Oxford Nanopore Technology-Based Identification of an Acanthamoeba castellanii Endosymbiosis in Microbial Keratitis
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Extraction
2.2. qPCR
2.3. Whole Genome Sequencing
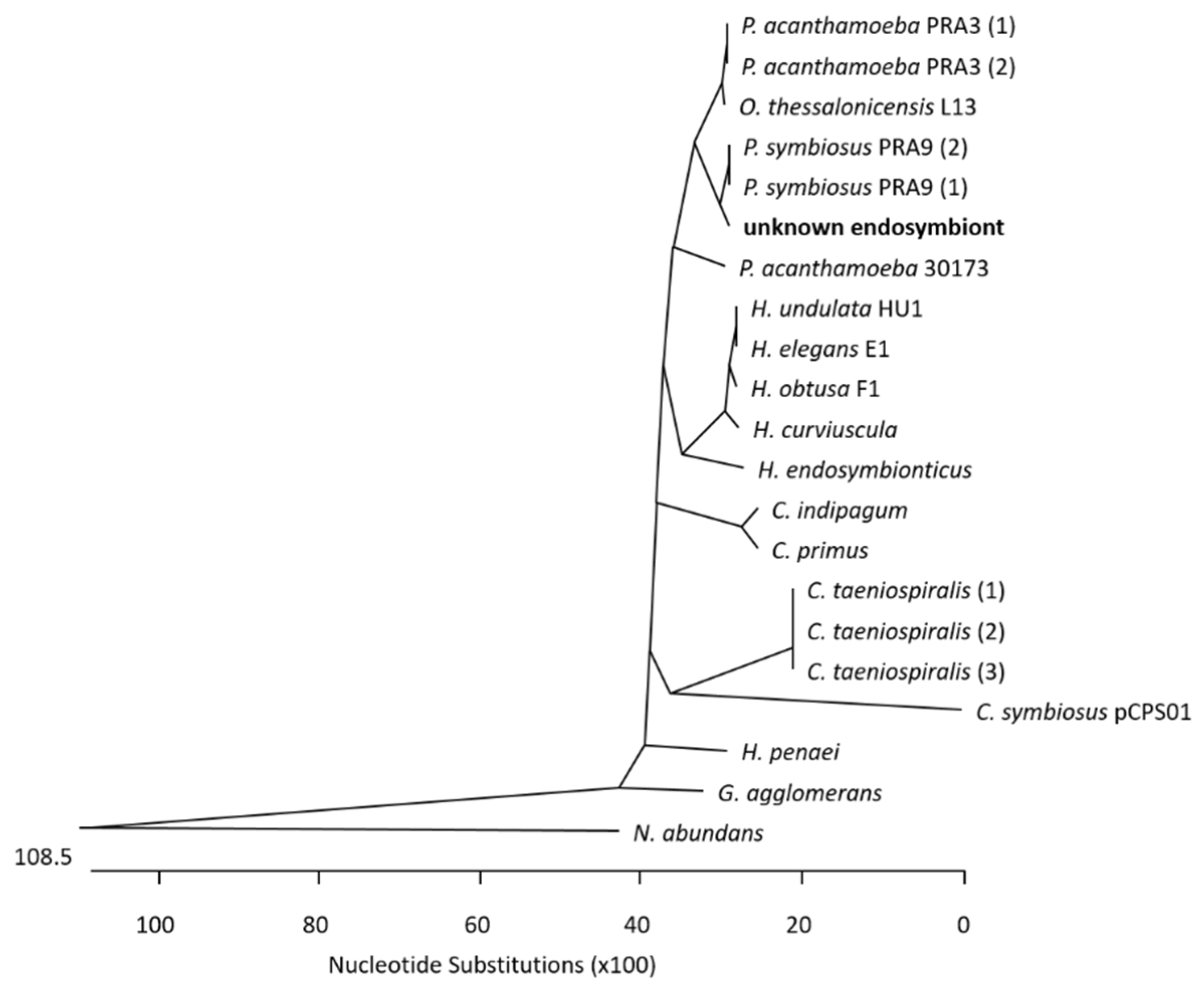
3. Results
3.1. Case Report
3.2. Microbiological Diagnostics
4. Discussion
4.1. Implications of Endosymbiosis for Pathogenicity
4.2. Comparison of Detection Methods
4.3. Concluding Considerations
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alkatan, H.M.; Al-Essa, R.S. Challenges in the diagnosis of microbial keratitis: A detailed review with update and general guidelines. Saudi J. Ophthalmol. 2019, 33, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Ting, D.S.J.; Gopal, B.P.; Deshmukh, R.; Seitzman, G.D.; Said, D.G.; Dua, H.S. Diagnostic armamentarium of infectious keratitis: A comprehensive review. Ocul. Surf. 2022, 23, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Sagerfors, S.; Edslev, S.; Lindblad, B.E.; Lilje, B.; Stegger, M.; Söderquist, B. In the eye of the ophthalmologist: The corneal microbiome in microbial keratitis. Graefe’s Arch. Clin. Exp. Ophthalmol. 2024, 262, 1579–1589. [Google Scholar] [CrossRef] [PubMed]
- Musa, F.; Tailor, R.; Gao, A.; Hutley, E.; Rauz, S.; Scott, R.A. Contact lens-related microbial keratitis in deployed British military personnel. Br. J. Ophthalmol. 2010, 94, 988–993. [Google Scholar] [CrossRef]
- Ng, J.K.; Fraunfelder, F.W.; Winthrop, K.L. Review and Update on the Epidemiology, Clinical Presentation, Diagnosis, and Treatment of Fungal Keratitis. Curr. Fungal Infect. Rep. 2013, 7, 293–300. [Google Scholar] [CrossRef]
- Fanselow, N.; Sirajuddin, N.; Yin, X.T.; Huang, A.J.W.; Stuart, P.M. Acanthamoeba Keratitis, Pathology, Diagnosis and Treatment. Pathogens 2021, 10, 323. [Google Scholar] [CrossRef]
- Lorenzo-Morales, J.; Khan, N.A.; Walochnik, J. An update on Acanthamoeba keratitis: Diagnosis, pathogenesis and treatment. Parasite 2015, 22, 10. [Google Scholar] [CrossRef]
- Paterson, G.N.; Rittig, M.; Siddiqui, R.; Khan, N.A. Is Acanthamoeba pathogenicity associated with intracellular bacteria? Exp. Parasitol. 2011, 129, 207–210. [Google Scholar] [CrossRef]
- Chan, L.L.; Mak, J.W.; Ambu, S.; Chong, P.Y. Identification and ultrastructural characterization of Acanthamoeba bacterial endocytobionts belonging to the Alphaproteobacteria class. PLoS ONE 2018, 13, e0204732. [Google Scholar] [CrossRef]
- Moon, E.K.; Park, S.M.; Chu, K.B.; Quan, F.S.; Kong, H.H. Differentially Expressed Gene Profile of Acanthamoeba castellanii Induced by an Endosymbiont Legionella pneumophila. Korean J. Parasitol. 2021, 59, 67–75. [Google Scholar] [CrossRef]
- Moon, E.K.; Kim, M.J.; Lee, H.A.; Quan, F.S.; Kong, H.H. Comparative analysis of differentially expressed genes in Acanthamoeba after ingestion of Legionella pneumophila and Escherichia coli. Exp. Parasitol. 2022, 232, 108188. [Google Scholar] [CrossRef] [PubMed]
- Marciano-Cabral, F.; Cabral, G. Acanthamoeba spp. as agents of disease in humans. Clin. Microbiol. Rev. 2003, 16, 273–307. [Google Scholar] [CrossRef] [PubMed]
- Alsam, S.; Jeong, S.R.; Sissons, J.; Dudley, R.; Kim, K.S.; Khan, N.A. Escherichia coli interactions with Acanthamoeba: A symbiosis with environmental and clinical implications. J. Med. Microbiol. 2006, 55, 689–694. [Google Scholar] [CrossRef] [PubMed]
- Haapanen, S.; Barker, H.; Carta, F.; Supuran, C.T.; Parkkila, S. Novel Drug Screening Assay for Acanthamoeba castellanii and the Anti-Amoebic Effect of Carbonic Anhydrase Inhibitors. J. Med. Chem. 2024, 67, 152–164. [Google Scholar] [CrossRef]
- Iovieno, A.; Ledee, D.R.; Miller, D.; Alfonso, E.C. Detection of bacterial endosymbionts in clinical acanthamoeba isolates. Ophthalmology 2010, 117, 445–452.e3. [Google Scholar] [CrossRef]
- Yildiz, E.H.; Abdalla, Y.F.; Elsahn, A.F.; Rapuano, C.J.; Hammersmith, K.M.; Laibson, P.R.; Cohen, E.J. Update on fungal keratitis from 1999 to 2008. Cornea 2010, 29, 1406–1411. [Google Scholar] [CrossRef]
- KrishnanNair Geetha, D.; Sivaraman, B.; Rammohan, R.; Venkatapathy, N.; Solai Ramatchandirane, P. A SYBR Green based multiplex Real-Time PCR assay for rapid detection and differentiation of ocular bacterial pathogens. J. Microbiol. Methods 2020, 171, 105875. [Google Scholar] [CrossRef]
- Wagner, K.; Springer, B.; Pires, V.P.; Keller, P.M. High-throughput screening of bacterial pathogens in clinical specimens using 16S rDNA qPCR and fragment analysis. Diagn. Microbiol. Infect. Dis. 2019, 93, 287–292. [Google Scholar] [CrossRef]
- Shimizu, D.; Miyazaki, D.; Ehara, F.; Shimizu, Y.; Uotani, R.; Inata, K.; Sasaki, S.I.; Inoue, Y. Effectiveness of 16S ribosomal DNA real-time PCR and sequencing for diagnosing bacterial keratitis. Graefe’s Arch. Clin. Exp. Ophthalmol. 2020, 258, 157–166. [Google Scholar] [CrossRef]
- Fang, P.C.; Chien, C.C.; Yu, H.J.; Ho, R.W.; Tseng, S.L.; Lai, Y.H.; Kuo, M.T. A dot hybridization assay for the diagnosis of bacterial keratitis. Mol. Vis. 2017, 23, 306–317. [Google Scholar]
- Panda, A.; Pal Singh, T.; Satpathy, G.; Wadhwani, M.; Monika, M. Comparison of polymerase chain reaction and standard microbiological techniques in presumed bacterial corneal ulcers. Int. Ophthalmol. 2015, 35, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Chowdhary, A.; Van Gelder, R.N.; Sundararajan, M. Methodologic Considerations for Studying the Ocular Surface Microbiome. Ophthalmol. Sci. 2023, 3, 100408. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Yi, S.; Wei, L. Ocular Microbiota and Intraocular Inflammation. Front. Immunol. 2020, 11, 609765. [Google Scholar] [CrossRef] [PubMed]
- Gomes JÁ, P.; Frizon, L.; Demeda, V.F. Ocular Surface Microbiome in Health and Disease. Asia-Pac. J. Ophthalmol. 2020, 9, 505–511. [Google Scholar] [CrossRef]
- Qvarnstrom, Y.; Visvesvara, G.S.; Sriram, R.; da Silva, A.J. Multiplex real-time PCR assay for simultaneous detection of Acanthamoeba spp., Balamuthia mandrillaris, and Naegleria fowleri. J. Clin. Microbiol. 2006, 44, 3589–3595. [Google Scholar] [CrossRef]
- Al-Sakati, H.; Kowollik, S.; Gabris, S.; Balasiu, A.; Ommerborn, M.; Pfeffer, K.; Henrich, B.; Raab, W.H. The benefit of culture-independent methods to detect bacteria and fungi in re-infected root filled teeth: A pilot study. Int. Endod. J. 2021, 54, 74–84. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 2021, 39, 1348–1365. [Google Scholar] [CrossRef]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Berrilli, F.; Montalbano Di Filippo, M.; Guadano-Procesi, I.; Ciavurro, M.; Di Cave, D. Acanthamoeba Sequence Types and Allelic Variations in Isolates from Clinical and Different Environmental Sources in Italy. Microorganisms 2024, 12, 544. [Google Scholar] [CrossRef]
- Hooshyar, H.; Hosseinbigi, B.; Saraei, M.; Alizadeh, S.; Eftakhar, M.; Rasti, S.; Khosro-Shahi, N. Genotyping of acanthamoeba isolated from surface and stagnant waters of qazvin, central iran. Iran. Red Crescent Med. J. 2013, 15, 536–538. [Google Scholar] [CrossRef] [PubMed]
- Schmitz-Esser, S.; Toenshoff, E.R.; Haider, S.; Heinz, E.; Hoenninger, V.M.; Wagner, M.; Horn, M. Diversity of Bacterial Endosymbionts of Environmental Acanthamoeba Isolates. Appl. Environ. Microbiol. 2008, 74, 5822–5831. [Google Scholar] [CrossRef] [PubMed]
- Hajialilo, E.; Rezaeian, M.; Niyyati, M.; Pourmand, M.R.; Mohebali, M.; Norouzi, M.; Razavi Pashabeyg, K.; Rezaie, S.; Khodavaisy, S. Molecular characterization of bacterial, viral and fungal endosymbionts of Acanthamoeba isolates in keratitis patients of Iran. Exp. Parasitol. 2019, 200, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Rayamajhee, B.; Sharma, S.; Willcox, M.; Henriquez, F.L.; Rajagopal, R.N.; Shrestha, G.S.; Subedi, D.; Bagga, B.; Carnt, N. Assessment of genotypes, endosymbionts and clinical characteristics of Acanthamoeba recovered from ocular infection. BMC Infect. Dis. 2022, 22, 757. [Google Scholar] [CrossRef]
- Dos Santos, D.L.; Virginio, V.G.; Berté, F.K.; Lorenzatto, K.R.; Marinho, D.R.; Kwitko, S.; Locatelli, C.I.; Freitas, E.C.; Rott, M.B. Clinical and molecular diagnosis of Acanthamoeba keratitis in contact lens wearers in southern Brazil reveals the presence of an endosymbiont. Parasitol. Res. 2022, 121, 1447–1454. [Google Scholar] [CrossRef]
- Cohen, G.; Hoffart, L.; La Scola, B.; Raoult, D.; Drancourt, M. Ameba-associated Keratitis, France. Emerg. Infect. Dis. 2011, 17, 1306–1308. [Google Scholar] [CrossRef]
- Rayamajhee, B.; Willcox, M.D.P.; Henriquez, F.L.; Petsoglou, C.; Subedi, D.; Carnt, N. Acanthamoeba, an environmental phagocyte enhancing survival and transmission of human pathogens. Trends Parasitol. 2022, 38, 975–990. [Google Scholar] [CrossRef]
- Hojo, F.; Osaki, T.; Yonezawa, H.; Hanawa, T.; Kurata, S.; Kamiya, S. Acanthamoeba castellanii supports extracellular survival of Helicobacter pylori in co-culture. J. Infect. Chemother. 2020, 26, 946–954. [Google Scholar] [CrossRef]
- Goh, J.W.Y.; Harrison, R.; Hau, S.; Alexander, C.L.; Tole, D.M.; Avadhanam, V.S. Comparison of In Vivo Confocal Microscopy, PCR and Culture of Corneal Scrapes in the Diagnosis of Acanthamoeba Keratitis. Cornea 2018, 37, 480–485. [Google Scholar] [CrossRef]
- Haapanen, S.; Patrikainen, M.S.; Parkkila, S. Ultrasensitive and rapid diagnostic tool for detection of Acanthamoeba castellanii. Diagn. Microbiol. Infect. Dis. 2023, 107, 116014. [Google Scholar] [CrossRef]
- Varacalli, G.; Di Zazzo, A.; Mori, T.; Dohlman, T.H.; Spelta, S.; Coassin, M.; Bonini, S. Challenges in Acanthamoeba Keratitis: A Review. J. Clin. Med. 2021, 10, 942. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Sun, G.; Sun, Y.; You, X.; Li, X.; Cheng, Y.; Xuan, Y. Identification and Genotypic Characterization of Potentially Pathogenic Acanthamoeba Isolated from Tap Water in Wuxi, China. Korean J. Parasitol. 2018, 56, 615–618. [Google Scholar] [CrossRef] [PubMed]
- Yera, H.; Ok, V.; Lee Koy Kuet, F.; Dahane, N.; Ariey, F.; Hasseine, L.; Delaunay, P.; Martiano, D.; Marty, P.; Bourges, J.L. PCR and culture for diagnosis of Acanthamoeba keratitis. Br. J. Ophthalmol. 2021, 105, 1302–1306. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, J.J.; Dart, J.K.G.; De, S.K.; Carnt, N.; Cleary, G.; Hau, S. Comparison of culture, confocal microscopy and PCR in routine hospital use for microbial keratitis diagnosis. Eye 2022, 36, 2172–2178. [Google Scholar] [CrossRef] [PubMed]
- Booton, G.C.; Visvesvara, G.S.; Byers, T.J.; Kelly, D.J.; Fuerst, P.A. Identification and distribution of Acanthamoeba species genotypes associated with nonkeratitis infections. J. Clin. Microbiol. 2005, 43, 1689–1693. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jiang, L.; Zhao, Y.; Ju, X.; Wang, L.; Jin, L.; Fine, R.D.; Li, M. Biological characteristics and pathogenicity of Acanthamoeba. Front. Microbiol. 2023, 14, 1147077. [Google Scholar] [CrossRef]
- Fritsche, T.R.; Horn, M.; Seyedirashti, S.; Gautom, R.K.; Schleifer, K.H.; Wagner, M. In situ detection of novel bacterial endosymbionts of Acanthamoeba spp. phylogenetically related to members of the order Rickettsiales. Appl. Environ. Microbiol. 1999, 65, 206–212. [Google Scholar] [CrossRef]
- Low, L.; Fuentes-Utrilla, P.; Hodson, J.; O’Neil, J.D.; Rossiter, A.E.; Begum, G.; Suleiman, K.; Murray, P.I.; Wallace, G.R.; Loman, N.J.; et al. Evaluation of full-length nanopore 16S sequencing for detection of pathogens in microbial keratitis. PeerJ 2021, 9, e10778. [Google Scholar] [CrossRef]
- Marić, J.; Križanović, K.; Riondet, S.; Nagarajan, N.; Šikić, M. Comparative analysis of metagenomic classifiers for long-read sequencing datasets. BMC Bioinform. 2024, 25, 15. [Google Scholar] [CrossRef]
- Chrisman, B.; He, C.; Jung, J.Y.; Stockham, N.; Paskov, K.; Washington, P.; Wall, D.P. The human “contaminome”: Bacterial, viral, and computational contamination in whole genome sequences from 1000 families. Sci. Rep. 2022, 12, 9863. [Google Scholar] [CrossRef]
- Peter, V.G.; Morandi, S.C.; Herzog, E.L.; Zinkernagel, M.S.; Zysset-Burri, D.C. Investigating the Ocular Surface Microbiome: What Can It Tell Us? Clin. Ophthalmol. 2023, 17, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Zhang, H.; Hu, M.; Ma, Y.; Chen, P.; Zhao, Z.; Li, J.; Ye, Y.; Zheng, M.; Lou, Y. Alterations in the Ocular Surface Microbiome in Traumatic Corneal Ulcer Patients. Investig. Ophthalmol. Vis. Sci. 2020, 61, 35. [Google Scholar] [CrossRef] [PubMed]
- Shigeyasu, C.; Yamada, M.; Aoki, K.; Ishii, Y.; Tateda, K.; Yaguchi, T.; Okajima, Y.; Hori, Y. Metagenomic analysis for detecting Fusarium solani in a case of fungal keratitis. J. Infect. Chemother. 2018, 24, 664–668. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.J.; Li, S.C.; Lin, W.C.; Huang, F.C. Intracellular Microbiome Profiling of the Acanthamoeba Clinical Isolates from Lens Associated Keratitis. Pathogens 2021, 10, 266. [Google Scholar] [CrossRef]
- Chuang, Y.H.; Wang, Y.C.; Yen, C.Y.; Lin, C.C.; Chen, C.C. Case Series: Unusual Presentation of Acanthamoeba Coinfection in the Cornea. Optom. Vis. Sci. 2022, 99, 605–611. [Google Scholar] [CrossRef]
- Szentmáry, N.; Daas, L.; Shi, L.; Laurik, K.L.; Lepper, S.; Milioti, G.; Seitz, B. Acanthamoeba keratitis-Clinical signs, differential diagnosis and treatment. J. Curr. Ophthalmol. 2019, 31, 16–23. [Google Scholar] [CrossRef]
- Raghavan, A.; Baidwal, S.; Venkatapathy, N.; Rammohan, R. The Acanthamoeba-Fungal Keratitis Study. Am. J. Ophthalmol. 2019, 201, 31–36. [Google Scholar] [CrossRef]
- Scruggs, B.A.; Quist, T.S.; Zimmerman, M.B.; Salinas, J.L.; Greiner, M.A. Risk factors, management, and outcomes of Acanthamoeba keratitis: A retrospective analysis of 110 cases. Am. J. Ophthalmol. Case Rep. 2022, 25, 101372. [Google Scholar] [CrossRef]
- Rammohan, R.; Baidwal, S.; Venkatapathy, N.; Lorenzo-Morales, J.; Raghavan, A. A 5-Year Review of Coinfections in Acanthamoeba keratitis from South India. Eye Contact Lens 2023, 49, 334–338. [Google Scholar] [CrossRef]
- Borroni, D.; Bonzano, C.; Sánchez-González, J.M.; Rachwani-Anil, R.; Zamorano-Martín, F.; Pereza-Nieves, J.; Traverso, C.E.; García Lorente, M.; Rodríguez-Calvo-de-Mora, M.; Esposito, A.; et al. Shotgun metagenomic sequencing in culture negative microbial keratitis. Eur. J. Ophthalmol. 2023, 33, 1589–1595. [Google Scholar] [CrossRef]
- Borroni, D. Granulicatella Adiacens as an Unusual Cause of Microbial Keratitis: A Metagenomic Approach. Ocul. Immunol. Inflamm. 2022, 30, 1550–1551. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Hao, Z.; Zhang, C.; Deng, A. Unveiling challenging corneal infections: A comprehensive etiological diagnosis through metagenomic next-generation sequencing (mNGS) of corneal tissue samples. Int. Ophthalmol. 2024, 44, 246. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Tian, L.; Gu, X.; Chen, Y.; Ma, X.; Lin, S.; Li, Z.; Lou, Y.; Zheng, M. Characterization of the Ocular Surface Microbiome in Keratitis Patients after Repeated Ophthalmic Antibiotic Exposure. Microbiol. Spectr. 2022, 10, e0216221. [Google Scholar] [CrossRef] [PubMed]

| Organism (NCBI ID) | No. of Reads (%) |
|---|---|
| Homo sapiens (9606) | 3,203,564 (99.92%) |
| Acanthamoeba castellanii str, Neff (1257118) | 1683 (0.05%) |
| Arthrobacter sp. KBS0702 (2578107) | 108 (<0.05%) |
| Plasmodium vivax (5855) | 35 (<0.05%) |
| Candidatus Paracaedibacter symbiosus (244582) | 32 (<0.05%) |
| Reads from Kraken2 Assigned Organism | Mapping Against Reference Genome 1 | Mapping Against Homo sapiens Genome 1 |
|---|---|---|
| Acanthamoeba castellanii | 933/1683 (55.4%) | 831/1683 (49.38%) |
| Arthrobacter sp. | 0/108 (0%) | 108/108 (100%) |
| Plasmodium vivax | 34/35 (97.14%) | 35/35 (100%) |
| Candidatus Paracaedibacter symbiosus | 29/32 (90.62%) | 0/32 (0%) |
| Acanthamoeba castellanii | 933/1683 (55.4%) | 831/1683 (49.38%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scharf, S.A.; Friedrichs, L.; Bock, R.; Borrelli, M.; MacKenzie, C.; Pfeffer, K.; Henrich, B. Oxford Nanopore Technology-Based Identification of an Acanthamoeba castellanii Endosymbiosis in Microbial Keratitis. Microorganisms 2024, 12, 2292. https://doi.org/10.3390/microorganisms12112292
Scharf SA, Friedrichs L, Bock R, Borrelli M, MacKenzie C, Pfeffer K, Henrich B. Oxford Nanopore Technology-Based Identification of an Acanthamoeba castellanii Endosymbiosis in Microbial Keratitis. Microorganisms. 2024; 12(11):2292. https://doi.org/10.3390/microorganisms12112292
Chicago/Turabian StyleScharf, Sebastian Alexander, Lennart Friedrichs, Robert Bock, Maria Borrelli, Colin MacKenzie, Klaus Pfeffer, and Birgit Henrich. 2024. "Oxford Nanopore Technology-Based Identification of an Acanthamoeba castellanii Endosymbiosis in Microbial Keratitis" Microorganisms 12, no. 11: 2292. https://doi.org/10.3390/microorganisms12112292
APA StyleScharf, S. A., Friedrichs, L., Bock, R., Borrelli, M., MacKenzie, C., Pfeffer, K., & Henrich, B. (2024). Oxford Nanopore Technology-Based Identification of an Acanthamoeba castellanii Endosymbiosis in Microbial Keratitis. Microorganisms, 12(11), 2292. https://doi.org/10.3390/microorganisms12112292






