Antimicrobial Susceptibility Profiles of Salmonella spp. Isolates from Clinical Cases of Waterfowl in Hungary Between 2022 and 2023
Abstract
1. Introduction
2. Materials and Methods
2.1. Origin of the Strains and Human Data
2.2. Determination of the Minimum Inhibitory Concentration (MIC)
2.3. Statistical Analysis
3. Results
3.1. The Distribution and Regional Origin of the Samples Received
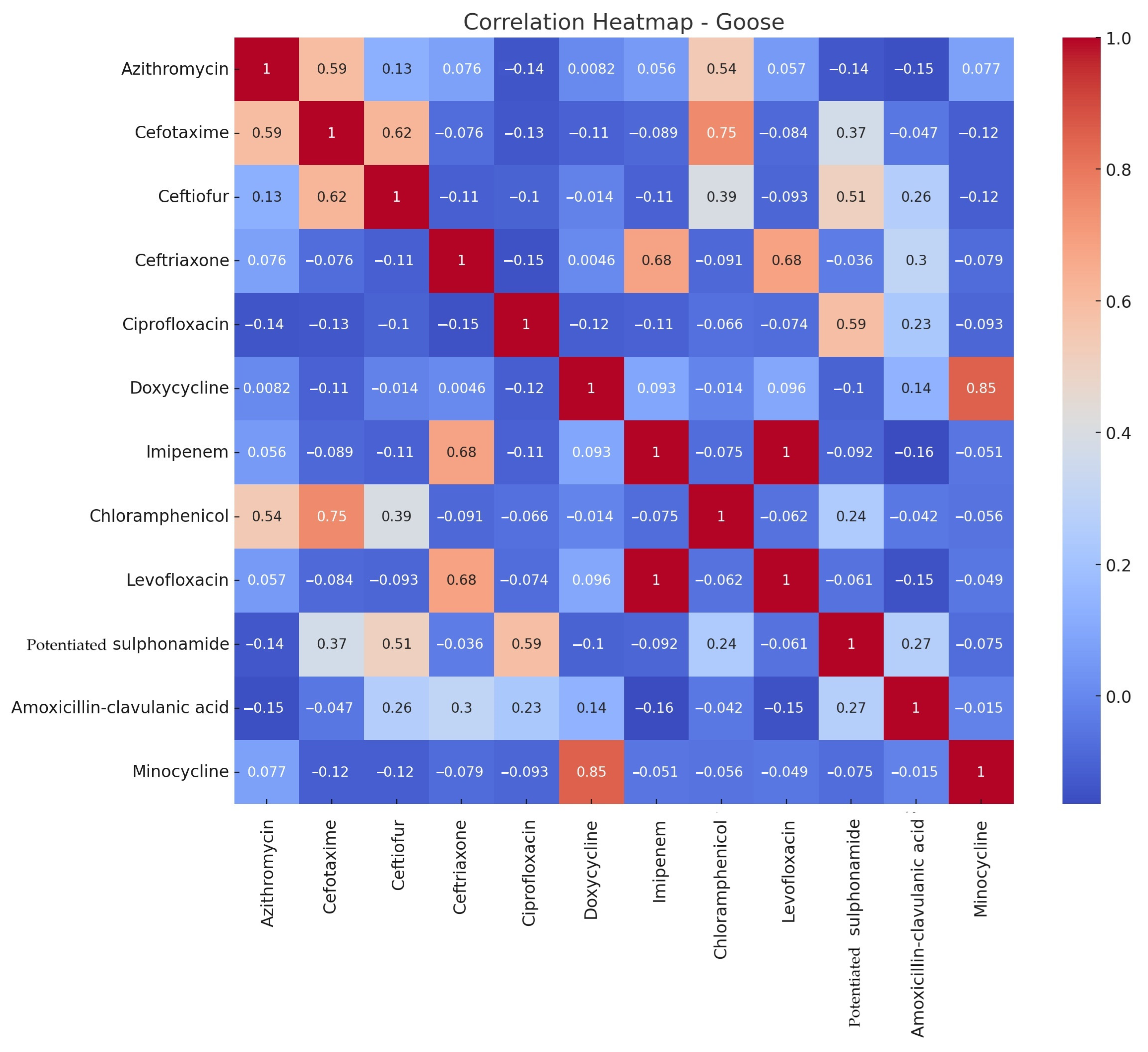
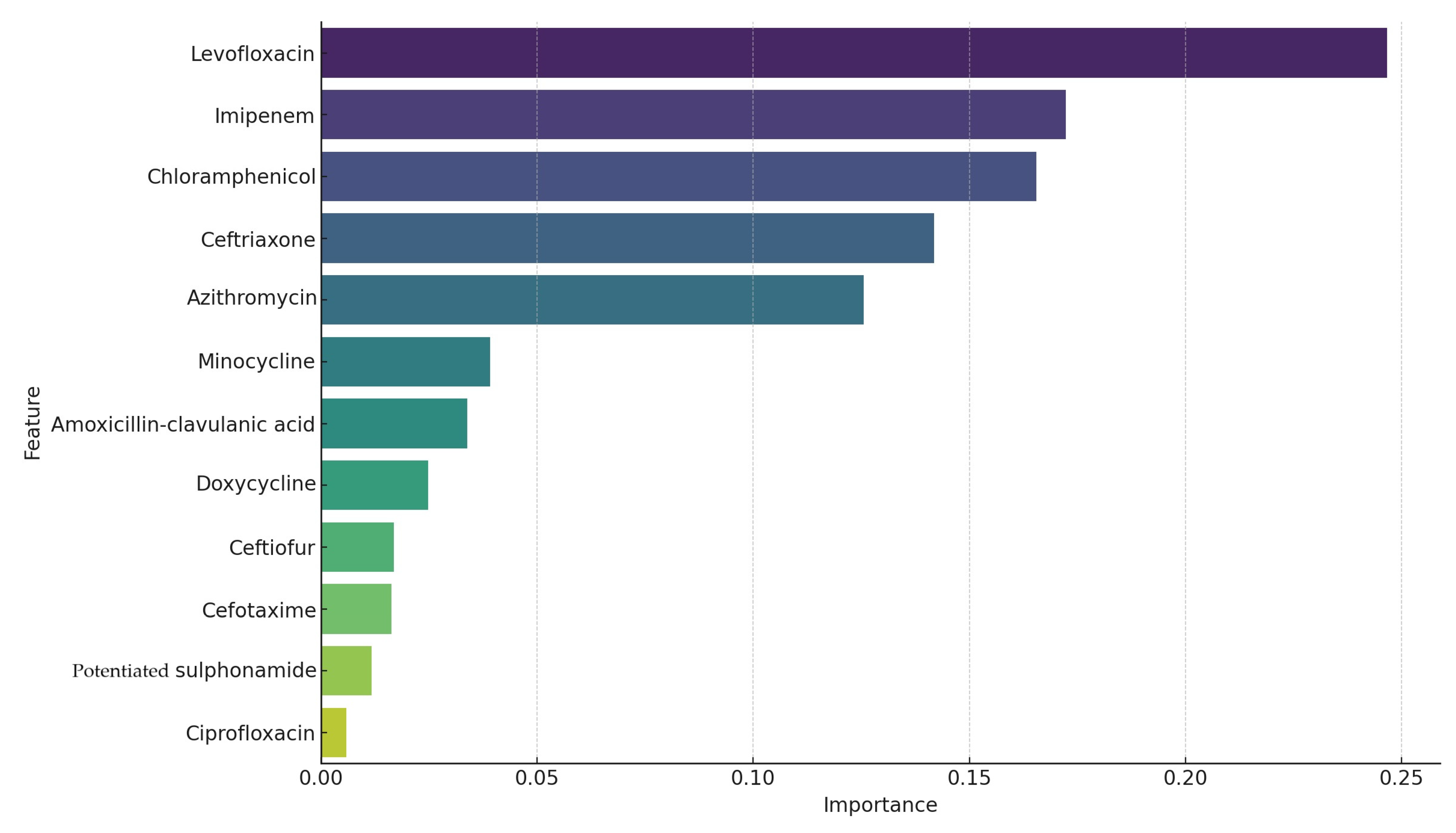
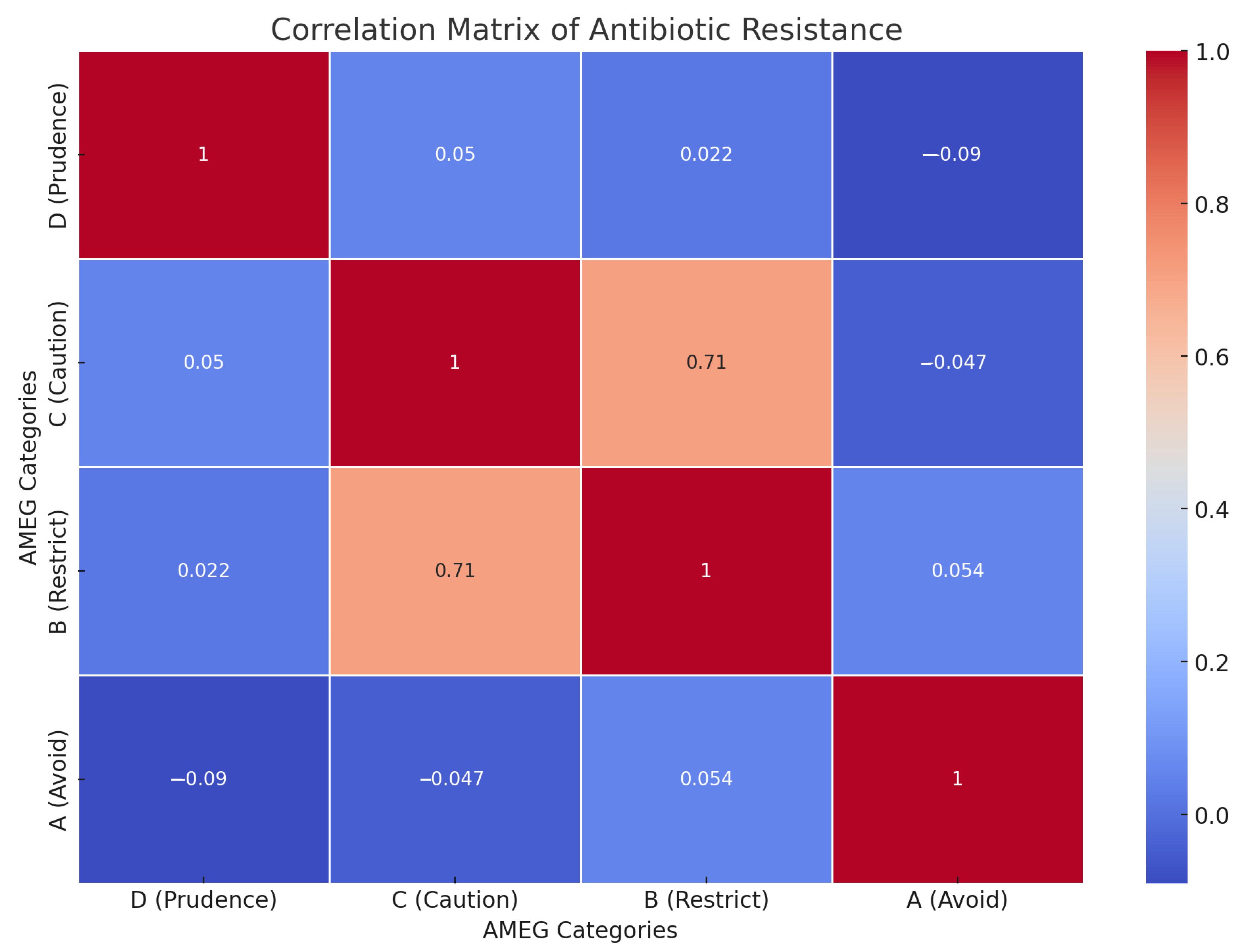
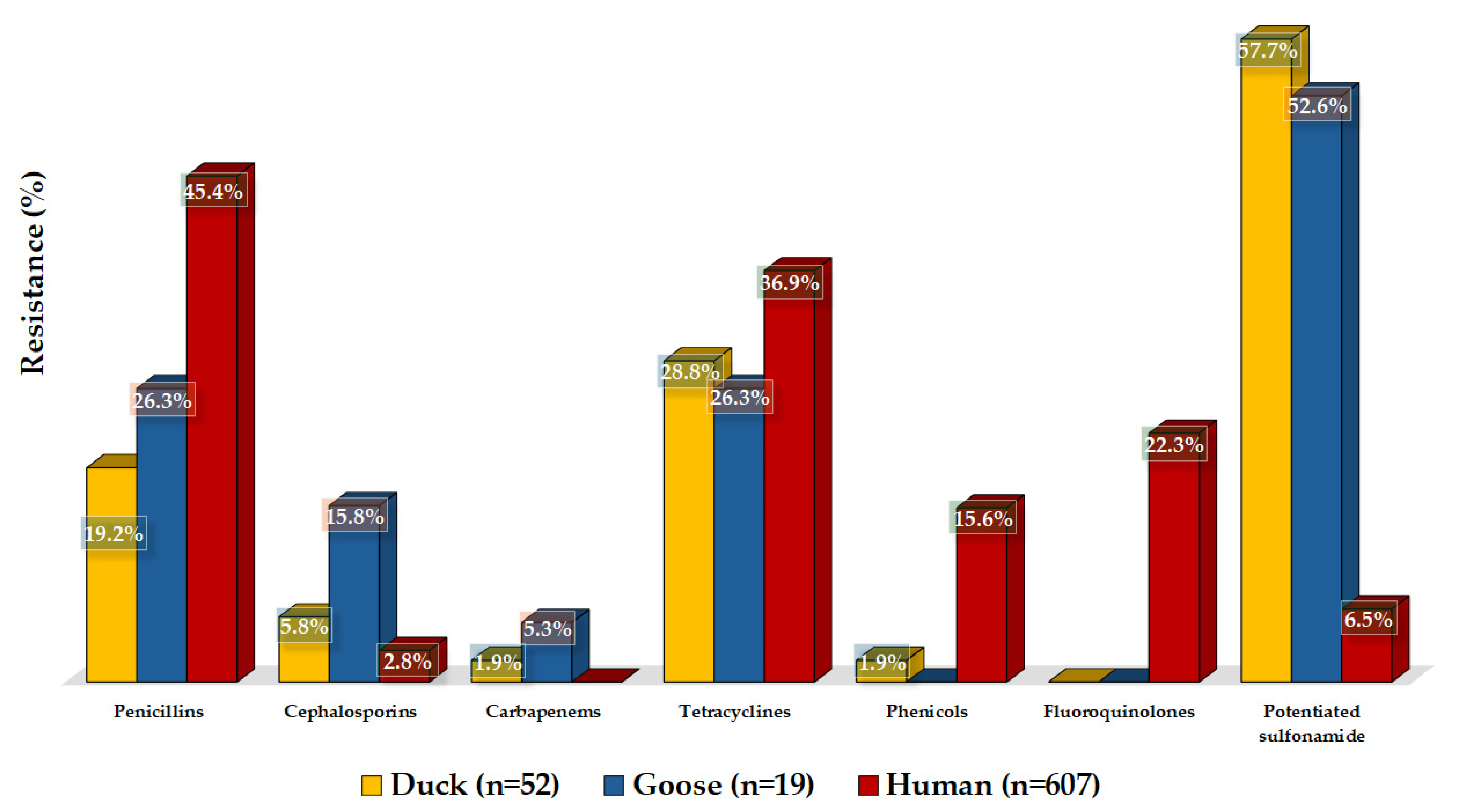
3.2. Antimicrobial Susceptibility Testing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, N.; Cheng, Z.; Zhang, X.; Lv, C.; Guo, C.; Liu, H.; Dong, K.; Zhang, Y.; Liu, C.; Chang, Y.-F.; et al. Global Antimicrobial Resistance: A System-Wide Comprehensive Investigation Using the Global One Health Index. Infect. Dis. Poverty 2022, 11, 92. [Google Scholar] [CrossRef] [PubMed]
- Bhargav, A.; Gupta, S.; Seth, S.; James, S.; Fatima, F.; Chaurasia, P.; Ramachandran, S. Knowledgebase of Potential Multifaceted Solutions to Antimicrobial Resistance. Comput. Biol. Chem. 2022, 101, 107772. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, B.A.; Bhat, B.A.; Mir, M.A. Antimicrobial Resistance: New Insights and Therapeutic Implications. Appl. Microbiol. Biotechnol. 2022, 106, 6427–6440. [Google Scholar] [CrossRef] [PubMed]
- Stanley, D.; Batacan, R.; Bajagai, Y.S. Rapid Growth of Antimicrobial Resistance: The Role of Agriculture in the Problem and the Solutions. Appl. Microbiol. Biotechnol. 2022, 106, 6953–6962. [Google Scholar] [CrossRef]
- Singhal, T. Antimicrobial Resistance: The “Other” Pandemic! Indian J. Pediatr. 2022, 89, 600–606. [Google Scholar] [CrossRef]
- Chattu, V.K.; Singh, B.; Pattanshetty, S.; Reddy, S. Access to Medicines through Global Health Diplomacy. Health Promot. Perspect. 2023, 13, 40–46. [Google Scholar] [CrossRef]
- Bjerke, L. Antibiotic Geographies and Access to Medicines: Tracing the Role of India’s Pharmaceutical Industry in Global Trade. Soc. Sci. Med. 2022, 312, 115386. [Google Scholar] [CrossRef]
- Despotovic, M.; de Nies, L.; Busi, S.B.; Wilmes, P. Reservoirs of Antimicrobial Resistance in the Context of One Health. Curr. Opin. Microbiol. 2023, 73, 102291. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; Flach, C.-F. Antibiotic Resistance in the Environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Barathe, P.; Kaur, K.; Reddy, S.; Shriram, V.; Kumar, V. Antibiotic Pollution and Associated Antimicrobial Resistance in the Environment. J. Hazard. Mater. Lett. 2024, 5, 100105. [Google Scholar] [CrossRef]
- Benmazouz, I.; Kövér, L.; Kardos, G. The Rise of Antimicrobial Resistance in Wild Birds: Potential AMR Sources and Wild Birds as AMR Reservoirs and Disseminators: Literature Review. Hung. Vet. J. 2024, 146, 91–105. [Google Scholar] [CrossRef]
- Yassin, A.K.; Gong, J.; Kelly, P.; Lu, G.; Guardabassi, L.; Wei, L.; Han, X.; Qiu, H.; Price, S.; Cheng, D.; et al. Antimicrobial Resistance in Clinical Escherichia coli Isolates from Poultry and Livestock, China. PLoS ONE 2017, 12, e0185326. [Google Scholar] [CrossRef] [PubMed]
- European Medicines Agency. Categorisation of Antibiotics in the European Union. EMA/CVMP/CHMP/682198/2017. Eur. Med. Agence 2020, 31, 1–73. [Google Scholar]
- Mag, P.; Németh, K.; Somogyi, Z.; Jerzsele, Á. Antibacterial therapy based on pharmacokinetic/ pharmacodynamic models in small animal medicine-1. Literature review. Hung. Vet. J. 2023, 145, 419–438. [Google Scholar] [CrossRef]
- Cha, S.-Y.; Kang, M.; Yoon, R.-H.; Park, C.-K.; Moon, O.-K.; Jang, H.-K. Prevalence and Antimicrobial Susceptibility of Salmonella Isolates in Pekin Ducks from South Korea. Comp. Immunol. Microbiol. Infect. Dis. 2013, 36, 473–479. [Google Scholar] [CrossRef]
- Sebők, C.; Márton, R.A.; Meckei, M.; Neogrády, Z.; Mátis, G. Antimicrobial Peptides as New Tools to Combat Infectious Diseases. Hung. Vet. J. 2024, 146, 181–191. [Google Scholar]
- Kovács, L.; Nagy, D.; Könyves, L.; Jerzsele, Á.; Kerek, Á. Antimicrobial Properties of Essential Oils—Animal Health Aspects. Hung. Vet. J. 2023, 145, 497–510. [Google Scholar]
- Kerek, Á.; Szabó, Á.; Dobra, P.F.; Bárdos, K.; Ózsvári, L.; Fehérvári, P.; Bata, Z.; Molnár-Nagy, V.; Jerzsele, Á. Determining the In Vivo Efficacy of Plant-Based and Probiotic-Based Antibiotic Alternatives against Mixed Infection with Salmonella enterica and Escherichia coli in Domestic Chickens. Vet. Sci. 2023, 10, 706. [Google Scholar] [CrossRef]
- Jerzsele, Á.; Somogyi, Z.; Szalai, M.; Kovács, D. Effects of Fermented Wheat Germ Extract on Artificial Salmonella Typhimurium Infection in Broiler Chickens. Hung. Vet. J. 2020, 142, 77–85. [Google Scholar]
- Olasz, Á.; Jerzsele, Á.; Balta, L.; Dobra, P.F.; Kerek, Á. In Vivo Efficacy of Different Extracts of Propolis in Broiler Salmonellosis. Hung. Vet. J. 2023, 145, 461–475. [Google Scholar]
- Kerek, Á.; Csanády, P.; Jerzsele, Á. Antibacterial Efficiency of Propolis—Part 1. Hung. Vet. J. 2022, 144, 285–298. [Google Scholar]
- Kerek, Á.; Csanády, P.; Tuska-Szalay, B.; Kovács, L.; Jerzsele, Á. In Vitro Efficacy of Hungarian Propolis against Bacteria, Yeast, and Trichomonas gallinae Isolated from Pigeons—A Possible Antibiotic Alternative? Resources 2023, 12, 101. [Google Scholar] [CrossRef]
- Kovács, D.; Palkovicsné Pézsa, N.; Farkas, O.; Jerzsele, Á. Usage of Antibiotic Alternatives in Pig Farming: Literature Review. Hung. Vet. J. 2021, 143, 281–282. [Google Scholar]
- Essősy, M.; Fodor, I.; Ihnáth, Z.; Karancsi, Z.; Kovács, D.; Szalai, K.V.; Szentmiklósi, D.; Jerzsele, Á. The Possibilities of Antibiotic-Free Broiler-Hen Fattening, with Special Reference to the Use of Pre- and Probiotics. Hung. Vet. J. 2020, 142, 397–407. [Google Scholar]
- KSH Baromfiállomány (19.1.1.29.). Available online: https://www.ksh.hu/stadat_files/mez/hu/mez0029.html (accessed on 18 May 2023).
- Xu, Z.; Wang, M.; Zhou, C.; Gu, G.; Liang, J.; Hou, X.; Wang, M.; Wei, P. Prevalence and Antimicrobial Resistance of Retail-Meat-Borne Salmonella in Southern China during the Years 2009–2016: The Diversity of Contamination and the Resistance Evolution of Multidrug-Resistant Isolates. Int. J. Food Microbiol. 2020, 333, 108790. [Google Scholar] [CrossRef]
- Shivaprasad, H.L. Fowl Typhoid and Pullorum Disease. Rev. Sci. Tech. 2000, 19, 405–424. [Google Scholar] [CrossRef]
- Newell, D.G.; Koopmans, M.; Verhoef, L.; Duizer, E.; Aidara-Kane, A.; Sprong, H.; Opsteegh, M.; Langelaar, M.; Threfall, J.; Scheutz, F.; et al. Food-Borne Diseases—The Challenges of 20 Years Ago Still Persist While New Ones Continue to Emerge. Int. J. Food Microbiol. 2010, 139 (Suppl. S1), S3–S15. [Google Scholar] [CrossRef]
- Gal-Mor, O.; Boyle, E.C.; Grassl, G.A. Same Species, Different Diseases: How and Why Typhoidal and Non-Typhoidal Salmonella enterica Serovars Differ. Front. Microbiol. 2014, 5, 391. [Google Scholar] [CrossRef]
- Tauxe, R.V.; Doyle, M.P.; Kuchenmüller, T.; Schlundt, J.; Stein, C.E. Evolving Public Health Approaches to the Global Challenge of Foodborne Infections. Int. J. Food Microbiol. 2010, 139 (Suppl. S1), S16–S28. [Google Scholar] [CrossRef]
- Authority (EFSA), E.F.S.; European Centre for Disease Prevention and Control (ECDC). The European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2020/2021. EFSA J. 2023, 21, e07867. [Google Scholar] [CrossRef]
- ISO 6579-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella spp. ISO: Geneva, Switzerland, 2017. Available online: https://www.iso.org/standard/56712.html (accessed on 22 November 2024).
- ISO 16140-7:2024; Microbiology of the Food Chain—Method Validation—Part 7: Protocol for the Validation of Identification Methods of Microorganisms. ISO: Geneva, Switzerland, 2024. Available online: https://www.iso.org/standard/79100.html (accessed on 22 November 2024).
- Dingle, T.C.; Butler-Wu, S.M. Maldi-Tof Mass Spectrometry for Microorganism Identification. Clin. Lab. Med. 2013, 33, 589–609. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; Volume CLSI standards M07. [Google Scholar]
- M100 Ed34; Performance Standards for Antimicrobial Susceptibility Testing. 34th ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA. Available online: https://clsi.org/standards/products/microbiology/documents/m100/ (accessed on 31 October 2024).
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria; European Environment Agency: Copenhagen, Denmark, 2020. [Google Scholar]
- Iwamoto, M.; Reynolds, J.; Karp, B.E.; Tate, H.; Fedorka-Cray, P.J.; Plumblee, J.R.; Hoekstra, R.M.; Whichard, J.M.; Mahon, B.E. Ceftriaxone-Resistant Nontyphoidal Salmonella from Humans, Retail Meats, and Food Animals in the United States, 1996–2013. Foodborne Pathog. Dis. 2017, 14, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Elkenany, R.M.; Eladl, A.H.; El-Shafei, R.A. Genetic Characterisation of Class 1 Integrons among Multidrug-Resistant Salmonella Serotypes in Broiler Chicken Farms. J. Glob. Antimicrob. Resist. 2018, 14, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Gebreyes, W.A.; Thakur, S. Multidrug-Resistant Salmonella enterica Serovar Muenchen from Pigs and Humans and Potential Interserovar Transfer of Antimicrobial Resistance. Antimicrob. Agents Chemother. 2005, 49, 503–511. [Google Scholar] [CrossRef]
- Global Animal Health Market Value 2002–2021. Available online: https://www.statista.com/statistics/260185/global-animal-health-market/ (accessed on 28 July 2024).
- Mehdi, Y.; Létourneau-Montminy, M.-P.; Gaucher, M.-L.; Chorfi, Y.; Suresh, G.; Rouissi, T.; Brar, S.K.; Côté, C.; Ramirez, A.A.; Godbout, S. Use of Antibiotics in Broiler Production: Global Impacts and Alternatives. Anim. Nutr. 2018, 4, 170–178. [Google Scholar] [CrossRef]
- Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 28 July 2024).
- Eid, H.M.; Algammal, A.M.; Elfeil, W.K.; Youssef, F.M.; Harb, S.M.; Abd-Allah, E.M. Prevalence, Molecular Typing, and Antimicrobial Resistance of Bacterial Pathogens Isolated from Ducks. Vet. World 2019, 12, 677–683. [Google Scholar] [CrossRef]
- Jamali, H.; Radmehr, B.; Ismail, S. Prevalence and Antimicrobial Resistance of Listeria, Salmonella, and Yersinia Species Isolates in Ducks and Geese. Poult. Sci. 2014, 93, 1023–1030. [Google Scholar] [CrossRef]
- Cao, Z.Z.; Xu, J.W.; Gao, M.; Li, X.S.; Zhai, Y.J.; Yu, K.; Wan, M.; Luan, X.H. Prevalence and Antimicrobial Resistance of Salmonella isolates from Goose Farms in Northeast China. Iran. J. Vet. Res. 2020, 21, 287–293. [Google Scholar]
- Guan, Y.; Li, Y.; Li, J.; Yang, Z.; Zhu, D.; Jia, R.; Liu, M.; Wang, M.; Chen, S.; Yang, Q.; et al. Phenotypic and Genotypic Characterization of Antimicrobial Resistance Profiles in Salmonella Isolated from Waterfowl in 2002–2005 and 2018–2020 in Sichuan, China. Front. Microbiol. 2022, 13, 987613. [Google Scholar] [CrossRef]
- Kerek, Á.; Török, B.; Laczkó, L.; Somogyi, Z.; Kardos, G.; Bányai, K.; Kaszab, E.; Bali, K.; Jerzsele, Á. In Vitro Microevolution and Co-Selection Assessment of Amoxicillin and Cefotaxime Impact on Escherichia coli Resistance Development. Antibiotics 2024, 13, 247. [Google Scholar] [CrossRef]
- Cao, T.-T.; Deng, G.-H.; Fang, L.-X.; Yang, R.-S.; Sun, J.; Liu, Y.-H.; Liao, X.-P. Characterization of Quinolone Resistance in Salmonella enterica from Farm Animals in China. J. Food Prot. 2017, 80, 1742–1748. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.-L.; Peng, J.-J.; Ming, Y.-Y.; Ma, Q.-C.; Liu, W.-C.; Ma, Y. Identification of Drug Resistance Genes and Drug Resistance Analysis of Salmonella in the Duck Farm Environment of Zhanjiang, China. Environ. Sci. Pollut. Res. Int. 2020, 27, 24999–25008. [Google Scholar] [CrossRef] [PubMed]
- Munanura, E.I.; Ntale, M.; Wasswa, J.; Kaggwa, B. Assessment of Enrofloxacin Usage and Residue Levels of Enrofloxacin-Ciprofloxacin in Breast and Liver Tissues of Commercial Broilers Sold in Kampala-Uganda. Infect. Drug Resist. 2023, 16, 7629–7639. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-S.; Kim, G.-S.; Son, J.-S.; Lai, V.D.; Mo, I.-P.; Jang, H. Prevalence, Biosecurity Factor, and Antimicrobial Susceptibility Analysis of Salmonella Species Isolated from Commercial Duck Farms in Korea. Poult. Sci. 2021, 100, 100893. [Google Scholar] [CrossRef]
- Yang, J.; Ju, Z.; Yang, Y.; Zhao, X.; Jiang, Z.; Sun, S. Serotype, Antimicrobial Susceptibility and Genotype Profiles of Salmonella Isolated from Duck Farms and a Slaughterhouse in Shandong Province, China. BMC Microbiol. 2019, 19, 202. [Google Scholar] [CrossRef]
- Vo, T.-T.; Dang, V.-T.; Le, D.-H.; Nguyen, T.-H. Identification, Serotyping, and Antimicrobial Susceptibility of Riemerella anatipestifer Isolated from Ducks in Vietnam. Open Vet. J. 2022, 12, 391–398. [Google Scholar] [CrossRef]
- Voss-Rech, D.; Potter, L.; Vaz, C.S.L.; Pereira, D.I.B.; Sangioni, L.A.; Vargas, Á.C.; de Avila Botton, S. Antimicrobial Resistance in Nontyphoidal Salmonella Isolated from Human and Poultry-Related Samples in Brazil: 20-Year Meta-Analysis. Foodborne Pathog. Dis. 2017, 14, 116–124. [Google Scholar] [CrossRef]
- Luo, Y.; Yi, W.; Yao, Y.; Zhu, N.; Qin, P. Characteristic Diversity and Antimicrobial Resistance of Salmonella from Gastroenteritis. J. Infect. Chemother. 2018, 24, 251–255. [Google Scholar] [CrossRef]
- Talukder, H.; Roky, S.A.; Debnath, K.; Sharma, B.; Ahmed, J.; Roy, S. Prevalence and Antimicrobial Resistance Profile of Salmonella Isolated from Human, Animal and Environment Samples in South Asia: A 10-Year Meta-Analysis. J. Epidemiol. Glob. Health 2023, 13, 637–652. [Google Scholar] [CrossRef]
- Chen, Z.; Bai, J.; Wang, S.; Zhang, X.; Zhan, Z.; Shen, H.; Zhang, H.; Wen, J.; Gao, Y.; Liao, M.; et al. Prevalence, Antimicrobial Resistance, Virulence Genes and Genetic Diversity of Salmonella Isolated from Retail Duck Meat in Southern China. Microorganisms 2020, 8, 444. [Google Scholar] [CrossRef]
- Han, X.; Peng, J.; Guan, X.; Li, J.; Huang, X.; Liu, S.; Wen, Y.; Zhao, Q.; Huang, X.; Yan, Q.; et al. Genetic and Antimicrobial Resistance Profiles of Salmonella spp. Isolated from Ducks along the Slaughter Line in Southwestern China. Food Control 2020, 107, 106805. [Google Scholar] [CrossRef]
- Aronin, S.I.; Gupta, V.; Dunne, M.W.; Watts, J.A.; Yu, K.C. Regional Differences in Antibiotic-Resistant Enterobacterales Urine Isolates in the United States: 2018–2020. Int. J. Infect. Dis. 2022, 119, 142–145. [Google Scholar] [CrossRef]
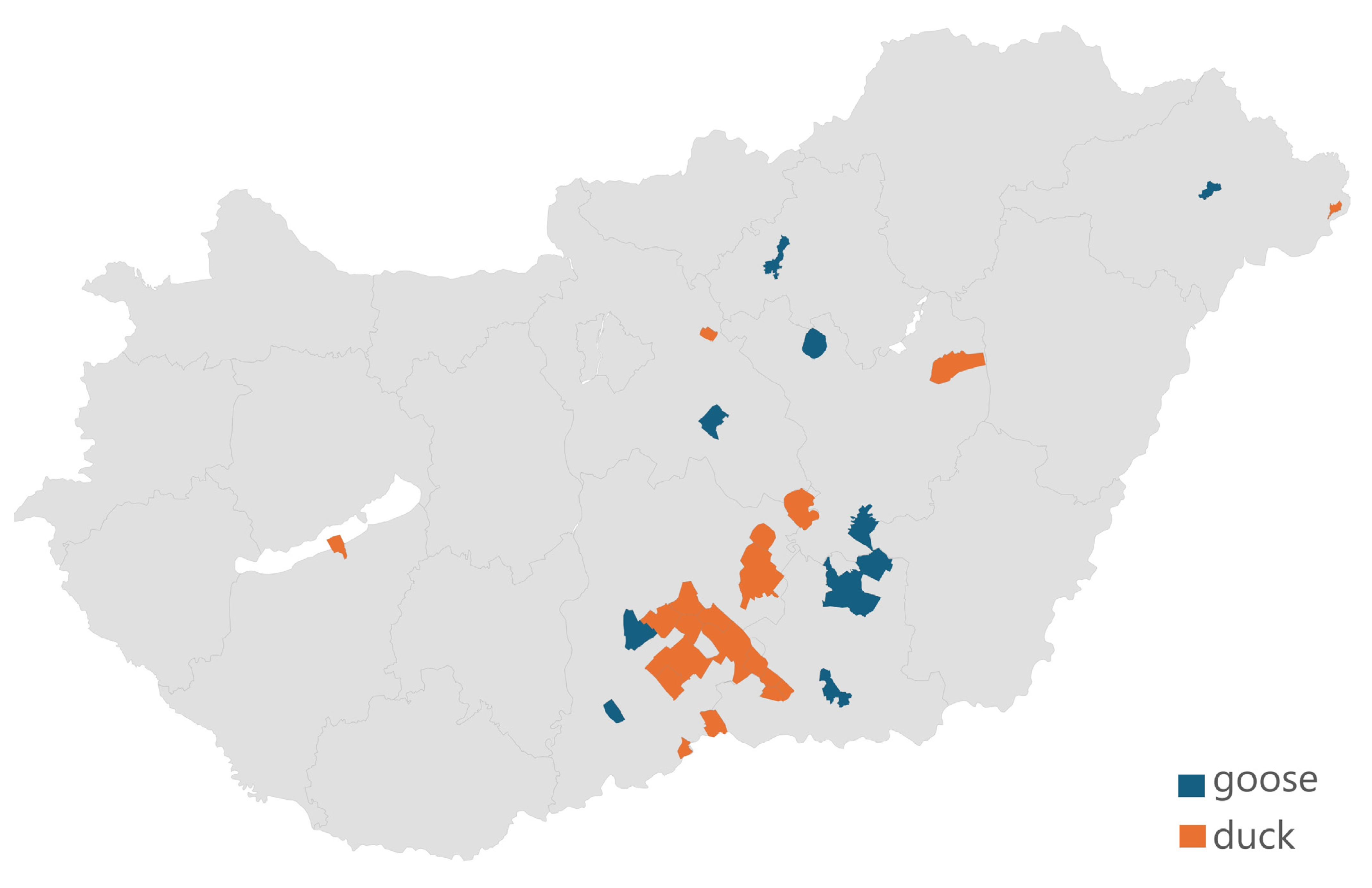
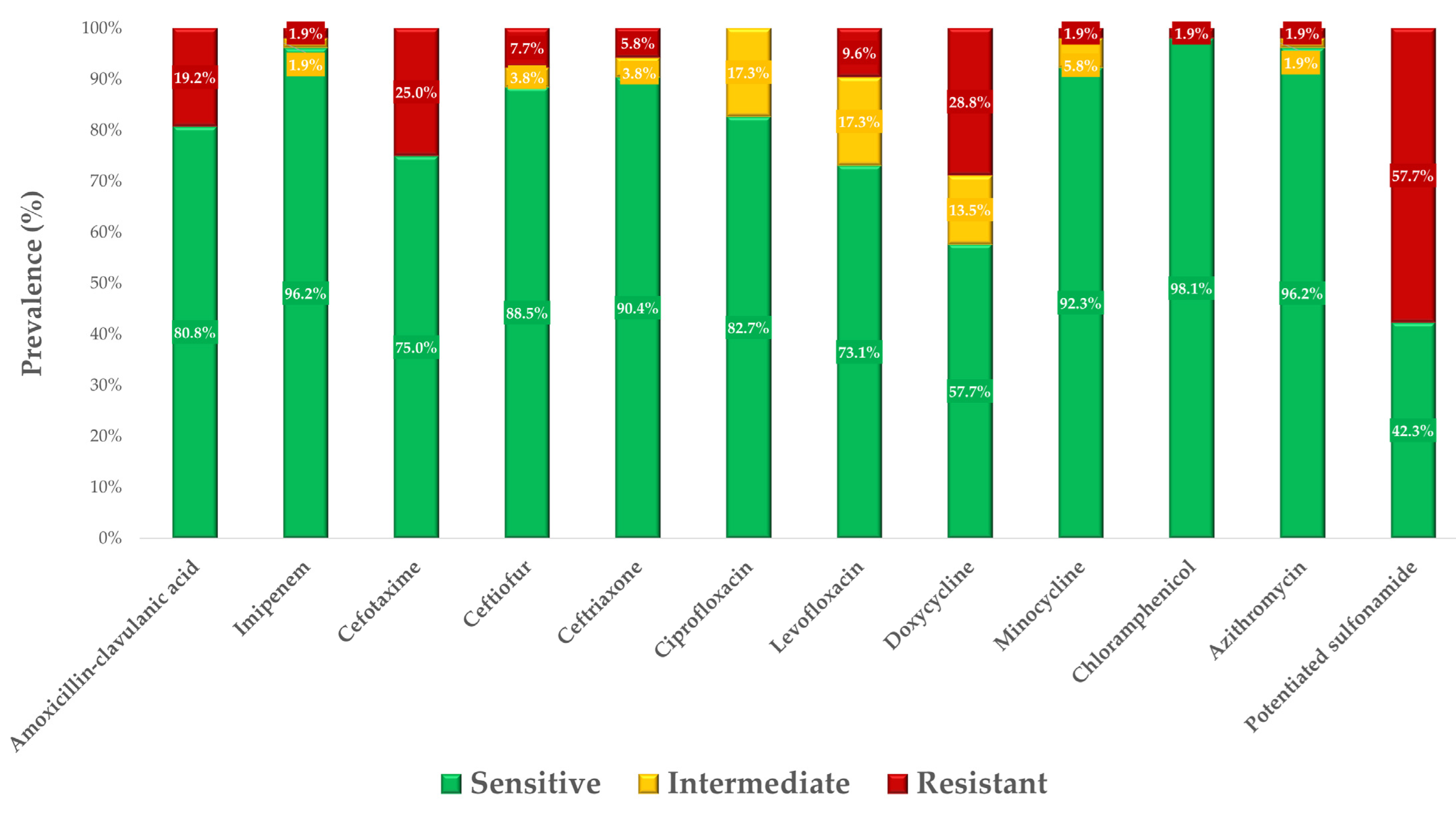
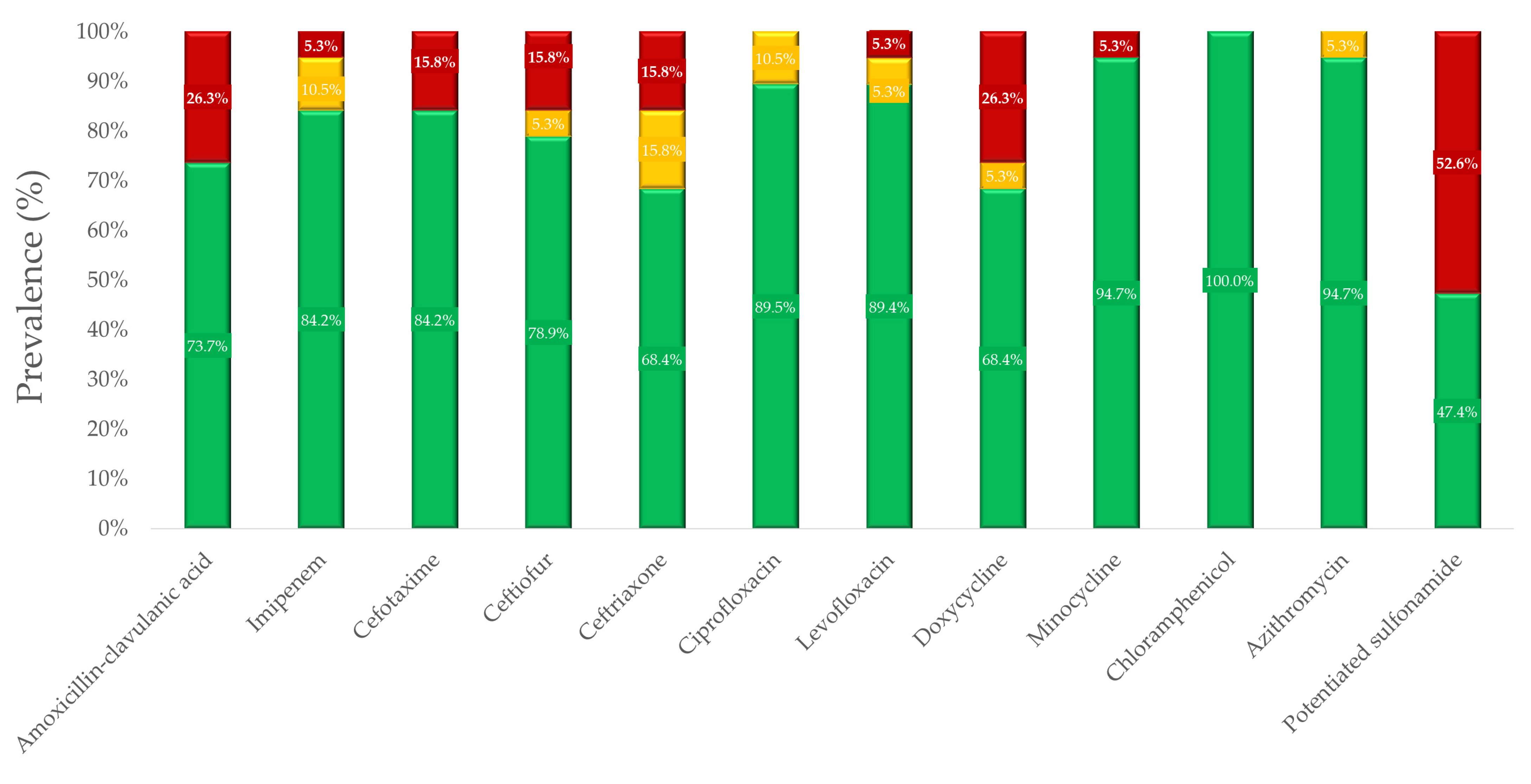

| Antibiotic | 1 BP * | 0.007 | 0.015 | 0.03 | 0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | 1024 | MIC50 | MIC90 | 2 ECOFF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| µg/mL | µg/mL | |||||||||||||||||||||
| Azithromycin | 32 | 7 | 43 | 1 | 0 | 0 | 0 | 0 | 1 | 8 | 8 | 16 | ||||||||||
| 13.5% | 82.7% | 1.9% | 0.0% | 0.0% | 0.0% | 0.0% | 1.9% | |||||||||||||||
| Cefotaxime | 4 | 1 | 0 | 3 | 11 | 5 | 7 | 8 | 3 | 0 | 0 | 7 | 6 | 0 | 1 | 0.25 | 16 | 4 | ||||
| 1.9% | 0.0% | 5.8% | 21.2% | 9.6% | 13.5% | 15.4% | 5.8% | 0.0% | 0.0% | 13.5% | 11.5% | 0.0% | 1.9% | |||||||||
| Ceftiofur | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 18 | 18 | 0 | 6 | 6 | 2 | 1 | 8 | 2 | ||||||
| 1.9% | 0.0% | 0.0% | 0.0% | 0.0% | 1.9% | 34.6% | 34.6% | 0.0% | 11.5% | 11.5% | 3.8% | |||||||||||
| Ceftriaxone | 1 | 1 | 6 | 17 | 9 | 5 | 3 | 5 | 2 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 1 | 0.125 | 2 | 0.25 | ||
| 1.9% | 11.5% | 32.7% | 17.3% | 9.6% | 5.8% | 9.6% | 3.8% | 0.0% | 0.0% | 1.9% | 3.8% | 0.0% | 0.0% | 0.0% | 1.9% | |||||||
| Ciprofloxacin | 0.06 | 20 | 10 | 4 | 8 | 4 | 2 | 3 | 1 | 0.015 | 0.25 | 0.125 | ||||||||||
| 38.5% | 19.2% | 7.7% | 15.4% | 7.7% | 3.8% | 5.8% | 1.9% | |||||||||||||||
| Doxycycline | 4 | 10 | 19 | 7 | 3 | 9 | 4 | 4 | 32 | 8 | ||||||||||||
| 19.2% | 36.5% | 13.5% | 5.8% | 17.3% | 7.7% | |||||||||||||||||
| Imipenem | 1 | 6 | 8 | 13 | 21 | 1 | 0 | 1 | 0 | 2 | 0.5 | 1 | 1 | |||||||||
| 11.5% | 15.4% | 25.0% | 40.4% | 1.9% | 0.0% | 1.9% | 0.0% | 3.8% | ||||||||||||||
| Chloramphenicol | 8 | 43 | 8 | 0 | 1 | 4 | 8 | 16 | ||||||||||||||
| 82.7% | 15.4% | 0.0% | 1.9% | |||||||||||||||||||
| Levofloxacin | 0.125 | 2 | 4 | 14 | 16 | 1 | 1 | 5 | 3 | 1 | 1 | 2 | 2 | 0.06 | 2 | 0.25 | ||||||
| 3.8% | 7.7% | 26.9% | 30.8% | 1.9% | 1.9% | 9.6% | 5.8% | 1.9% | 1.9% | 3.8% | 3.8% | |||||||||||
| 3 Potentiated sulfonamide | 4/76 | 5 | 2 | 14 | 8 | 4 | 5 | 2 | 3 | 3 | 4 | 1 | 1 | 4 | 256 | 1 | ||||||
| 9.6% | 3.8% | 26.9% | 15.4% | 7.7% | 9.6% | 3.8% | 5.8% | 5.8% | 7.7% | 1.9% | 1.9% | |||||||||||
| Antibiotic | 1 BP * | 0.007 | 0.015 | 0.03 | 0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | 1024 | MIC50 | MIC90 | 2 ECOFF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| µg/mL | µg/mL | |||||||||||||||||||||
| Azithromycin | 32 | 1 | 0 | 4 | 13 | 1 | 8 | 8 | 16 | |||||||||||||
| 5.3% | 0.0% | 21.1% | 68.4% | 5.3% | ||||||||||||||||||
| Cefotaxime | 4 | 2 | 5 | 1 | 1 | 3 | 4 | 0 | 1 | 2 | 0.5 | 4 | 4 | |||||||||
| 10.5% | 26.3% | 5.3% | 5.3% | 15.8% | 21.1% | 0.0% | 5.3% | 10.5% | ||||||||||||||
| Ceftiofur | 8 | 3 | 0 | 1 | 0 | 1 | 3 | 5 | 2 | 0 | 1 | 2 | 1 | 0.5 | 8 | 2 | ||||||
| 15.8% | 0.0% | 5.3% | 0.0% | 5.3% | 15.8% | 26.3% | 10.5% | 0.0% | 5.3% | 10.5% | 5.3% | |||||||||||
| Ceftriaxone | 4 | 1 | 2 | 4 | 4 | 1 | 1 | 1 | 3 | 0 | 0 | 0 | 2 | 0.125 | 2 | 0.25 | ||||||
| 5.3% | 10.5% | 21.1% | 21.1% | 5.3% | 5.3% | 5.3% | 15.8% | 0.0% | 0.0% | 0.0% | 10.5% | |||||||||||
| Ciprofloxacin | 1 | 11 | 4 | 1 | 1 | 1 | 1 | 0.007 | 0.06 | 0.125 | ||||||||||||
| 57.9% | 21.1% | 5.3% | 5.3% | 5.3% | 5.3% | |||||||||||||||||
| Doxycycline | 16 | 1 | 0 | 2 | 3 | 7 | 1 | 3 | 1 | 1 | 4 | 16 | 8 | |||||||||
| 5.3% | 0.0% | 10.5% | 15.8% | 36.8% | 5.3% | 15.8% | 5.3% | 5.3% | ||||||||||||||
| Imipenem | 4 | 1 | 2 | 11 | 1 | 2 | 0 | 0 | 0 | 1 | 0.5 | 2 | 1 | |||||||||
| 5.3% | 10.5% | 57.9% | 5.3% | 10.5% | 0.0% | 0.0% | 0.0% | 5.3% | ||||||||||||||
| Chloramphenicol | 32 | 2 | 0 | 13 | 4 | 4 | 8 | 16 | ||||||||||||||
| 10.5% | 0.0% | 68.4% | 21.1% | |||||||||||||||||||
| Levofloxacin | 2 | 3 | 4 | 6 | 3 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0.03 | 0.125 | 0.25 | ||||||
| 15.8% | 21.1% | 31.6% | 15.8% | 5.3% | 0.0% | 5.3% | 0.0% | 0.0% | 0.0% | 0.0% | 5.3% | |||||||||||
| 3 Potentiated sulfonamide | 4/76 | 4 | 0 | 5 | 3 | 4 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 4 | 128 | 1 | ||||||
| 21.1% | 0.0% | 26.3% | 15.8% | 21.1% | 0.0% | 0.0% | 0.0% | 5.3% | 0.0% | 0.0% | 10.5% | |||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kerek, Á.; Szabó, Á.; Jerzsele, Á. Antimicrobial Susceptibility Profiles of Salmonella spp. Isolates from Clinical Cases of Waterfowl in Hungary Between 2022 and 2023. Microorganisms 2024, 12, 2462. https://doi.org/10.3390/microorganisms12122462
Kerek Á, Szabó Á, Jerzsele Á. Antimicrobial Susceptibility Profiles of Salmonella spp. Isolates from Clinical Cases of Waterfowl in Hungary Between 2022 and 2023. Microorganisms. 2024; 12(12):2462. https://doi.org/10.3390/microorganisms12122462
Chicago/Turabian StyleKerek, Ádám, Ábel Szabó, and Ákos Jerzsele. 2024. "Antimicrobial Susceptibility Profiles of Salmonella spp. Isolates from Clinical Cases of Waterfowl in Hungary Between 2022 and 2023" Microorganisms 12, no. 12: 2462. https://doi.org/10.3390/microorganisms12122462
APA StyleKerek, Á., Szabó, Á., & Jerzsele, Á. (2024). Antimicrobial Susceptibility Profiles of Salmonella spp. Isolates from Clinical Cases of Waterfowl in Hungary Between 2022 and 2023. Microorganisms, 12(12), 2462. https://doi.org/10.3390/microorganisms12122462






