; Shrestha, A.; Reddy, K.R.; Wijewardane, N.K.; Reddy, K.N.; Bheemanahalli, R. Resilience of soybean cultivars to drought stress during flowering and early-seed setting stages. Sci. Rep. 2023, 13, 1277. [Google Scholar] [CrossRef]
- Kumar, N.; Mishra, B.K.; Liu, J.; Mohan, B.; Thingujam, D.; Pajerowska-Mukhtar, K.M.; Mukhtar, M.S. Network Biology Analyses and Dynamic Modeling of Gene Regulatory Networks under Drought Stress Reveal Major Transcriptional Regulators in Arabidopsis. Int. J. Mol. Sci. 2023, 24, 7349. [Google Scholar] [CrossRef] [PubMed]
- Ayilara, M.S.; Adeleke, B.S.; Babalola, O.O. Bioprospecting and Challenges of Plant Microbiome Research for Sustainable Agriculture, a Review on Soybean Endophytic Bacteria. Microb. Ecol. 2023, 85, 1113–1135. [Google Scholar] [CrossRef] [PubMed]
- Petrushin, I.S.; Vasilev, I.A.; Markova, Y.A. Drought Tolerance of Legumes: Physiology and the Role of the Microbiome. Curr. Issues Mol. Biol. 2023, 45, 6311–6324. [Google Scholar] [CrossRef] [PubMed]
- Parasar, B.J.; Kashyap, S.; Sharma, I.; Marme, S.; Das, P.; Agarwala, N. Microbe mediated alleviation of drought and heat stress in plants- current understanding and future prospects. Discov. Plants 2024, 1, 26. [Google Scholar] [CrossRef]
- Ali, S.; Tyagi, A.; Mir, R.A.; Rather, I.A.; Anwar, Y.; Mahmoudi, H. Plant beneficial microbiome a boon for improving multiple stress tolerance in plants. Front. Plant Sci. 2023, 14, 1266182. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Mazahar, S.; Chapadgaonkar, S.S.; Giri, P.; Shourie, A. Phyto-microbiome to mitigate abiotic stress in crop plants. Front. Microbiol. 2023, 14, 1210890. [Google Scholar] [CrossRef]
- Timmusk, S.; Abd El-Daim, I.A.; Copolovici, L.; Tanilas, T.; Kannaste, A.; Behers, L.; Nevo, E.; Seisenbaeva, G.; Stenstrom, E.; Niinemets, U. Drought-tolerance of wheat improved by rhizosphere bacteria from harsh environments: Enhanced biomass production and reduced emissions of stress volatiles. PLoS ONE 2014, 9, e96086. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, A.; Mavrodi, O.; Bhowmik, N.; Weller, D.; Thomashow, L.; Mavrodi, D. Bacterial biofilms as an essential component of rhizosphere plant-microbe interactions. Methods Microbiol. 2023, 53, 3–48. [Google Scholar]
- Naylor, D.; DeGraaf, S.; Purdom, E.; Coleman-Derr, D. Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J. 2017, 11, 2691–2704. [Google Scholar] [CrossRef] [PubMed]
- Chieb, M.; Gachomo, E.W. The role of plant growth promoting rhizobacteria in plant drought stress responses. BMC Plant Biol. 2023, 23, 407. [Google Scholar] [CrossRef] [PubMed]
- Mayhood, P.; Mirza, B.S. Soybean Root Nodule and Rhizosphere Microbiome: Distribution of Rhizobial and Nonrhizobial Endophytes. Appl. Environ. Microbiol. 2021, 87, e02884-20. [Google Scholar] [CrossRef]
- Matchado, M.S.; Lauber, M.; Reitmeier, S.; Kacprowski, T.; Baumbach, J.; Haller, D.; List, M. Network analysis methods for studying microbial communities: A mini review. Comput. Struct. Biotechnol. J. 2021, 19, 2687–2698. [Google Scholar] [CrossRef]
- Guo, B.; Zhang, L.; Sun, H.; Gao, M.; Yu, N.; Zhang, Q.; Mou, A.; Liu, Y. Microbial co-occurrence network topological properties link with reactor parameters and reveal importance of low-abundance genera. NPJ Biofilms Microbiomes 2022, 8, 3. [Google Scholar] [CrossRef]
- Kumar, N.; Mukhtar, M.S. Ranking Plant Network Nodes Based on Their Centrality Measures. Entropy 2023, 25, 676. [Google Scholar] [CrossRef]
- Ahmed, H.; Howton, T.C.; Sun, Y.; Weinberger, N.; Belkhadir, Y.; Mukhtar, M.S. Network biology discovers pathogen contact points in host protein-protein interactomes. Nat. Commun. 2018, 9, 2312. [Google Scholar] [CrossRef]
- Apprill, A.; McNally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Robeson, M.S., 2nd; O’Rourke, D.R.; Kaehler, B.D.; Ziemski, M.; Dillon, M.R.; Foster, J.T.; Bokulich, N.A. RESCRIPt: Reproducible sequence taxonomy reference database management. PLoS Comput. Biol. 2021, 17, e1009581. [Google Scholar] [CrossRef]
- Barnett, D.; Arts, I.; Penders, J. microViz: An R package for microbiome data visualization and statistics. J. Open Source Softw. 2021, 6, 3201. [Google Scholar] [CrossRef]
- Bisanz, J.E. qiime2R: Importing QIIME2 Artifacts and Associated Data into R Sessions; Version 0.99; Github: San Francisco, CA, USA, 2018; Volume 13. [Google Scholar]
- McMurdie, P.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer International Publishing: Cham, Switzerland, 2016. [Google Scholar]
- Wen, T.; Xie, P.; Yang, S.; Niu, G.; Liu, X.; Ding, Z.; Xue, C.; Liu, Y.X.; Shen, Q.; Yuan, J. ggClusterNet: An R package for microbiome network analysis and modularity-based multiple network layouts. Imeta 2022, 1, e32. [Google Scholar] [CrossRef]
- Louden, B.C.; Haarmann, D.; Lynne, A.M. Use of Blue Agar CAS Assay for Siderophore Detection. J. Microbiol. Biol. Educ. 2011, 12, 51–53. [Google Scholar] [CrossRef] [PubMed]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- AlAli, H.A.; Khalifa, A.; Almalki, M. Plant Growth-Promoting Bacterium from Non-Agricultural Soil Improves Okra Plant Growth. Agriculture 2022, 12, 873. [Google Scholar] [CrossRef]
- Kayasth, M.; Gera, R.; Dudeja, S.S.; Sharma, P.K.; Kumar, V. Studies on salinization in Haryana soils on free-living nitrogen-fixing bacterial populations and their activity. J. Basic. Microbiol. 2014, 54, 170–179. [Google Scholar] [CrossRef]
- Ramachandran, K.; Srinivasan, V.; Srambikkal, H.; Muthuswamy, A. Phosphate solubilizing bacteria isolated from the rhizosphere soil and its growth promotion on black pepper (Piper nigrum L.) cuttings. In First International Meeting on Microbial Phosphate Solubilization. Developments in Plant and Soil Sciences; Springer: Berlin/Heidelberg, Germany, 2007; Volume 102, pp. 325–331. [Google Scholar]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Saura-Mas, S.; Lloret, F. Leaf and shoot water content and leaf dry matter content of Mediterranean woody species with different post-fire regenerative strategies. Ann. Bot. 2007, 99, 545–554. [Google Scholar] [CrossRef]
- Engelbrecht, B.M.J.; Tyree, M.T.; Kursar, T.A. Visual assessment of wilting as a measure of leaf water potential and seedling drought survival. J. Trop. Ecol. 2007, 23, 497–500. [Google Scholar] [CrossRef]
- Sintaha, M.; Man, C.K.; Yung, W.S.; Duan, S.; Li, M.W.; Lam, H.M. Drought Stress Priming Improved the Drought Tolerance of Soybean. Plants 2022, 11, 2954. [Google Scholar] [CrossRef]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef] [PubMed]
- Shaffique, S.; Imran, M.; Kang, S.M.; Khan, M.A.; Asaf, S.; Kim, W.C.; Lee, I.J. Seed Bio-priming of wheat with a novel bacterial strain to modulate drought stress in Daegu, South Korea. Front. Plant Sci. 2023, 14, 1118941. [Google Scholar] [CrossRef] [PubMed]
- Zboralski, A.; Filion, M. Pseudomonas spp. can help plants face climate change. Front. Microbiol. 2023, 14, 1198131. [Google Scholar] [CrossRef]
- Vaishnav, A.; Kumari, S.; Jain, S.; Varma, A.; Choudhary, D.K. Putative bacterial volatile-mediated growth in soybean (Glycine max L. Merrill) and expression of induced proteins under salt stress. J. Appl. Microbiol. 2015, 119, 539–551. [Google Scholar] [CrossRef] [PubMed]
- Zboralski, A.; Filion, M. Genetic factors involved in rhizosphere colonization by phytobeneficial Pseudomonas spp. Comput. Struct. Biotechnol. J. 2020, 18, 3539–3554. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Nishu, S.D.; No, J.H.; Lee, T.K. Transcriptional Response and Plant Growth Promoting Activity of Pseudomonas fluorescens DR397 under Drought Stress Conditions. Microbiol. Spectr. 2022, 10, e0097922. [Google Scholar] [CrossRef]
- Kang, S.-M.; Radhakrishnan, R.; Khan, A.L.; Kim, M.-J.; Park, J.-M.; Kim, B.-R.; Shin, D.-H.; Lee, I.-J. Gibberellin secreting rhizobacterium, Pseudomonas putida H-2-3 modulates the hormonal and stress physiology of soybean to improve the plant growth under saline and drought conditions. Plant Physiol. Biochem. 2014, 84, 115–124. [Google Scholar] [CrossRef]
- Calderon, R.B.; Ontoy, J.C.; Barpharga, I.; Ham, J.H. Characterization of the growth promotion and defense enhancement of soybean plants driven by seed treatment of multiple soybean-associated beneficial bacteria. bioRxiv 2024. [Google Scholar] [CrossRef]
- Amellal, N.; Burtin, G.; Bartoli, F.; Heulin, T. Colonization of wheat roots by an exopolysaccharide-producing pantoea agglomerans strain and its effect on rhizosphere soil aggregation. Appl. Environ. Microbiol. 1998, 64, 3740–3747. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Xin, K.; Liu, H.; Cheng, J.; Shen, X.; Wang, Y.; Zhang, L. Pantoea alhagi, a novel endophytic bacterium with ability to improve growth and drought tolerance in wheat. Sci. Rep. 2017, 7, 41564. [Google Scholar] [CrossRef]
- Sun, L.; Yang, Y.; Wang, R.; Li, S.; Qiu, Y.; Lei, P.; Gao, J.; Xu, H.; Zhang, F.; Lv, Y. Effects of exopolysaccharide derived from Pantoea alhagi NX-11 on drought resistance of rice and its efficient fermentation preparation. Int. J. Biol. Macromol. 2020, 162, 946–955. [Google Scholar] [CrossRef]
- Wang, S.; Wang, J.; Zhou, Y.; Huang, Y.; Tang, X. Isolation, Classification, and Growth-Promoting Effects of Pantoea sp. YSD J2 from the Aboveground Leaves of Cyperus Esculentus L. var. sativus. Curr. Microbiol. 2022, 79, 66. [Google Scholar] [CrossRef]
- Mayfield, C.I.; Williams, S.T.; Ruddick, S.M.; Hatfield, H.L. Studies on the ecology of actinomycetes in soil IV. Observations on the form and growth of streptomycetes in soil. Soil. Biol. Biochem. 1972, 4, 79–91. [Google Scholar] [CrossRef]
- Shepherdson, E.M.; Baglio, C.R.; Elliot, M.A. Streptomyces behavior and competition in the natural environment. Curr. Opin. Microbiol. 2023, 71, 102257. [Google Scholar] [CrossRef]
- Arun, B.; Gopinath, B.; Sharma, S. Plant growth promoting potential of bacteria isolated on N free media from rhizosphere of Cassia occidentalis. World J. Microbiol. Biotechnol. 2012, 28, 2849–2857. [Google Scholar] [CrossRef]
- Lafi, F.F.; Ramirez-Prado, J.S.; Alam, I.; Bajic, V.B.; Hirt, H.; Saad, M.M. Draft Genome Sequence of Plant Growth-Promoting Micrococcus luteus Strain K39 Isolated from Cyperus conglomeratus in Saudi Arabia. Genome Announc. 2017, 5, e01520-16. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Zhang, M.Y.; Wang, W.X.; Manikprabhu, D.; Salam, N.; Zhang, T.Y.; Wu, Y.Y.; Li, W.J.; Zhang, Y.X. Luteimonas notoginsengisoli sp. nov. isolated from rhizosphere. Int. J. Syst. Evol. Microbiol. 2016, 66, 946–950. [Google Scholar] [CrossRef]
- Belimov, A.A.; Dodd, I.C.; Hontzeas, N.; Theobald, J.C.; Safronova, V.I.; Davies, W.J. Rhizosphere bacteria containing 1-aminocyclopropane-1-carboxylate deaminase increase yield of plants grown in drying soil via both local and systemic hormone signalling. New Phytol. 2009, 181, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Martins, S.J.; Rocha, G.A.; de Melo, H.C.; de Castro Georg, R.; Ulhoa, C.J.; de Campos Dianese, E.; Oshiquiri, L.H.; da Cunha, M.G.; da Rocha, M.R.; de Araujo, L.G.; et al. Plant-associated bacteria mitigate drought stress in soybean. Environ. Sci. Pollut. Res. Int. 2018, 25, 13676–13686. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Mishra, S.; Dixit, V.; Kumar, M.; Agarwal, L.; Chauhan, P.S.; Nautiyal, C.S. Synergistic effect of Pseudomonas putida and Bacillus amyloliquefaciens ameliorates drought stress in chickpea (Cicer arietinum L.). Plant Signal Behav. 2016, 11, e1071004. [Google Scholar] [CrossRef]
- Liu, J.; Fimognari, L.; de Almeida, J.; Jensen, C.N.G.; Compant, S.; Oliveira, T.; Baelum, J.; Pastar, M.; Sessitsch, A.; Moelbak, L.; et al. Effect of Bacillus paralicheniformis on soybean (Glycine max) roots colonization, nutrient uptake and water use efficiency under drought stress. J. Agron. Crop Sci. 2023, 209, 547–565. [Google Scholar] [CrossRef]
- Shaffique, S.; Imran, M.; Adhikari, A.; Aaqil Khan, M.; Rahim, W.; Alomrani, S.O.; Yun, B.-W.; Kang, S.-M.; Lee, I.-J. A newly isolated Bacillus pumilus strain SH-9 modulates response to drought stress in soybean via endogenous phytohormones and gene expression (Daegu, South Korea). Plant Stress. 2023, 10, 100279. [Google Scholar] [CrossRef]
- de Lima, B.C.; Moro, A.L.; Santos, A.C.P.; Bonifacio, A.; Araujo, A.S.F.; de Araujo, F.F. Bacillus subtilis ameliorates water stress tolerance in maize and common bean. J. Plant Interact. 2019, 14, 432–439. [Google Scholar] [CrossRef]
- Kour, D.; Rana, K.L.; Yadav, A.N.; Yadav, N.; Kumar, V.; Kumar, A.; Sayyed, R.Z.; Hesham, A.E.-L.; Dhaliwal, H.S.; Saxena, A.K. Drought-Tolerant Phosphorus-Solubilizing Microbes: Biodiversity and Biotechnological Applications for Alleviation of Drought Stress in Plants. In Plant Growth Promoting Rhizobacteria for Sustainable Stress Management: Volume 1: Rhizobacteria in Abiotic Stress Management; Sayyed, R.Z., Arora, N.K., Reddy, M.S., Eds.; Springer: Singapore, 2019; pp. 255–308. [Google Scholar]
- Yadav, A.N.; Kumar, V.; Dhaliwal, H.S.; Prasad, R.; Saxena, A.K. Chapter 15—Microbiome in Crops: Diversity, Distribution, and Potential Role in Crop Improvement. In Crop Improvement through Microbial Biotechnology; Prasad, R., Gill, S.S., Tuteja, N., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 305–332. [Google Scholar]
- Shankar, A.; Prasad, V. Potential of desiccation-tolerant plant growth-promoting rhizobacteria in growth augmentation of wheat (Triticum aestivum L.) under drought stress. Front. Microbiol. 2023, 14, 1017167. [Google Scholar] [CrossRef]
- Kumar, S.; Diksha; Sindhu, S.S.; Kumar, R. Biofertilizers: An ecofriendly technology for nutrient recycling and environmental sustainability. Curr. Res. Microb. Sci. 2022, 3, 100094. [Google Scholar] [PubMed]




| Endophytic Network | Rhizospheric Network | |||||||
|---|---|---|---|---|---|---|---|---|
| Centrality Features | Surviving (Plot A) | Non-Surviving (Plot A) | Surviving (Plot B) | Non-Surviving (Plot B) | Surviving (Plot A) | Non-Surviving (Plot A | Surviving (Plot B) | Non-Surviving (Plot B) |
| Nodes | 476 | 648 | 327 | 406 | 1094 | 1011 | 872 | 861 |
| Edges | 6449 | 10208 | 3840 | 5474 | 38632 | 32640 | 21395 | 20119 |
| Positive Edges | 6320 | 10050 | 3765 | 5426 | 38590 | 32543 | 21352 | 19992 |
| Negative Edges | 129 | 158 | 75 | 48 | 42 | 97 | 43 | 127 |
| Number of Clusters | 73 | 92 | 52 | 61 | 90 | 92 | 73 | 93 |
| Connectance (Edge Density) | 0.05704556 | 0.048695785 | 0.07204368 | 0.06658152 | 0.06461595 | 0.063930429 | 0.0563388 | 0.054341896 |
| Average degree (Average K) | 27.0966387 | 31.50617284 | 23.4862385 | 26.9655172 | 70.6252285 | 64.56973294 | 49.071101 | 46.7340302 |
| Average Path Length | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Diameter | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Mean Clustering Coefficient (Average CC) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Centralization Degree | 0.06927023 | 0.064132654 | 0.1334778 | 0.06181354 | 0.08817454 | 0.095475512 | 0.0791377 | 0.0712395 |
| RM (Relative Modularity) | 5.2693558 | 6.040679146 | 3.17505499 | 4.79215729 | 12.0494888 | 10.78020977 | 8.7625972 | 8.561277694 |
| Bacterial Isolates | Mucoidal Appearance | Siderophore Production | Nitrogen Fixation | Phosphate Solubilization |
|---|---|---|---|---|
| DRS1 | +++ | +++ | +++ | +++ |
| DRS2 | ++ | +++ | + | +++ |
| DRS3 | +++ | +++ | + | +++ |
| DRS4 | +++ | +++ | +++ | +++ |
| DRS5 | +++ | +++ | +++ | + |
| DES1 | +++ | +++ | +++ | + |
| DES2 | +++ | +++ | +++ | − |
| DES3 | +++ | +++ | +++ | ++ |
| DES4 | +++ | +++ | +++ | − |
| DES5 | ++ | + | + | − |
| Isolate | Gram +/− | Probable Identity with Highest Homology Match | % Similarity |
|---|---|---|---|
| DRS1 | − | Pseudomonas lini | 99.18 |
| DRS2 | − | Acinetobacter pittii | 99.73 |
| DRS3 | − | Pseudomonas sp. | 98.63 |
| DRS4 | − | Pseudomonas sp. | 96.37 |
| DRS5 | − | Pseudomonas sp. | 99.86 |
| DES1 | − | Pseudomonas sp. | 97.75 |
| DES2 | − | Enterobacter ludwigii | 85.82 |
| DES3 | − | Pseudomonas sp. | 99.33 |
| DES4 | * | * | No similarity found |
| DES5 | − | Stenotrophomonas sp. | 96.8 |
| Bacterial Seed Treatments | Root Length (cm) | Shoot Length (cm) | Root Fresh Weight (g) | Shoot Fresh Weight (g) | Root dry Weight (g) | Shoot Dry Weight (g) | Root Water Content (%) | Shoot Water Content (%) |
|---|---|---|---|---|---|---|---|---|
| DRS1 | 20.63 cd | 24.30 ab | 0.150 bcde | 0.77 bcd | 0.051 cde | 0.253 bcd | 66.37 bcd | 66.69 bc |
| DRS2 | 25.58 a | 26.01 a | 0.188 ab | 1.16 a | 0.072 ab | 0.348 a | 60.82 cde | 69.48 abc |
| DRS3 | 23.49 ab | 23.67 abc | 0.194 a | 0.71 cd | 0.038 e | 0.218 cde | 79.87 a | 68.04 abc |
| DRS4 | 22.80 abc | 24.16 ab | 0.204 a | 0.97 ab | 0.056 bcd | 0.298 ab | 72.23 ab | 68.81 abc |
| DRS5 | 19.86 d | 23.74 abc | 0.172 abcd | 0.69 d | 0.045 cde | 0.212 cde | 74.10 ab | 67.75 abc |
| DES1 | 19.06 d | 24.70 ab | 0.139 cde | 0.58 d | 0.052 cde | 0.204 cde | 63.02 cde | 64.14 bc |
| DES2 | 20.46 cd | 20.95 d | 0.134 de | 0.61 d | 0.040 de | 0.226 cde | 68.79 bc | 61.38 c |
| DES3 | 24.74 a | 24.19 ab | 0.181 abc | 0.94 abc | 0.079 a | 0.262 bc | 55.52 e | 71.65 ab |
| DES4 | 20.06 cd | 23.47 bc | 0.148 bcde | 0.58 d | 0.037 e | 0.180 e | 74.20 ab | 67.96 abc |
| DES5 | 21.15 bcd | 24.77 ab | 0.108 e | 0.77 bcd | 0.039 de | 0.221 cde | 62.53 cde | 70.16 ab |
| CMC | 18.81 d | 23.80 abc | 0.146 bcde | 1.01 ab | 0.060 bc | 0.246 bcde | 58.69 de | 75.64 a |
| Utr | 18.76 d | 21.69 cd | 0.118 e | 0.65 d | 0.045 cde | 0.188 de | 60.01 cde | 70.51 ab |
| Treatment effect (ANOVA) | p < 0.0001 | |||||||
| Bacterial Seed Treatments | Drought Stress Index (DSI) | SPAD Reading |
|---|---|---|
| DRS1 | 6.0 a | 37.8 a |
| DRS2 | 3.0 b | 39.6 a |
| DRS3 | 4.4 ab | 38.4 a |
| DRS4 | 5.6 a | 38.4 a |
| DRS5 | 5.4 ab | 39.6 a |
| DES1 | 4.8 ab | 35.2 a |
| DES2 | 4.2 ab | 35.7 a |
| DES3 | 5.6 a | 36.4 a |
| DES4 | 5.8 a | 35.7 a |
| DES5 | 6.2 a | 36.9 a |
| CMC | 6.2 a | 38.7 a |
| Utr | 6.6 a | 36.0 a |
| Treatment effect (ANOVA) | p < 0.0001 | p = 0.045 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gouli, S.; Majeed, A.; Liu, J.; Moseley, D.; Mukhtar, M.S.; Ham, J.H. Microbiome Structures and Beneficial Bacteria in Soybean Roots Under Field Conditions of Prolonged High Temperatures and Drought Stress. Microorganisms 2024, 12, 2630. https://doi.org/10.3390/microorganisms12122630
Gouli S, Majeed A, Liu J, Moseley D, Mukhtar MS, Ham JH. Microbiome Structures and Beneficial Bacteria in Soybean Roots Under Field Conditions of Prolonged High Temperatures and Drought Stress. Microorganisms. 2024; 12(12):2630. https://doi.org/10.3390/microorganisms12122630
Chicago/Turabian StyleGouli, Sandeep, Aqsa Majeed, Jinbao Liu, David Moseley, M. Shahid Mukhtar, and Jong Hyun Ham. 2024. "Microbiome Structures and Beneficial Bacteria in Soybean Roots Under Field Conditions of Prolonged High Temperatures and Drought Stress" Microorganisms 12, no. 12: 2630. https://doi.org/10.3390/microorganisms12122630
APA StyleGouli, S., Majeed, A., Liu, J., Moseley, D., Mukhtar, M. S., & Ham, J. H. (2024). Microbiome Structures and Beneficial Bacteria in Soybean Roots Under Field Conditions of Prolonged High Temperatures and Drought Stress. Microorganisms, 12(12), 2630. https://doi.org/10.3390/microorganisms12122630






