Probiotic Mixtures Consisting of Representatives of Bacteroidetes and Selenomonadales Increase Resistance of Newly Hatched Chicks to Salmonella Enteritidis Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Salmonella and Campylobacter Culture
2.3. Analysis of Gut Microbiota by Sequencing of V3/V4 Variable Region of 16S rRNA Gene
2.4. Statistics
2.5. Ethical Statement
3. Results
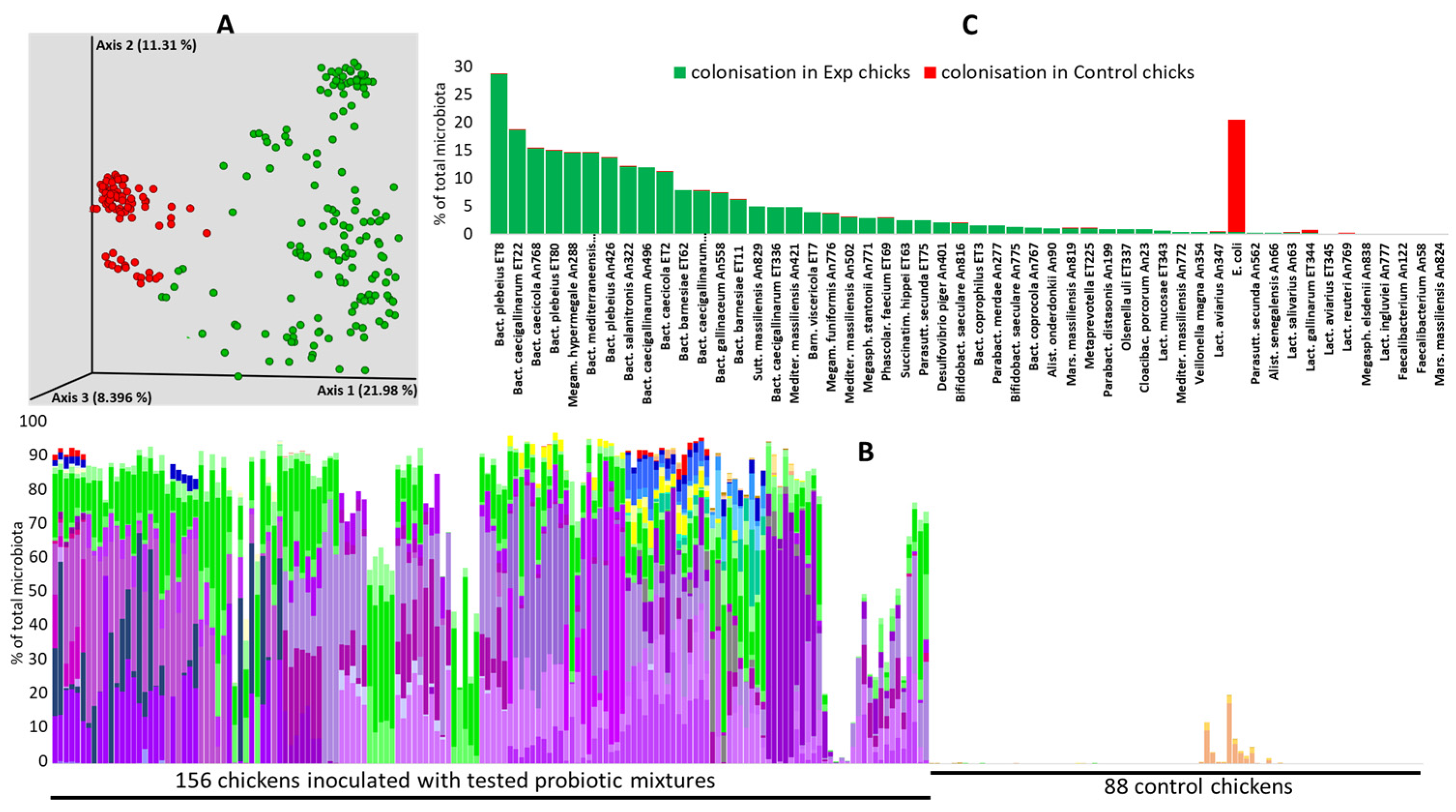
3.1. Colonisation Ability
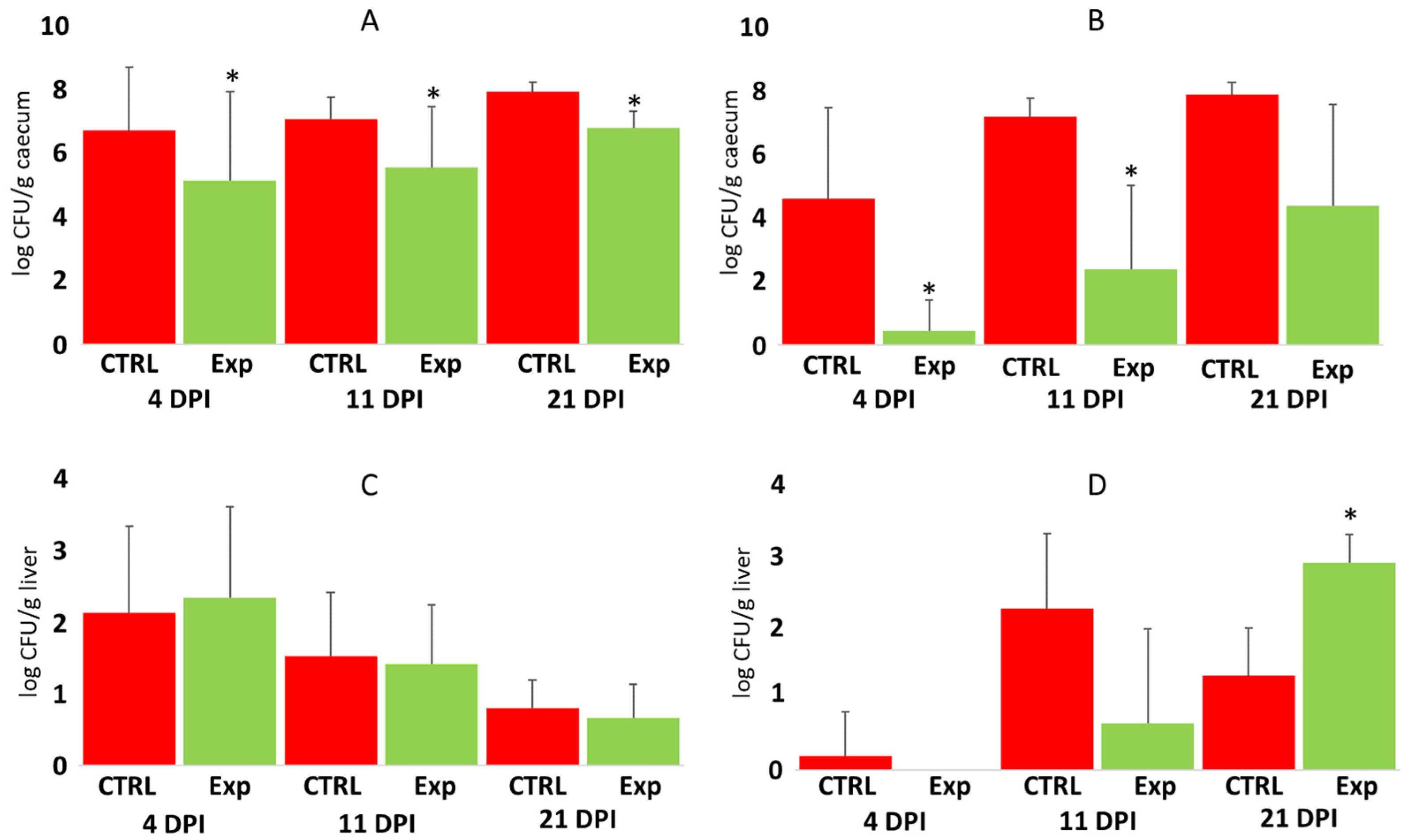
3.2. The Protective Effect of Defined Bacterial Mixtures Against Salmonella Enteritidis Challenges
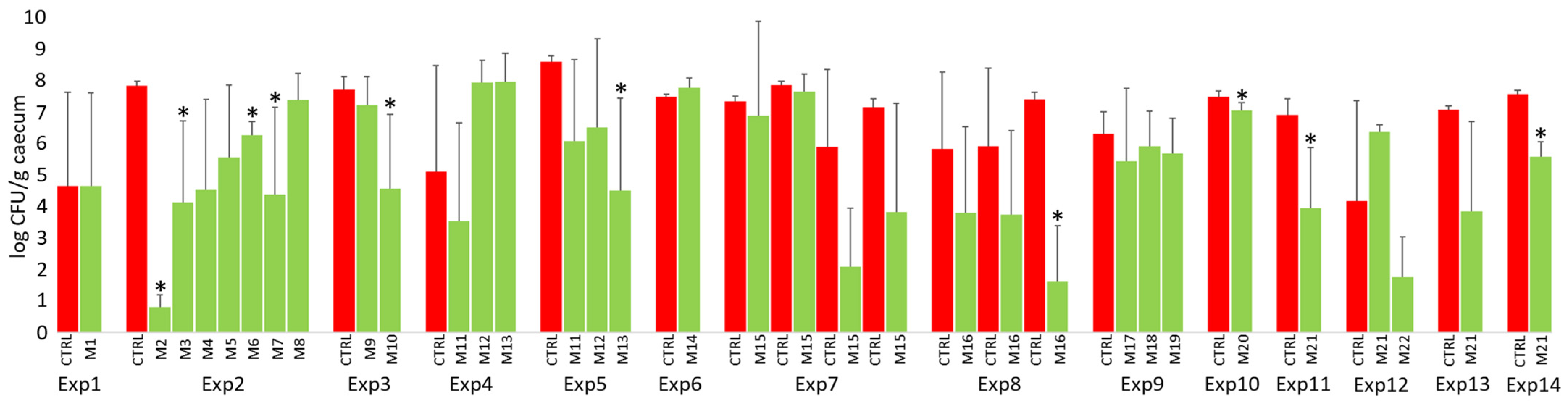
3.3. Performance of Probiotic Mixtures in Individual Experiments
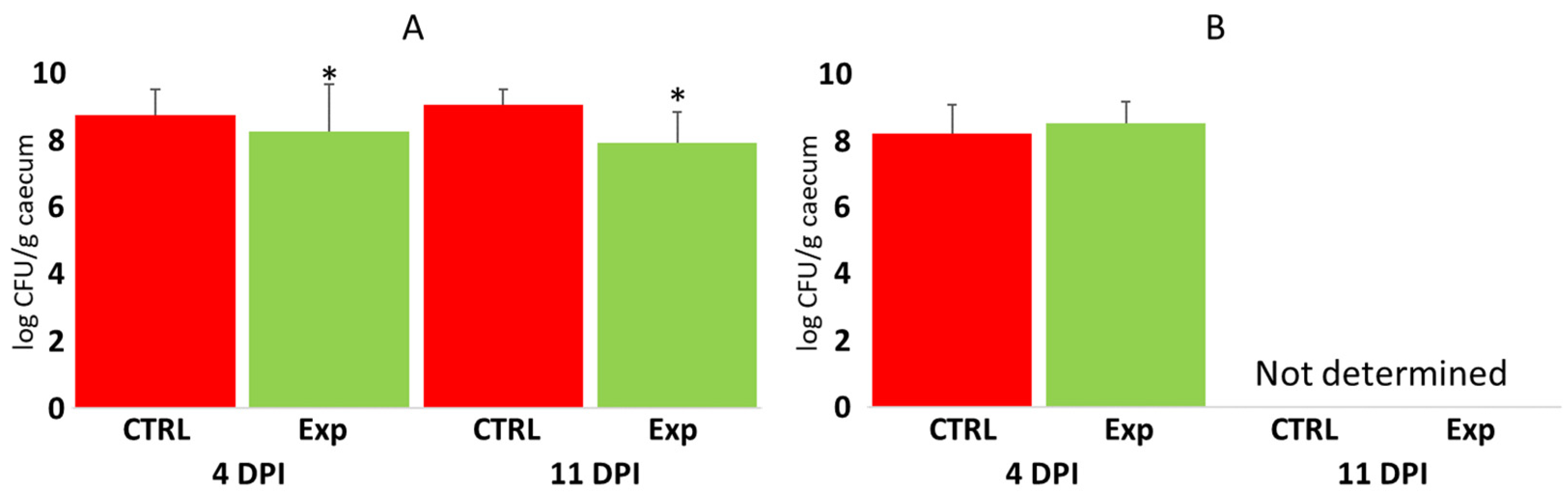
3.4. Protective Effect of Administering Defined Mixtures Against Campylobacter jejuni Challenges
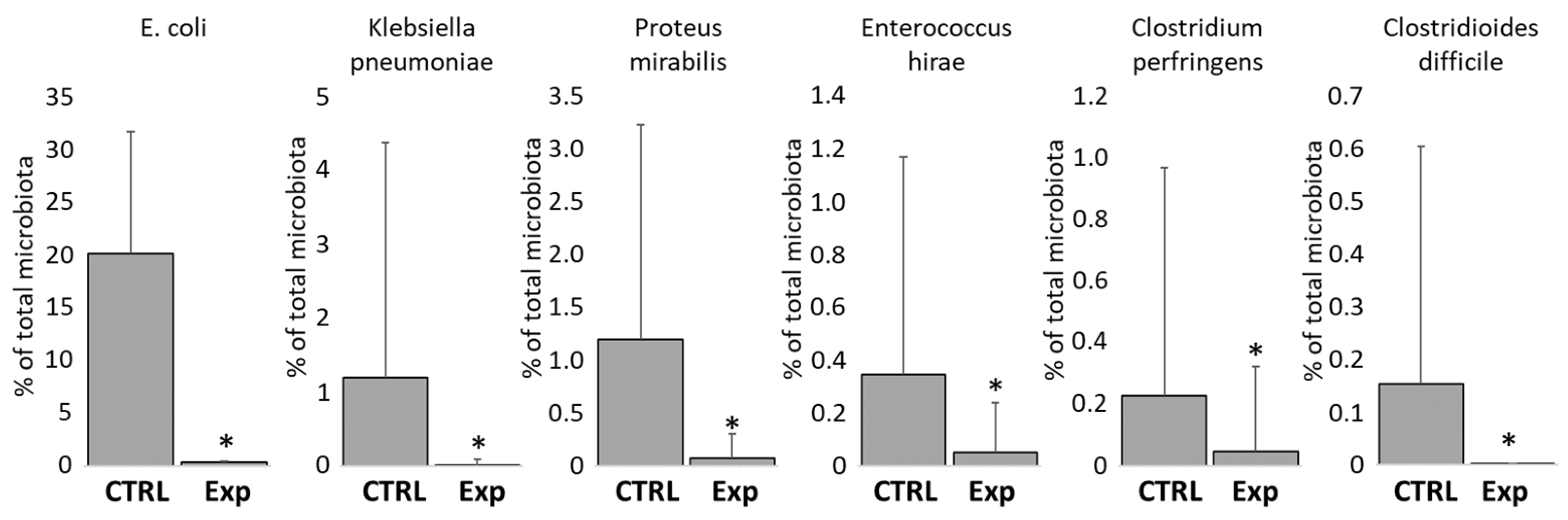
3.5. Protective Effect of Administration of Defined Mixtures Against Opportunistic Pathogens
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Varmuzova, K.; Kubasova, T.; Davidova-Gerzova, L.; Sisak, F.; Havlickova, H.; Sebkova, A.; Faldynova, M.; Rychlik, I. Composition of Gut Microbiota Influences Resistance of Newly Hatched Chickens to Salmonella Enteritidis Infection. Front. Microbiol. 2016, 7, 957. [Google Scholar] [CrossRef]
- Kubasova, T.; Kollarcikova, M.; Crhanova, M.; Karasova, D.; Cejkova, D.; Sebkova, A.; Matiasovicova, J.; Faldynova, M.; Pokorna, A.; Cizek, A.; et al. Contact with adult hen affects development of caecal microbiota in newly hatched chicks. PLoS ONE 2019, 14, e0212446. [Google Scholar] [CrossRef] [PubMed]
- Marcolla, C.S.; Ju, T.; Willing, B.P. Cecal Microbiota Development and Physiological Responses of Broilers Following Early Life Microbial Inoculation Using Different Delivery Methods and Microbial Sources. Appl. Environ. Microbiol. 2023, 89, e0027123. [Google Scholar] [CrossRef] [PubMed]
- Videnska, P.; Sedlar, K.; Lukac, M.; Faldynova, M.; Gerzova, L.; Cejkova, D.; Sisak, F.; Rychlik, I. Succession and replacement of bacterial populations in the caecum of egg laying hens over their whole life. PLoS ONE 2014, 9, e115142. [Google Scholar] [CrossRef]
- Stanley, D.; Geier, M.S.; Hughes, R.J.; Denman, S.E.; Moore, R.J. Highly variable microbiota development in the chicken gastrointestinal tract. PLoS ONE 2013, 8, e84290. [Google Scholar] [CrossRef]
- Xi, Y.; Shuling, N.; Kunyuan, T.; Qiuyang, Z.; Hewen, D.; ChenCheng, G.; Tianhe, Y.; Liancheng, L.; Xin, F. Characteristics of the intestinal flora of specific pathogen free chickens with age. Microb. Pathog. 2019, 132, 325–334. [Google Scholar] [CrossRef]
- Sergeant, M.J.; Constantinidou, C.; Cogan, T.A.; Bedford, M.R.; Penn, C.W.; Pallen, M.J. Extensive microbial and functional diversity within the chicken cecal microbiome. PLoS ONE 2014, 9, e91941. [Google Scholar] [CrossRef]
- Beal, R.K.; Wigley, P.; Powers, C.; Hulme, S.D.; Barrow, P.A.; Smith, A.L. Age at primary infection with Salmonella enterica serovar Typhimurium in the chicken influences persistence of infection and subsequent immunity to re-challenge. Vet. Immunol. Immunopathol. 2004, 100, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Juricova, H.; Matiasovicova, J.; Faldynova, M.; Sebkova, A.; Kubasova, T.; Prikrylova, H.; Karasova, D.; Crhanova, M.; Havlickova, H.; Rychlik, I. Probiotic Lactobacilli Do Not Protect Chickens against Salmonella Enteritidis Infection by Competitive Exclusion in the Intestinal Tract but in Feed, Outside the Chicken Host. Microorganisms 2022, 10, 219. [Google Scholar] [CrossRef]
- Rantala, M.; Nurmi, E. Prevention of the growth of Salmonella infantis in chicks by the flora of the alimentary tract of chickens. Br. Poult. Sci. 1973, 14, 627–630. [Google Scholar] [CrossRef]
- Methner, U.; Barrow, P.A.; Martin, G.; Meyer, H. Comparative study of the protective effect against Salmonella colonisation in newly hatched SPF chickens using live, attenuated Salmonella vaccine strains, wild-type Salmonella strains or a competitive exclusion product. Int. J. Food Microbiol. 1997, 35, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Faldynova, M.; Prikrylova, H.; Sebkova, A.; Volf, J.; Karasova, D.; Crhanova, M.; Babak, V.; Rychlik, I. Contact with adult hens affects the composition of skin and respiratory tract microbiota in newly hatched chicks. Poult. Sci. 2024, 103, 103302. [Google Scholar] [CrossRef] [PubMed]
- Kubasova, T.; Seidlerova, Z.; Rychlik, I. Ecological Adaptations of Gut Microbiota Members and Their Consequences for Use as a New Generation of Probiotics. Int. J. Mol. Sci. 2021, 22, 5471. [Google Scholar] [CrossRef]
- Karasova, D.; Faldynova, M.; Matiasovicova, J.; Sebkova, A.; Crhanova, M.; Kubasova, T.; Seidlerova, Z.; Prikrylova, H.; Volf, J.; Zeman, M.; et al. Host Species Adaptation of Obligate Gut Anaerobes Is Dependent on Their Environmental Survival. Microorganisms 2022, 10, 1085. [Google Scholar] [CrossRef]
- Papouskova, A.; Rychlik, I.; Harustiakova, D.; Cizek, A. Research Note: A mixture of Bacteroides spp. and other probiotic intestinal anaerobes reduces colonization by pathogenic E. coli strain O78:H4-ST117 in newly hatched chickens. Poult. Sci. 2023, 102, 102529. [Google Scholar] [CrossRef]
- Medvecky, M.; Cejkova, D.; Polansky, O.; Karasova, D.; Kubasova, T.; Cizek, A.; Rychlik, I. Whole genome sequencing and function prediction of 133 gut anaerobes isolated from chicken caecum in pure cultures. BMC Genom. 2018, 19, 561. [Google Scholar] [CrossRef]
- Kubasova, T.; Kollarcikova, M.; Crhanova, M.; Karasova, D.; Cejkova, D.; Sebkova, A.; Matiasovicova, J.; Faldynova, M.; Sisak, F.; Babak, V.; et al. Gut anaerobes capable of chicken caecum colonisation. Microorganisms 2019, 7, 597. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Girardot, C.; Scholtalbers, J.; Sauer, S.; Su, S.Y.; Furlong, E.E. Je, a versatile suite to handle multiplexed NGS libraries with unique molecular identifiers. BMC Bioinform. 2016, 17, 419. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Rajova, J.; Zeman, M.; Seidlerova, Z.; Vlasatikova, L.; Matiasovicova, J.; Sebkova, A.; Faldynova, M.; Prikrylova, H.; Karasova, D.; Crhanova, M.; et al. In Vivo Expression of Chicken Gut Anaerobes Identifies Carbohydrate- or Amino Acid-Utilising, Motile or Type VI Secretion System-Expressing Bacteria. Int. J. Mol. Sci. 2024, 25, 6505. [Google Scholar] [CrossRef]
- Vlasatikova, L.; Zeman, M.; Crhanova, M.; Matiasovicova, J.; Karasova, D.; Faldynova, M.; Prikrylova, H.; Sebkova, A.; Rychlik, I. Colonization of chickens with competitive exclusion products results in extensive differences in metabolite composition in cecal digesta. Poult. Sci. 2024, 103, 103217. [Google Scholar] [CrossRef]
- Hapfelmeier, S.; Lawson, M.A.; Slack, E.; Kirundi, J.K.; Stoel, M.; Heikenwalder, M.; Cahenzli, J.; Velykoredko, Y.; Balmer, M.L.; Endt, K.; et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 2010, 328, 1705–1709. [Google Scholar] [CrossRef]
- Takeuchi, T.; Miyauchi, E.; Kanaya, T.; Kato, T.; Nakanishi, Y.; Watanabe, T.; Kitami, T.; Taida, T.; Sasaki, T.; Negishi, H.; et al. Acetate differentially regulates IgA reactivity to commensal bacteria. Nature 2021, 595, 560–564. [Google Scholar] [CrossRef]
- Ty, M.; Taha-Abdelaziz, K.; Demey, V.; Castex, M.; Sharif, S.; Parkinson, J. Performance of distinct microbial based solutions in a Campylobacter infection challenge model in poultry. Anim. Microbiome 2022, 4, 2. [Google Scholar] [CrossRef]
- Polansky, O.; Sekelova, Z.; Faldynova, M.; Sebkova, A.; Sisak, F.; Rychlik, I. Important Metabolic Pathways and Biological Processes Expressed by Chicken Cecal Microbiota. Appl. Environ. Microbiol. 2015, 82, 1569–1576. [Google Scholar] [CrossRef]
- Iqbal, M.; Philbin, V.J.; Withanage, G.S.; Wigley, P.; Beal, R.K.; Goodchild, M.J.; Barrow, P.; McConnell, I.; Maskell, D.J.; Young, J.; et al. Identification and functional characterization of chicken toll-like receptor 5 reveals a fundamental role in the biology of infection with Salmonella enterica serovar typhimurium. Infect. Immun. 2005, 73, 2344–2350. [Google Scholar] [CrossRef]
- Hayashi, F.; Smith, K.D.; Ozinsky, A.; Hawn, T.R.; Yi, E.C.; Goodlett, D.R.; Eng, J.K.; Akira, S.; Underhill, D.M.; Aderem, A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 2001, 410, 1099–1103. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.S.; Kim, I.H. Effect of Bacillus subtilis C-3102 spores as a probiotic feed supplement on growth performance, noxious gas emission, and intestinal microflora in broilers. Poult. Sci. 2014, 93, 3097–3103. [Google Scholar] [CrossRef] [PubMed]
- Menconi, A.; Morgan, M.J.; Pumford, N.R.; Hargis, B.M.; Tellez, G. Physiological Properties and Salmonella Growth Inhibition of Probiotic Bacillus Strains Isolated from Environmental and Poultry Sources. Int. J. Bacteriol. 2013, 2013, 958408. [Google Scholar] [CrossRef] [PubMed]
- Coyne, M.J.; Roelofs, K.G.; Comstock, L.E. Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genom. 2016, 17, 58. [Google Scholar] [CrossRef]
- Verster, A.J.; Ross, B.D.; Radey, M.C.; Bao, Y.; Goodman, A.L.; Mougous, J.D.; Borenstein, E. The Landscape of Type VI Secretion across Human Gut Microbiomes Reveals Its Role in Community Composition. Cell Host Microbe 2017, 22, 411–419.e4. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Volf, J.; Faldynova, M.; Matiasovicova, J.; Sebkova, A.; Karasova, D.; Prikrylova, H.; Havlickova, H.; Rychlik, I. Probiotic Mixtures Consisting of Representatives of Bacteroidetes and Selenomonadales Increase Resistance of Newly Hatched Chicks to Salmonella Enteritidis Infection. Microorganisms 2024, 12, 2145. https://doi.org/10.3390/microorganisms12112145
Volf J, Faldynova M, Matiasovicova J, Sebkova A, Karasova D, Prikrylova H, Havlickova H, Rychlik I. Probiotic Mixtures Consisting of Representatives of Bacteroidetes and Selenomonadales Increase Resistance of Newly Hatched Chicks to Salmonella Enteritidis Infection. Microorganisms. 2024; 12(11):2145. https://doi.org/10.3390/microorganisms12112145
Chicago/Turabian StyleVolf, Jiri, Marcela Faldynova, Jitka Matiasovicova, Alena Sebkova, Daniela Karasova, Hana Prikrylova, Hana Havlickova, and Ivan Rychlik. 2024. "Probiotic Mixtures Consisting of Representatives of Bacteroidetes and Selenomonadales Increase Resistance of Newly Hatched Chicks to Salmonella Enteritidis Infection" Microorganisms 12, no. 11: 2145. https://doi.org/10.3390/microorganisms12112145
APA StyleVolf, J., Faldynova, M., Matiasovicova, J., Sebkova, A., Karasova, D., Prikrylova, H., Havlickova, H., & Rychlik, I. (2024). Probiotic Mixtures Consisting of Representatives of Bacteroidetes and Selenomonadales Increase Resistance of Newly Hatched Chicks to Salmonella Enteritidis Infection. Microorganisms, 12(11), 2145. https://doi.org/10.3390/microorganisms12112145







