Spillover: Mechanisms, Genetic Barriers, and the Role of Reservoirs in Emerging Pathogens
Abstract
1. Introduction
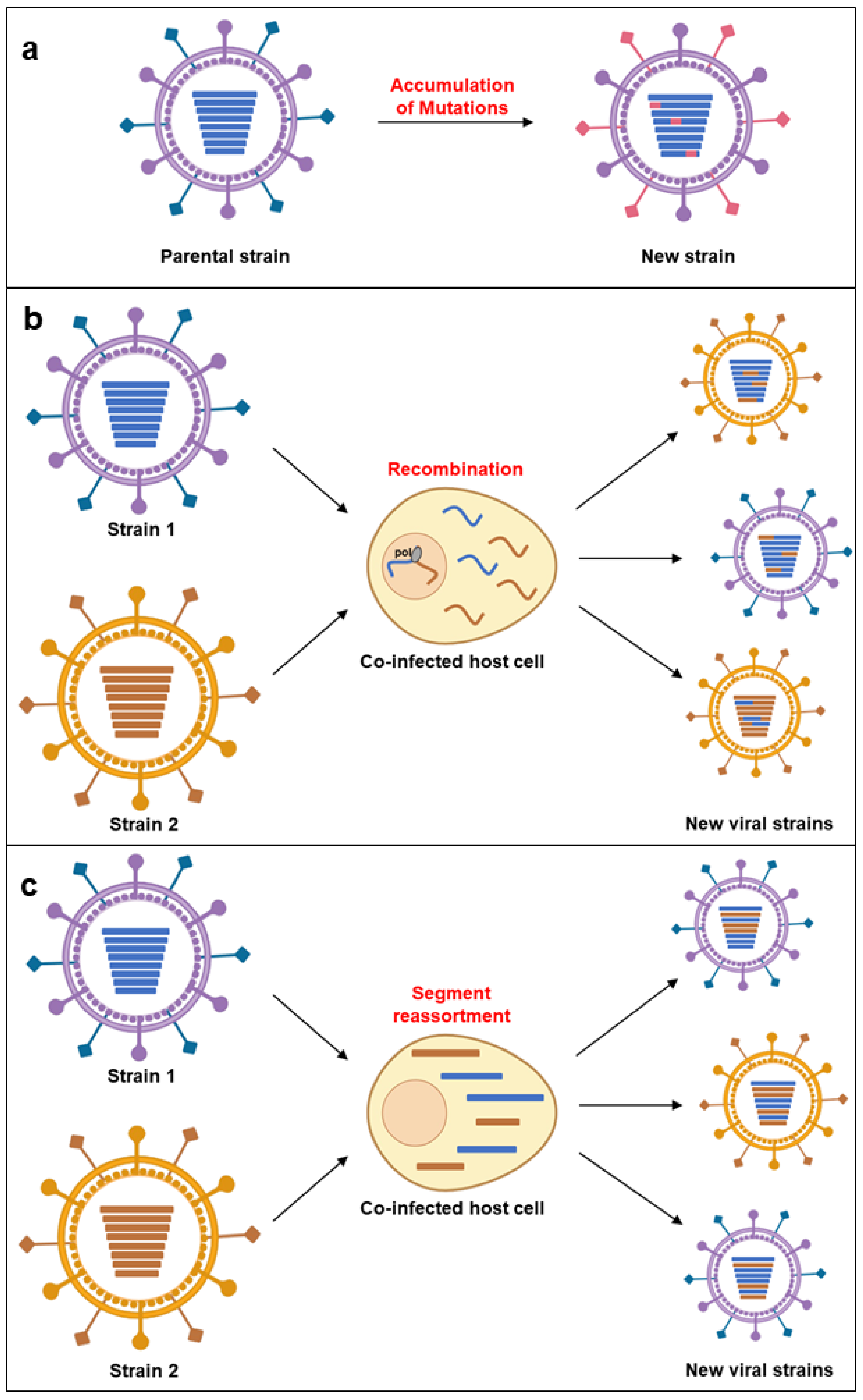
2. General Aspects of Emerging Pathogens
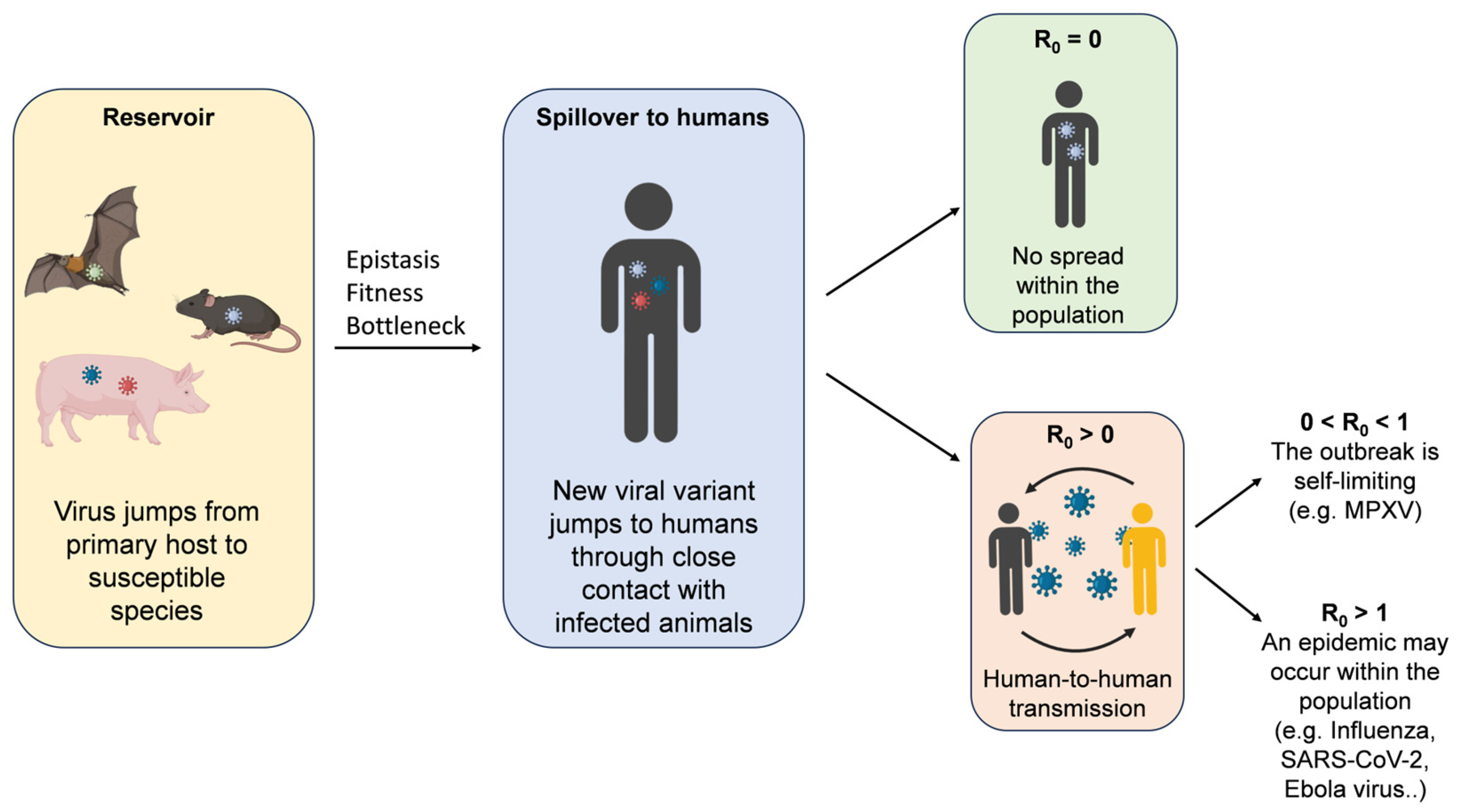
3. Factors That Favor Spillover
- (1)
- Eshel’s model: the initial strain cannot survive without mutations; therefore, the optimal mutation rate is strictly positive. In the case ω ≥ (R2 − 1)/R2 the fit strain will go extinct with certainty, so the optimal mutation rate is bounded below this value (ω = mutation rate; R1 = initial strain fitness; R2 = reproductive number of the strain) [63];
- (2)
- Alexander and Day’s model, which explores mutation rates in relation to fitness [64].
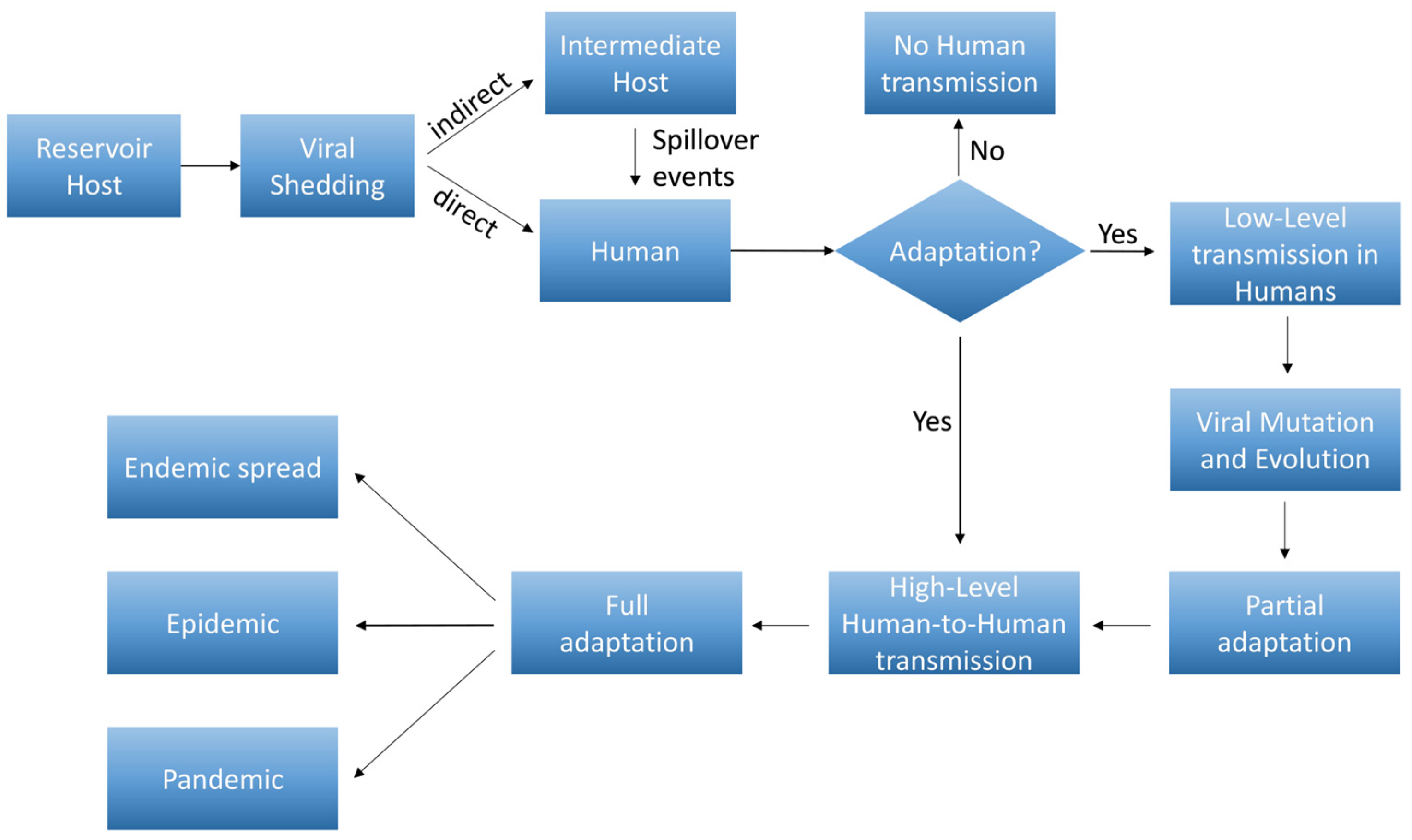
4. Pathogen Adaptation in a New Host
5. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Glossary
References
- Gavotte, L.; Gaucherel, C.; Frutos, R. Environmental Spillover of Emerging Viruses: Is It True? Environ. Res. 2023, 233, 116416. [Google Scholar] [CrossRef]
- Morris, J.G. Cholera—Modern Pandemic Disease of Ancient Lineage. Emerg. Infect. Dis. 2011, 17, 2099–2104. [Google Scholar] [CrossRef]
- Alexander, K.A.; Lewis, B.L.; Marathe, M.; Eubank, S.; Blackburn, J.K. Modeling of Wildlife-Associated Zoonoses: Applications and Caveats. Vector-Borne Zoonotic Dis. 2012, 12, 1005–1018. [Google Scholar] [CrossRef]
- Chinery, W.A. Impact of Rapid Urbanization on Mosquitoes and Their Disease Transmission Potential in Accra and Tema, Ghana. Afr. J. Med. Med. Sci. 1995, 24, 179–188. [Google Scholar]
- Daszak, P.; Cunningham, A.A.; Hyatt, A.D. Emerging Infectious Diseases of Wildlife—Threats to Biodiversity and Human Health. Science 2000, 287, 443–449. [Google Scholar] [CrossRef]
- Power, A.G.; Mitchell, C.E. Pathogen Spillover in Disease Epidemics. Am. Nat. 2004, 164 (Suppl. S5), S79–S89. [Google Scholar] [CrossRef]
- Borremans, B.; Faust, C.; Manlove, K.R.; Sokolow, S.H.; Lloyd-Smith, J.O. Cross-Species Pathogen Spillover across Ecosystem Boundaries: Mechanisms and Theory. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20180344. [Google Scholar] [CrossRef]
- Wilkinson, D.A.; Marshall, J.C.; French, N.P.; Hayman, D.T.S. Habitat Fragmentation, Biodiversity Loss and the Risk of Novel Infectious Disease Emergence. J. R. Soc. Interface 2018, 15, 20180403. [Google Scholar] [CrossRef]
- Wood, C.L.; Lafferty, K.D.; DeLeo, G.; Young, H.S.; Hudson, P.J.; Kuris, A.M. Does Biodiversity Protect Humans against Infectious Disease? Ecology 2014, 95, 817–832. [Google Scholar] [CrossRef]
- Keesing, F.; Belden, L.K.; Daszak, P.; Dobson, A.; Harvell, C.D.; Holt, R.D.; Hudson, P.; Jolles, A.; Jones, K.E.; Mitchell, C.E.; et al. Impacts of Biodiversity on the Emergence and Transmission of Infectious Diseases. Nature 2010, 468, 647–652. [Google Scholar] [CrossRef]
- Roche, B.; Dobson, A.P.; Guégan, J.-F.; Rohani, P. Linking Community and Disease Ecology: The Impact of Biodiversity on Pathogen Transmission. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 2807–2813. [Google Scholar] [CrossRef]
- Murray, K.A.; Daszak, P. Human Ecology in Pathogenic Landscapes: Two Hypotheses on How Land Use Change Drives Viral Emergence. Curr. Opin. Virol. 2013, 3, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Plowright, R.K.; Parrish, C.R.; McCallum, H.; Hudson, P.J.; Ko, A.I.; Graham, A.L.; Lloyd-Smith, J.O. Pathways to Zoonotic Spillover. Nat. Rev. Microbiol. 2017, 15, 502–510. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Mortality and Global Health Estimates. Available online: https://www.who.int/data/gho/data/themes/mortality-and-global-health-estimates (accessed on 19 August 2024).
- Dhama, K.; Khan, S.; Tiwari, R.; Sircar, S.; Bhat, S.; Malik, Y.S.; Singh, K.P.; Chaicumpa, W.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. Coronavirus Disease 2019-COVID-19. Clin. Microbiol. Rev. 2020, 33, e00028-20. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global Trends in Emerging Infectious Diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef] [PubMed]
- Barro, R.J.; Ursúa, J.F. Macroeconomics of the Great Influenza Pandemic, 1918–1920. Res. Econ. Ric. Econ. 2022, 76, 21–29. [Google Scholar] [CrossRef]
- Naseer, S.; Khalid, S.; Parveen, S.; Abbass, K.; Song, H.; Achim, M.V. COVID-19 Outbreak: Impact on Global Economy. Front. Public Health 2023, 10, 1009393. [Google Scholar] [CrossRef]
- Carroll, D.; Daszak, P.; Wolfe, N.D.; Gao, G.F.; Morel, C.M.; Morzaria, S.; Pablos-Méndez, A.; Tomori, O.; Mazet, J.A.K. The Global Virome Project. Science 2018, 359, 872–874. [Google Scholar] [CrossRef]
- Carlson, C.J.; Zipfel, C.M.; Garnier, R.; Bansal, S. Global Estimates of Mammalian Viral Diversity Accounting for Host Sharing. Nat. Ecol. Evol. 2019, 3, 1070–1075. [Google Scholar] [CrossRef]
- Wacharapluesadee, S.; Tan, C.W.; Maneeorn, P.; Duengkae, P.; Zhu, F.; Joyjinda, Y.; Kaewpom, T.; Chia, W.N.; Ampoot, W.; Lim, B.L.; et al. Evidence for SARS-CoV-2 Related Coronaviruses Circulating in Bats and Pangolins in Southeast Asia. Nat. Commun. 2021, 12, 972. [Google Scholar] [CrossRef]
- Latinne, A.; Hu, B.; Olival, K.J.; Zhu, G.; Zhang, L.; Li, H.; Chmura, A.A.; Field, H.E.; Zambrana-Torrelio, C.; Epstein, J.H.; et al. Origin and Cross-Species Transmission of Bat Coronaviruses in China. Nat. Commun. 2020, 11, 4235. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.A.L.; Andrade, A.C.D.S.P.; Boratto, P.V.d.M.; Trindade, G.d.S.; Kroon, E.G.; Abrahão, J.S. An Anthropocentric View of the Virosphere-Host Relationship. Front. Microbiol. 2017, 8, 1673. [Google Scholar] [CrossRef] [PubMed]
- Taylor, L.H.; Latham, S.M.; Woolhouse, M.E. Risk Factors for Human Disease Emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001, 356, 983–989. [Google Scholar] [CrossRef]
- Woolhouse, M.E.J.; Gowtage-Sequeria, S. Host Range and Emerging and Reemerging Pathogens. Emerg. Infect. Dis. 2005, 11, 1842–1847. [Google Scholar] [CrossRef]
- Woolhouse, M.; Gaunt, E. Ecological Origins of Novel Human Pathogens. Crit. Rev. Microbiol. 2007, 33, 231–242. [Google Scholar] [CrossRef]
- Olival, K.J.; Hosseini, P.R.; Zambrana-Torrelio, C.; Ross, N.; Bogich, T.L.; Daszak, P. Host and Viral Traits Predict Zoonotic Spillover from Mammals. Nature 2017, 546, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Cleaveland, S.; Laurenson, M.K.; Taylor, L.H. Diseases of Humans and Their Domestic Mammals: Pathogen Characteristics, Host Range and the Risk of Emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001, 356, 991–999. [Google Scholar] [CrossRef]
- Luis, A.D.; Hayman, D.T.S.; O’Shea, T.J.; Cryan, P.M.; Gilbert, A.T.; Pulliam, J.R.C.; Mills, J.N.; Timonin, M.E.; Willis, C.K.R.; Cunningham, A.A.; et al. A Comparison of Bats and Rodents as Reservoirs of Zoonotic Viruses: Are Bats Special? Proc. Biol. Sci. 2013, 280, 20122753. [Google Scholar] [CrossRef]
- Han, B.A.; Kramer, A.M.; Drake, J.M. Global Patterns of Zoonotic Disease in Mammals. Trends Parasitol. 2016, 32, 565–577. [Google Scholar] [CrossRef]
- Brook, C.E.; Dobson, A.P. Bats as “special” Reservoirs for Emerging Zoonotic Pathogens. Trends Microbiol. 2015, 23, 172–180. [Google Scholar] [CrossRef]
- Brook, C.E.; Boots, M.; Chandran, K.; Dobson, A.P.; Drosten, C.; Graham, A.L.; Grenfell, B.T.; Müller, M.A.; Ng, M.; Wang, L.-F.; et al. Accelerated Viral Dynamics in Bat Cell Lines, with Implications for Zoonotic Emergence. eLife 2020, 9, e48401. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Baker, M.L.; Kulcsar, K.; Misra, V.; Plowright, R.; Mossman, K. Novel Insights Into Immune Systems of Bats. Front. Immunol. 2020, 11, 26. [Google Scholar] [CrossRef]
- Zhang, G.; Cowled, C.; Shi, Z.; Huang, Z.; Bishop-Lilly, K.A.; Fang, X.; Wynne, J.W.; Xiong, Z.; Baker, M.L.; Zhao, W.; et al. Comparative Analysis of Bat Genomes Provides Insight into the Evolution of Flight and Immunity. Science 2013, 339, 456–460. [Google Scholar] [CrossRef]
- Irving, A.T.; Ahn, M.; Goh, G.; Anderson, D.E.; Wang, L.-F. Lessons from the Host Defences of Bats, a Unique Viral Reservoir. Nature 2021, 589, 363–370. [Google Scholar] [CrossRef]
- Nunes, H.; Rocha, F.L.; Cordeiro-Estrela, P. Bats in Urban Areas of Brazil: Roosts, Food Resources and Parasites in Disturbed Environments. Urban Ecosyst. 2017, 20, 953–969. [Google Scholar] [CrossRef]
- Eskew, E.A.; Bird, B.H.; Ghersi, B.M.; Bangura, J.; Basinski, A.J.; Amara, E.; Bah, M.A.; Kanu, M.C.; Kanu, O.T.; Lavalie, E.G.; et al. Reservoir Displacement by an Invasive Rodent Reduces Lassa Virus Zoonotic Spillover Risk. Nat. Commun. 2024, 15, 3589. [Google Scholar] [CrossRef] [PubMed]
- Williams, E.P.; Spruill-Harrell, B.M.; Taylor, M.K.; Lee, J.; Nywening, A.V.; Yang, Z.; Nichols, J.H.; Camp, J.V.; Owen, R.D.; Jonsson, C.B. Common Themes in Zoonotic Spillover and Disease Emergence: Lessons Learned from Bat- and Rodent-Borne RNA Viruses. Viruses 2021, 13, 1509. [Google Scholar] [CrossRef] [PubMed]
- Mollentze, N.; Babayan, S.A.; Streicker, D.G. Identifying and Prioritizing Potential Human-Infecting Viruses from Their Genome Sequences. PLoS Biol. 2021, 19, e3001390. [Google Scholar] [CrossRef]
- Halsby, K.D.; Walsh, A.L.; Smith, R.; Said, B.; Kirkbride, H.; Smyth, B.; Browning, L.; Larkin, L.; Morgan, D. The Health Burden of Orphan Zoonotic Disease in the United Kingdom, 2005–2009. Zoonoses Public Health 2014, 61, 39–47. [Google Scholar] [CrossRef]
- Ge, X.-Y.; Li, J.-L.; Yang, X.-L.; Chmura, A.A.; Zhu, G.; Epstein, J.H.; Mazet, J.K.; Hu, B.; Zhang, W.; Peng, C.; et al. Isolation and Characterization of a Bat SARS-like Coronavirus That Uses the ACE2 Receptor. Nature 2013, 503, 535–538. [Google Scholar] [CrossRef]
- Eco Health Alliance PREDICT. Available online: https://www.ecohealthalliance.org/program/predict (accessed on 19 August 2024).
- Lloyd-Smith, J.O. Infectious Diseases: Predictions of Virus Spillover Across Species. Nature 2017, 546, 603–604. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.; Shackelton, L.A.; Holmes, E.C. Rates of Evolutionary Change in Viruses: Patterns and Determinants. Nat. Rev. Genet. 2008, 9, 267–276. [Google Scholar] [CrossRef]
- Jenkins, G.M.; Rambaut, A.; Pybus, O.G.; Holmes, E.C. Rates of Molecular Evolution in RNA Viruses: A Quantitative Phylogenetic Analysis. J. Mol. Evol. 2002, 54, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Eigen, M.; Winkler-Oswatitsch, R.; Woolley, P. Steps Towards Life: A Perspective on Evolution; Oxford university press: Oxford, UK; New York, NY, USA; Tokyo, Japan, 1992; ISBN 978-0-19-854751-8. [Google Scholar]
- Holmes, E.C. Error Thresholds and the Constraints to RNA Virus Evolution. Trends Microbiol. 2003, 11, 543–546. [Google Scholar] [CrossRef]
- Rambaut, A.; Holmes, E. The Early Molecular Epidemiology of the Swine-Origin A/H1N1 Human Influenza Pandemic. PLoS Curr. 2009, 1, RRN1003. [Google Scholar] [CrossRef]
- Robson, F.; Khan, K.S.; Le, T.K.; Paris, C.; Demirbag, S.; Barfuss, P.; Rocchi, P.; Ng, W.-L. Coronavirus RNA Proofreading: Molecular Basis and Therapeutic Targeting. Mol. Cell 2020, 79, 710–727. [Google Scholar] [CrossRef] [PubMed]
- Konstantoulas, C.J.; Lamp, B.; Rumenapf, T.H.; Indik, S. Single Amino Acid Substitution (G42E) in the Receptor Binding Domain of Mouse Mammary Tumour Virus Envelope Protein Facilitates Infection of Non-Murine Cells in a Transferrin Receptor 1-Independent Manner. Retrovirology 2015, 12, 43. [Google Scholar] [CrossRef][Green Version]
- Baranowski, E.; Ruiz-Jarabo, C.M.; Pariente, N.; Verdaguer, N.; Domingo, E. Evolution of Cell Recognition by Viruses: A Source of Biological Novelty with Medical Implications. Adv. Virus Res. 2003, 62, 19–111. [Google Scholar] [CrossRef]
- Brault, A.C.; Powers, A.M.; Ortiz, D.; Estrada-Franco, J.G.; Navarro-Lopez, R.; Weaver, S.C. Venezuelan Equine Encephalitis Emergence: Enhanced Vector Infection from a Single Amino Acid Substitution in the Envelope Glycoprotein. Proc. Natl. Acad. Sci. USA 2004, 101, 11344–11349. [Google Scholar] [CrossRef]
- Sanjuán, R. Mutational Fitness Effects in RNA and Single-Stranded DNA Viruses: Common Patterns Revealed by Site-Directed Mutagenesis Studies. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 1975–1982. [Google Scholar] [CrossRef]
- Krakauer, D.C.; Komarova, N.L. Levels of Selection in Positive-Strand Virus Dynamics. J. Evol. Biol. 2003, 16, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.; Turner, P.E.; Burch, C.L. Pleiotropic Costs of Niche Expansion in the RNA Bacteriophage Phi 6. Genetics 2006, 172, 751–757. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, N.D.; Dunavan, C.P.; Diamond, J. Origins of Major Human Infectious Diseases. Nature 2007, 447, 279–283. [Google Scholar] [CrossRef]
- Woelk, C.H.; Holmes, E.C. Reduced Positive Selection in Vector-Borne RNA Viruses. Mol. Biol. Evol. 2002, 19, 2333–2336. [Google Scholar] [CrossRef]
- Baranowski, E.; Ruiz-Jarabo, C.M.; Domingo, E. Evolution of Cell Recognition by Viruses. Science 2001, 292, 1102–1105. [Google Scholar] [CrossRef]
- Wichman, H.A.; Badgett, M.R.; Scott, L.A.; Boulianne, C.M.; Bull, J.J. Different Trajectories of Parallel Evolution during Viral Adaptation. Science 1999, 285, 422–424. [Google Scholar] [CrossRef]
- Fares, M.A.; Moya, A.; Escarmís, C.; Baranowski, E.; Domingo, E.; Barrio, E. Evidence for Positive Selection in the Capsid Protein-Coding Region of the Foot-and-Mouth Disease Virus (FMDV) Subjected to Experimental Passage Regimens. Mol. Biol. Evol. 2001, 18, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Loverdo, C.; Lloyd-Smith, J.O. Evolutionary Invasion and Escape in the Presence of Deleterious Mutations. PLoS ONE 2013, 8, e68179. [Google Scholar] [CrossRef][Green Version]
- Lynch, M. Mutation Pressure, Drift, and the Pace of Molecular Coevolution. Proc. Natl. Acad. Sci. USA 2023, 120, e2306741120. [Google Scholar] [CrossRef]
- Eshel, I. Clone-Selection and Optimal Rates of Mutation. J. Appl. ProbaB 1973, 10, 728–738. [Google Scholar] [CrossRef]
- Alexander, H.K.; Day, T. Risk Factors for the Evolutionary Emergence of Pathogens. J. R. Soc. Interface 2010, 7, 1455–1474. [Google Scholar] [CrossRef] [PubMed]
- Cleaveland, S.; Haydon, D.T.; Taylor, L. Overviews of Pathogen Emergence: Which Pathogens Emerge, When and Why? Curr. Top. Microbiol. Immunol. 2007, 315, 85–111. [Google Scholar] [CrossRef] [PubMed]
- Sturtevant, A.H. Essays on Evolution. I. On the Effects of Selection on Mutation Rate. Q. Rev. Biol. 1937, 12, 464–467. [Google Scholar] [CrossRef]
- Sniegowski, P.D.; Gerrish, P.J.; Johnson, T.; Shaver, A. The Evolution of Mutation Rates: Separating Causes from Consequences. BioEssays News Rev. Mol. Cell. Dev. Biol. 2000, 22, 1057–1066. [Google Scholar] [CrossRef]
- Furió, V.; Moya, A.; Sanjuán, R. The Cost of Replication Fidelity in an RNA Virus. Proc. Natl. Acad. Sci. USA 2005, 102, 10233–10237. [Google Scholar] [CrossRef]
- Regoes, R.R.; Hamblin, S.; Tanaka, M.M. Viral Mutation Rates: Modelling the Roles of within-Host Viral Dynamics and the Trade-off between Replication Fidelity and Speed. Proc. Biol. Sci. 2013, 280, 20122047. [Google Scholar] [CrossRef]
- Denamur, E.; Matic, I. Evolution of Mutation Rates in Bacteria. Mol. Microbiol. 2006, 60, 820–827. [Google Scholar] [CrossRef]
- André, J.-B.; Godelle, B. The Evolution of Mutation Rate in Finite Asexual Populations. Genetics 2006, 172, 611–626. [Google Scholar] [CrossRef][Green Version]
- Leigh, E.G. Natural Selection and Mutability. Am. Nat. 1970, 104, 301–305. [Google Scholar] [CrossRef]
- Baer, C.F.; Miyamoto, M.M.; Denver, D.R. Mutation Rate Variation in Multicellular Eukaryotes: Causes and Consequences. Nat. Rev. Genet. 2007, 8, 619–631. [Google Scholar] [CrossRef]
- Bonhoeffer, S.; Nowak, M.A. Intra-Host versus Inter-Host Selection: Viral Strategies of Immune Function Impairment. Proc. Natl. Acad. Sci. USA 1994, 91, 8062–8066. [Google Scholar] [CrossRef] [PubMed]
- MacArthur, R.H. Geographical Ecology: Patterns in the Distribution of Species; Princeton University Press: Princeton, NJ, USA, 1984; ISBN 978-0-691-08353-7. [Google Scholar]
- Via, S. Ecological Genetics and Host Adaptation in Herbivorous Insects: The Experimental Study of Evolution in Natural and Agricultural Systems. Annu. Rev. Entomol. 1990, 35, 421–446. [Google Scholar] [CrossRef] [PubMed]
- Lalić, J.; Cuevas, J.M.; Elena, S.F. Effect of Host Species on the Distribution of Mutational Fitness Effects for an RNA Virus. PLoS Genet. 2011, 7, e1002378. [Google Scholar] [CrossRef]
- Whitlock, M.C.; Phillips, P.C.; Moore, F.B.-G.; Tonsor, S.J. Multiple Fitness Peaks and Epistasis. Annu. Rev. Ecol. Syst. 1995, 26, 601–629. [Google Scholar] [CrossRef]
- Poelwijk, F.J.; Tănase-Nicola, S.; Kiviet, D.J.; Tans, S.J. Reciprocal Sign Epistasis Is a Necessary Condition for Multi-Peaked Fitness Landscapes. J. Theor. Biol. 2011, 272, 141–144. [Google Scholar] [CrossRef]
- Bedhomme, S.; Hillung, J.; Elena, S.F. Emerging Viruses: Why They Are Not Jacks of All Trades? Curr. Opin. Virol. 2015, 10, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Devi, S. First Fatality from Alaskapox Virus. Lancet Infect. Dis. 2024, 24, e282. [Google Scholar] [CrossRef]
- Phillips, P.C. Epistasis--the Essential Role of Gene Interactions in the Structure and Evolution of Genetic Systems. Nat. Rev. Genet. 2008, 9, 855–867. [Google Scholar] [CrossRef]
- Wolf, J.B.; Wade, M.J.; Brodie, E.D. Epistasis and the Evolutionary Process; Oxford University Press: Oxford, UK, 2000; ISBN 978-0-19-512806-2. [Google Scholar]
- Phillips, P.C. The Language of Gene Interaction. Genetics 1998, 149, 1167–1171. [Google Scholar] [CrossRef]
- Fisher, R.A. The Genetical Theory of Natural Selection; Dover Pubblications: New York, NY, USA, 1958. [Google Scholar]
- Martin, G.; Elena, S.F.; Lenormand, T. Distributions of Epistasis in Microbes Fit Predictions from a Fitness Landscape Model. Nat. Genet. 2007, 39, 555–560. [Google Scholar] [CrossRef]
- Kondrashov, F.A.; Kondrashov, A.S. Multidimensional Epistasis and the Disadvantage of Sex. Proc. Natl. Acad. Sci. USA 2001, 98, 12089–12092. [Google Scholar] [CrossRef] [PubMed]
- Remold, S. Understanding Specialism When the Jack of All Trades Can Be the Master of All. Proc. Biol. Sci. 2012, 279, 4861–4869. [Google Scholar] [CrossRef] [PubMed]
- Lehner, B. Molecular Mechanisms of Epistasis within and between Genes. Trends Genet. TIG 2011, 27, 323–331. [Google Scholar] [CrossRef]
- McCallum, M.; Walls, A.C.; Sprouse, K.R.; Bowen, J.E.; Rosen, L.E.; Dang, H.V.; De Marco, A.; Franko, N.; Tilles, S.W.; Logue, J.; et al. Molecular Basis of Immune Evasion by the Delta and Kappa SARS-CoV-2 Variants. Science 2021, 374, 1621–1626. [Google Scholar] [CrossRef]
- Carabelli, A.M.; Peacock, T.P.; Thorne, L.G.; Harvey, W.T.; Hughes, J.; COVID-19 Genomics UK Consortium; De Silva, T.I.; Peacock, S.J.; Barclay, W.S.; De Silva, T.I.; et al. SARS-CoV-2 Variant Biology: Immune Escape, Transmission and Fitness. Nat. Rev. Microbiol. 2023, 21, 162–177. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Arenas, M.; Galán, J.C.; Palero, F.; González-Candelas, F. Recombination in Viruses: Mechanisms, Methods of Study, and Evolutionary Consequences. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2015, 30, 296–307. [Google Scholar] [CrossRef]
- Martin, D.P.; Biagini, P.; Lefeuvre, P.; Golden, M.; Roumagnac, P.; Varsani, A. Recombination in Eukaryotic Single Stranded DNA Viruses. Viruses 2011, 3, 1699–1738. [Google Scholar] [CrossRef] [PubMed]
- Simon-Loriere, E.; Holmes, E.C. Why Do RNA Viruses Recombine? Nat. Rev. Microbiol. 2011, 9, 617–626. [Google Scholar] [CrossRef]
- Stedman, K.M. Deep Recombination: RNA and ssDNA Virus Genes in DNA Virus and Host Genomes. Annu. Rev. Virol. 2015, 2, 203–217. [Google Scholar] [CrossRef]
- Domingo, E. Mechanisms of Viral Emergence. Vet. Res. 2010, 41, 38. [Google Scholar] [CrossRef]
- Smith, G.J.D.; Vijaykrishna, D.; Bahl, J.; Lycett, S.J.; Worobey, M.; Pybus, O.G.; Ma, S.K.; Cheung, C.L.; Raghwani, J.; Bhatt, S.; et al. Origins and Evolutionary Genomics of the 2009 Swine-Origin H1N1 Influenza A Epidemic. Nature 2009, 459, 1122–1125. [Google Scholar] [CrossRef] [PubMed]
- Stern, A.; Bianco, S.; Yeh, M.T.; Wright, C.; Butcher, K.; Tang, C.; Nielsen, R.; Andino, R. Costs and Benefits of Mutational Robustness in RNA Viruses. Cell Rep. 2014, 8, 1026–1036. [Google Scholar] [CrossRef] [PubMed]
- Domingo, E. Virology Evolution. In Fields virology; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; ISBN 978-0-7817-6060-7. [Google Scholar]
- Thébaud, G.; Chadoeuf, J.; Morelli, M.J.; McCauley, J.W.; Haydon, D.T. The Relationship Between Mutation Frequency and Replication Strategy in Positive-Sense Single-Stranded RNA Viruses. Proc. Biol. Sci. 2010, 277, 809–817. [Google Scholar] [CrossRef]
- Crow, J.F.; Kimura, M. An Introduction to Population Genetics Theory; Harper&Row: New York, NY, USA; Scient. Publ.: New York, NY, USA, 1970. [Google Scholar]
- Smith, J.M. Natural Selection and the Concept of a Protein Space. Nature 1970, 225, 563–564. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. Model of Effectively Neutral Mutations in Which Selective Constraint Is Incorporated. Proc. Natl. Acad. Sci. USA 1979, 76, 3440–3444. [Google Scholar] [CrossRef]
- Gillespie, J.H. The Causes of Molecular Evolution; Oxford Series in Ecology and Evolution; Oxford University Press: New York, NY, USA, 1991; ISBN 978-0-19-506883-2. [Google Scholar]
- Gumbel, E.J. Statistics of Extremes; Columbia University Press: New York, NY, USA, 1958. [Google Scholar]
- Leadbetter, M.R.; Lindgren, G.; Rootzén, H. Extremes and Related Properties of Random Sequences and Processes; Springer Series in Statistics; Springer: New York, NY, USA; Berlin/Heidelberg, Germany, 1983; ISBN 978-1-4612-5451-5. [Google Scholar]
- Glidden, C.K.; Nova, N.; Kain, M.P.; Lagerstrom, K.M.; Skinner, E.B.; Mandle, L.; Sokolow, S.H.; Plowright, R.K.; Dirzo, R.; De Leo, G.A.; et al. Human-Mediated Impacts on Biodiversity and the Consequences for Zoonotic Disease Spillover. Curr. Biol. CB 2021, 31, R1342–R1361. [Google Scholar] [CrossRef]
- Ellwanger, J.H.; Chies, J.A.B. Zoonotic Spillover: Understanding Basic Aspects for Better Prevention. Genet. Mol. Biol. 2021, 44, e20200355. [Google Scholar] [CrossRef]
- Lehmer, E.M.; Korb, J.; Bombaci, S.; McLean, N.; Ghachu, J.; Hart, L.; Kelly, A.; Jara-Molinar, E.; O’Brien, C.; Wright, K. The Interplay of Plant and Animal Disease in a Changing Landscape: The Role of Sudden Aspen Decline in Moderating Sin Nombre Virus Prevalence in Natural Deer Mouse Populations. EcoHealth 2012, 9, 205–216. [Google Scholar] [CrossRef]
- Field, H.; Young, P.; Yob, J.M.; Mills, J.; Hall, L.; Mackenzie, J. The Natural History of Hendra and Nipah Viruses. Microbes Infect. 2001, 3, 307–314. [Google Scholar] [CrossRef]
- Tewari, P.; Guo, P.; Dickens, B.; Ma, P.; Bansal, S.; Lim, J.T. Associations Between Dengue Incidence, Ecological Factors, and Anthropogenic Factors in Singapore. Viruses 2023, 15, 1917. [Google Scholar] [CrossRef]
- Giesen, C.; Herrador, Z.; Fernandez-Martinez, B.; Figuerola, J.; Gangoso, L.; Vazquez, A.; Gómez-Barroso, D. A Systematic Review of Environmental Factors Related to WNV Circulation in European and Mediterranean Countries. One Health 2023, 16, 100478. [Google Scholar] [CrossRef]
- Fuller, T.L.; Calvet, G.; Genaro Estevam, C.; Rafael Angelo, J.; Abiodun, G.J.; Halai, U.-A.; De Santis, B.; Carvalho Sequeira, P.; Machado Araujo, E.; Alves Sampaio, S.; et al. Behavioral, Climatic, and Environmental Risk Factors for Zika and Chikungunya Virus Infections in Rio de Janeiro, Brazil, 2015–2016. PLoS ONE 2017, 12, e0188002. [Google Scholar] [CrossRef]
- Düx, A.; Lequime, S.; Patrono, L.V.; Vrancken, B.; Boral, S.; Gogarten, J.F.; Hilbig, A.; Horst, D.; Merkel, K.; Prepoint, B.; et al. Measles Virus and Rinderpest Virus Divergence Dated to the Sixth Century BCE. Science 2020, 368, 1367–1370. [Google Scholar] [CrossRef]
- Morand, S. Emerging Diseases, Livestock Expansion and Biodiversity Loss Are Positively Related at Global Scale. Biol. Conserv. 2020, 248, 108707. [Google Scholar] [CrossRef]
- Lemey, P.; Rambaut, A.; Pybus, O.G. HIV Evolutionary Dynamics within and among Hosts. AIDS Rev. 2006, 8, 125–140. [Google Scholar]
- Holmes, E.C. Viral Evolution in the Genomic Age. PLoS Biol. 2007, 5, e278. [Google Scholar] [CrossRef][Green Version]
- Poon, L.L.M.; Song, T.; Rosenfeld, R.; Lin, X.; Rogers, M.B.; Zhou, B.; Sebra, R.; Halpin, R.A.; Guan, Y.; Twaddle, A.; et al. Quantifying Influenza Virus Diversity and Transmission in Humans. Nat. Genet. 2016, 48, 195–200. [Google Scholar] [CrossRef]
- Boeras, D.I.; Hraber, P.T.; Hurlston, M.; Evans-Strickfaden, T.; Bhattacharya, T.; Giorgi, E.E.; Mulenga, J.; Karita, E.; Korber, B.T.; Allen, S.; et al. Role of Donor Genital Tract HIV-1 Diversity in the Transmission Bottleneck. Proc. Natl. Acad. Sci. USA 2011, 108, E1156–E1163. [Google Scholar] [CrossRef]
- Wilker, P.R.; Dinis, J.M.; Starrett, G.; Imai, M.; Hatta, M.; Nelson, C.W.; O’Connor, D.H.; Hughes, A.L.; Neumann, G.; Kawaoka, Y.; et al. Selection on Haemagglutinin Imposes a Bottleneck during Mammalian Transmission of Reassortant H5N1 Influenza Viruses. Nat. Commun. 2013, 4, 2636. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Moncla, L.H.; Zhong, G.; Nelson, C.W.; Dinis, J.M.; Mutschler, J.; Hughes, A.L.; Watanabe, T.; Kawaoka, Y.; Friedrich, T.C. Selective Bottlenecks Shape Evolutionary Pathways Taken during Mammalian Adaptation of a 1918-like Avian Influenza Virus. Cell Host Microbe 2016, 19, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Alizon, S.; Fraser, C. Within-Host and Between-Host Evolutionary Rates across the HIV-1 Genome. Retrovirology 2013, 10, 49. [Google Scholar] [CrossRef]
- Carlson, J.M.; Schaefer, M.; Monaco, D.C.; Batorsky, R.; Claiborne, D.T.; Prince, J.; Deymier, M.J.; Ende, Z.S.; Klatt, N.R.; DeZiel, C.E.; et al. HIV Transmission. Selection Bias at the Heterosexual HIV-1 Transmission Bottleneck. Science 2014, 345, 1254031. [Google Scholar] [CrossRef]
- Gog, J.R.; Pellis, L.; Wood, J.L.N.; McLean, A.R.; Arinaminpathy, N.; Lloyd-Smith, J.O. Seven Challenges in Modeling Pathogen Dynamics Within-Host and across Scales. Epidemics 2015, 10, 45–48. [Google Scholar] [CrossRef]
- Schreiber, S.J.; Ke, R.; Loverdo, C.; Park, M.; Ahsan, P.; Lloyd-Smith, J.O. Cross-Scale Dynamics and the Evolutionary Emergence of Infectious Diseases. Virus Evol. 2021, 7, veaa105. [Google Scholar] [CrossRef]
- Antia, R.; Regoes, R.R.; Koella, J.C.; Bergstrom, C.T. The Role of Evolution in the Emergence of Infectious Diseases. Nature 2003, 426, 658–661. [Google Scholar] [CrossRef]
- Park, M.; Loverdo, C.; Schreiber, S.J.; Lloyd-Smith, J.O. Multiple Scales of Selection Influence the Evolutionary Emergence of Novel Pathogens. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20120333. [Google Scholar] [CrossRef]
- André, J.-B.; Day, T. The Effect of Disease Life History on the Evolutionary Emergence of Novel Pathogens. Proc. R. Soc. B Biol. Sci. 2005, 272, 1949–1956. [Google Scholar] [CrossRef]
- Anderson, R.M.; May, R.M. Directly Transmitted Infections Diseases: Control by Vaccination. Science 1982, 215, 1053–1060. [Google Scholar] [CrossRef]
- Woolhouse, M.E.J.; Brierley, L.; McCaffery, C.; Lycett, S. Assessing the Epidemic Potential of RNA and DNA Viruses. Emerg. Infect. Dis. 2016, 22, 2037–2044. [Google Scholar] [CrossRef]
- Gostic, K.M.; Ambrose, M.; Worobey, M.; Lloyd-Smith, J.O. Potent Protection against H5N1 and H7N9 Influenza via Childhood Hemagglutinin Imprinting. Science 2016, 354, 722–726. [Google Scholar] [CrossRef]
- Fine, P.E.M.; Jezek, Z.; Grab, B.; Dixon, H. The Transmission Potential of Monkeypox Virus in Human Populations. Int. J. Epidemiol. 1988, 17, 643–650. [Google Scholar] [CrossRef]
- Mummah, R.O.; Hoff, N.A.; Rimoin, A.W.; Lloyd-Smith, J.O. Controlling Emerging Zoonoses at the Animal-Human Interface. One Health Outlook 2020, 2, 17. [Google Scholar] [CrossRef]
- Zwart, M.P.; Elena, S.F. Matters of Size: Genetic Bottlenecks in Virus Infection and Their Potential Impact on Evolution. Annu. Rev. Virol. 2015, 2, 161–179. [Google Scholar] [CrossRef]
- McCrone, J.T.; Lauring, A.S. Genetic Bottlenecks in Intraspecies Virus Transmission. Curr. Opin. Virol. 2018, 28, 20–25. [Google Scholar] [CrossRef]
- Feng, Z.; Hensley, L.; McKnight, K.L.; Hu, F.; Madden, V.; Ping, L.; Jeong, S.-H.; Walker, C.; Lanford, R.E.; Lemon, S.M. A Pathogenic Picornavirus Acquires an Envelope by Hijacking Cellular Membranes. Nature 2013, 496, 367–371. [Google Scholar] [CrossRef]
- Arantes, T.S.; Rodrigues, R.A.L.; Dos Santos Silva, L.K.; Oliveira, G.P.; de Souza, H.L.; Khalil, J.Y.B.; de Oliveira, D.B.; Torres, A.A.; da Silva, L.L.; Colson, P.; et al. The Large Marseillevirus Explores Different Entry Pathways by Forming Giant Infectious Vesicles. J. Virol. 2016, 90, 5246–5255. [Google Scholar] [CrossRef]
- Santiana, M.; Ghosh, S.; Ho, B.A.; Rajasekaran, V.; Du, W.-L.; Mutsafi, Y.; De Jésus-Diaz, D.A.; Sosnovtsev, S.V.; Levenson, E.A.; Parra, G.I.; et al. Vesicle-Cloaked Virus Clusters Are Optimal Units for Inter-Organismal Viral Transmission. Cell Host Microbe 2018, 24, 208–220.e8. [Google Scholar] [CrossRef]
- Lago, M.; Rodríguez, J.F.; Bandín, I.; Dopazo, C.P. Aquabirnavirus Polyploidy: A New Strategy to Modulate Virulence? J. Gen. Virol. 2016, 97, 1168–1177. [Google Scholar] [CrossRef]
- Vignuzzi, M.; Stone, J.K.; Arnold, J.J.; Cameron, C.E.; Andino, R. Quasispecies Diversity Determines Pathogenesis Through Cooperative Interactions in a Viral Population. Nature 2006, 439, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Andino, R.; Domingo, E. Viral Quasispecies. Virology 2015, 479–480, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Leeks, A.; Segredo-Otero, E.A.; Sanjuán, R.; West, S.A. Beneficial Coinfection Can Promote Within-Host Viral Diversity. Virus Evol. 2018, 4, vey028. [Google Scholar] [CrossRef]
- Lauring, A.S.; Andino, R. Quasispecies Theory and the Behavior of RNA Viruses. PLoS Pathog. 2010, 6, e1001005. [Google Scholar] [CrossRef]
- Domingo, E.; Perales, C. Quasispecies and Virus. Eur. Biophys. J. EBJ 2018, 47, 443–457. [Google Scholar] [CrossRef]
- Chen, Y.-H.; Du, W.; Hagemeijer, M.C.; Takvorian, P.M.; Pau, C.; Cali, A.; Brantner, C.A.; Stempinski, E.S.; Connelly, P.S.; Ma, H.-C.; et al. Phosphatidylserine Vesicles Enable Efficient En Bloc Transmission of Enteroviruses. Cell 2015, 160, 619–630. [Google Scholar] [CrossRef]
- Aguilera, E.R.; Erickson, A.K.; Jesudhasan, P.R.; Robinson, C.M.; Pfeiffer, J.K. Plaques Formed by Mutagenized Viral Populations Have Elevated Coinfection Frequencies. mBio 2017, 8, e02020-16. [Google Scholar] [CrossRef]
- Manzoni, T.B.; López, C.B. Defective (Interfering) Viral Genomes Re-Explored: Impact on Antiviral Immunity and Virus Persistence. Future Virol. 2018, 13, 493–503. [Google Scholar] [CrossRef]
- Brooke, C.B. Population Diversity and Collective Interactions during Influenza Virus Infection. J. Virol. 2017, 91, e01164-17. [Google Scholar] [CrossRef]
- Díaz-Muñoz, S.L. Viral Coinfection Is Shaped by Host Ecology and Virus-Virus Interactions across Diverse Microbial Taxa and Environments. Virus Evol. 2017, 3, vex011. [Google Scholar] [CrossRef]
- Chao, L.; Elena, S.F. Nonlinear Trade-Offs Allow the Cooperation Game to Evolve from Prisoner’s Dilemma to Snowdrift. Proc. Biol. Sci. 2017, 284, 20170228. [Google Scholar] [CrossRef]
- Segredo-Otero, E.; Sanjuán, R. The Effect of Genetic Complementation on the Fitness and Diversity of Viruses Spreading as Collective Infectious Units. Virus Res. 2019, 267, 41–48. [Google Scholar] [CrossRef]
- Bull, J.J.; Meyers, L.A.; Lachmann, M. Quasispecies Made Simple. PLoS Comput. Biol. 2005, 1, e61. [Google Scholar] [CrossRef]
- Domingo, E.; Escarmís, C.; Lázaro, E.; Manrubia, S.C. Quasispecies Dynamics and RNA Virus Extinction. Virus Res. 2005, 107, 129–139. [Google Scholar] [CrossRef]
- Becker, D.J.; Washburne, A.D.; Faust, C.L.; Pulliam, J.R.C.; Mordecai, E.A.; Lloyd-Smith, J.O.; Plowright, R.K. Dynamic and Integrative Approaches to Understanding Pathogen Spillover. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20190014. [Google Scholar] [CrossRef]
- Tsetsarkin, K.A.; Chen, R.; Weaver, S.C. Interspecies Transmission and Chikungunya Virus Emergence. Curr. Opin. Virol. 2016, 16, 143–150. [Google Scholar] [CrossRef]
- Daszak, P.; Plowright, R.K.; Epstein, J.H.; Pulliam, J.; Abdul Rahman, S.; Field, H.E.; Jamaluddin, A.; Sharifah, S.H.; Smith, C.S.; Olival, K.J.; et al. The Emergence of Nipah and Hendra Virus: Pathogen Dynamics across a Wildlife-Livestock-Human Continuum. In Disease Ecology: Community Structure and Pathogen Dynamics; Collinge, S.K., Ray, C., Eds.; Oxford University Press: Oxford, UK, 2006; pp. 186–201. ISBN 978-0-19-856708-0. [Google Scholar]
- Takata, M.A.; Gonçalves-Carneiro, D.; Zang, T.M.; Soll, S.J.; York, A.; Blanco-Melo, D.; Bieniasz, P.D. CG Dinucleotide Suppression Enables Antiviral Defence Targeting Non-Self RNA. Nature 2017, 550, 124–127. [Google Scholar] [CrossRef]
- Kustin, T.; Stern, A. Biased Mutation and Selection in RNA Viruses. Mol. Biol. Evol. 2021, 38, 575–588. [Google Scholar] [CrossRef]
- Greninger, A.L.; Naccache, S.N.; Federman, S.; Yu, G.; Mbala, P.; Bres, V.; Stryke, D.; Bouquet, J.; Somasekar, S.; Linnen, J.M.; et al. Rapid Metagenomic Identification of Viral Pathogens in Clinical Samples by Real-Time Nanopore Sequencing Analysis. Genome Med. 2015, 7, 99. [Google Scholar] [CrossRef]
- Quick, J.; Loman, N.J.; Duraffour, S.; Simpson, J.T.; Severi, E.; Cowley, L.; Bore, J.A.; Koundouno, R.; Dudas, G.; Mikhail, A.; et al. Real-Time, Portable Genome Sequencing for Ebola Surveillance. Nature 2016, 530, 228–232. [Google Scholar] [CrossRef]
- Ding, X.; Yin, K.; Li, Z.; Lalla, R.V.; Ballesteros, E.; Sfeir, M.M.; Liu, C. Ultrasensitive and Visual Detection of SARS-CoV-2 Using All-in-One Dual CRISPR-Cas12a Assay. Nat. Commun. 2020, 11, 4711. [Google Scholar] [CrossRef]
- Alizon, S. Predicting the Virulence of Future Emerging Zoonotic Viruses. PLoS Biol. 2023, 21, e3002286. [Google Scholar] [CrossRef]
- Brook, C.E.; Rozins, C.; Guth, S.; Boots, M. Reservoir Host Immunology and Life History Shape Virulence Evolution in Zoonotic Viruses. PLoS Biol. 2023, 21, e3002268. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pauciullo, S.; Zulian, V.; La Frazia, S.; Paci, P.; Garbuglia, A.R. Spillover: Mechanisms, Genetic Barriers, and the Role of Reservoirs in Emerging Pathogens. Microorganisms 2024, 12, 2191. https://doi.org/10.3390/microorganisms12112191
Pauciullo S, Zulian V, La Frazia S, Paci P, Garbuglia AR. Spillover: Mechanisms, Genetic Barriers, and the Role of Reservoirs in Emerging Pathogens. Microorganisms. 2024; 12(11):2191. https://doi.org/10.3390/microorganisms12112191
Chicago/Turabian StylePauciullo, Silvia, Verdiana Zulian, Simone La Frazia, Paola Paci, and Anna Rosa Garbuglia. 2024. "Spillover: Mechanisms, Genetic Barriers, and the Role of Reservoirs in Emerging Pathogens" Microorganisms 12, no. 11: 2191. https://doi.org/10.3390/microorganisms12112191
APA StylePauciullo, S., Zulian, V., La Frazia, S., Paci, P., & Garbuglia, A. R. (2024). Spillover: Mechanisms, Genetic Barriers, and the Role of Reservoirs in Emerging Pathogens. Microorganisms, 12(11), 2191. https://doi.org/10.3390/microorganisms12112191







