Revolutionizing Malaria Vector Control: The Importance of Accurate Species Identification through Enhanced Molecular Capacity
Abstract
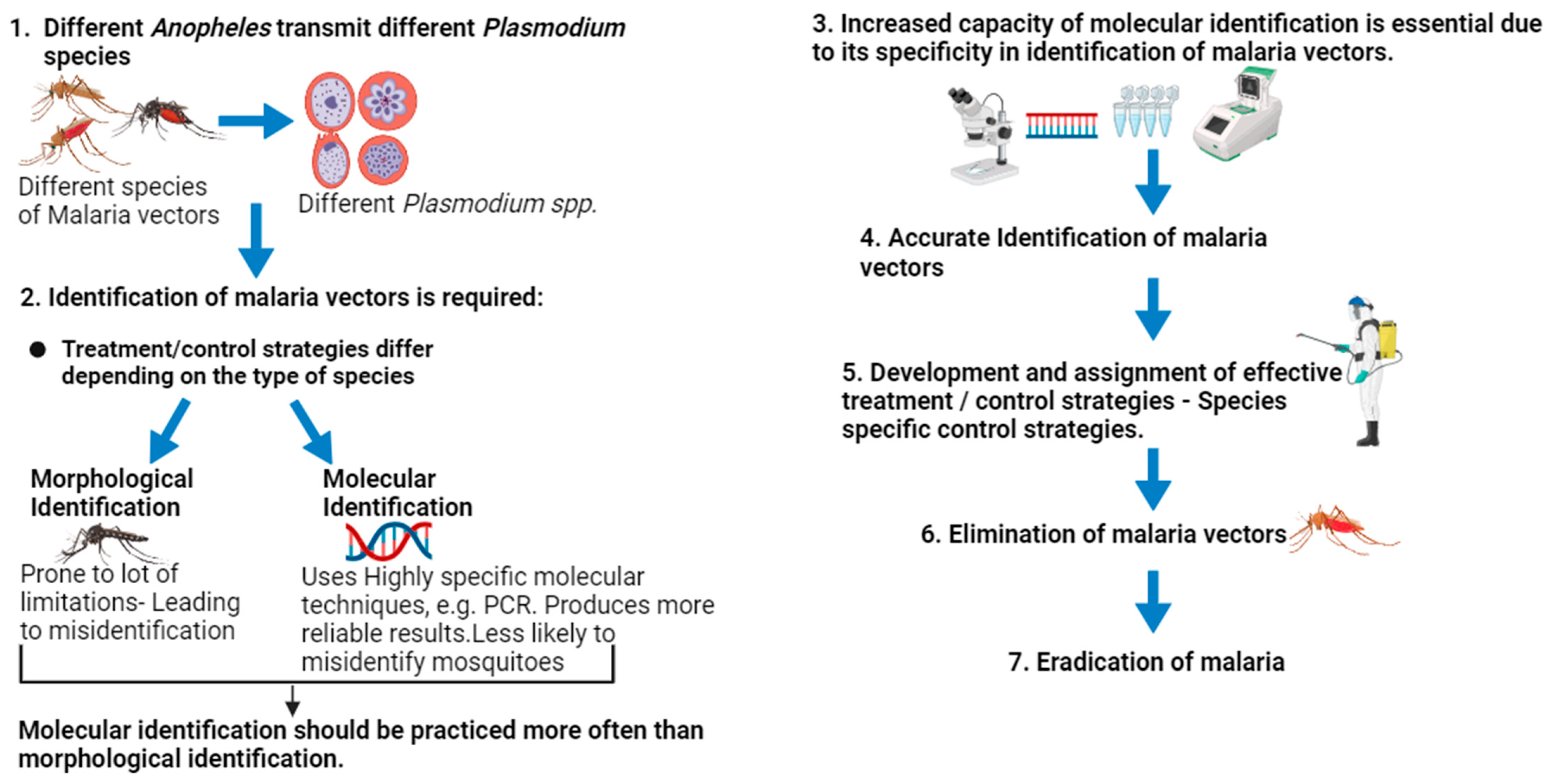
1. Introduction
Vectorial Biology and Behavioral Patterns That Enhance Malaria Transmission
2. Distribution, Prevalence, and Control of Malaria Vectors and Challenges with Malaria Identification
2.1. Lessons Learned from Europe
2.2. Lessons Learned from America
2.3. Lessons Learned from Asia
2.4. Lessons Learned from Africa
3. Mosquito Control Strategies
3.1. Mosquito Control Strategies in Africa
3.2. Mosquito Control Strategies in Asia
3.3. Mosquito Control Strategies in the USA and Europe
4. Identification and Characterization of Malaria Vectors
4.1. Morphological Identification of Malaria Vectors
4.2. Classification of Mosquitoes
4.3. Molecular Identification of Mosquito Species
4.4. Taxonomic Characterization of Malaria Vector Species
4.5. Challenges Associated with Structural Species Identification
4.6. Molecular Characterization of Malaria Vectors
4.7. Sequencing Techniques Normally Used in Population Genetics Studies, Their Strengths, and Limitations
4.7.1. Next-Generation Sequencing (NGS)
4.7.2. Genotyping-by-Sequencing (GBS)
4.7.3. Restriction Site-Associated DNA Sequencing (RAD-Seq)
4.7.4. RNA Sequencing (RNA-Seq)
4.8. Capacity for Molecular Identification and Characterization of Malaria Vectors in Sub-Saharan Africa
5. Evidence Supporting the Superiority of DNA-Based Identification of Malaria Vectors over Morphological Identification
6. Routes to Be Taken to Advance Malaria Vector Control Strategies in Different Endemic Regions and Basic Solutions to Overcome Insecticides’ Resistance and Imported Cases
7. Future Insight and Prediction
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organisation (WHO). Malaria. 2021. Available online: https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2021 (accessed on 1 February 2022).
- Talapko, J.; Škrlec, I.; Alebić, T.; Jukić, M.; Včev, A. Malaria. The past and the present. Microorganisms 2019, 7, 179. [Google Scholar] [CrossRef] [PubMed]
- Haldar, K.; Bhattacharjee, S.; Safeukui, I. Drug resistance in Plasmodium. Nat. Rev. Microbiol. 2018, 16, 156–170. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.B. Encyclopedia of Geology; Selley, R.C., Cocks, L.R.M., Plimer, I.R., Eds.; Elsevier: Oxford, UK, 2005; pp. 334–341. [Google Scholar]
- Sato, S. Plasmodium—A brief introduction to the parasites causing human malaria and their basic biology. J. Physiol. Anthr. 2021, 40, 1. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Malaria Surveillance, Monitoring & Evaluation: A Reference Manual; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Coetzee, M. Key to the females of Afrotropical Anopheles mosquitoes (Diptera: Culicidae). Malar. J. 2020, 19, 70. [Google Scholar] [CrossRef] [PubMed]
- Kahamba, N.F.; Finda, M.; Ngowo, H.S.; Msugupakulya, B.J.; Baldini, F.; Koekemoer, L.L.; Ferguson, H.M.; Okumu, F.O. Using ecological observations to improve malaria control in areas where Anopheles funestus is the dominant vector. Malar. J. 2022, 21, 158. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.A.; Brogdon, W.G.; Collins, F.H. Identification of single specimens of the Anopheles gambiae complex by the polymerase chains reaction. Am. J. Trop. Med. Hyg. 1993, 49, 520–529. [Google Scholar] [CrossRef]
- Douglas, E.N. Genetic markers for study of the anopheline vectors of human malaria. D.E. Norris/Int. J. Parasitol. 2002, 32, 1607–1615. [Google Scholar]
- Collins, F.H.; Paskewitz, S.M. A review of the use of ribosomal DNA (rDNA) to differentiate among cryptic Anopheles species. Insect Mol. Biol. 1996, 5, 1–9. [Google Scholar] [CrossRef]
- Favia, G.; Dimopoulos, G.; Torre, A.D.; Touré, Y.T.; Coluzzi, M.; Louis, C. Polymorphisms detected by random PCR distinguish between different chromosomal forms of Anopheles gambiae. Proc. Natl. Acad. Sci. USA 1994, 91, 10315–10319. [Google Scholar] [CrossRef]
- Erlank, E.; Koekemoer, L.L.; Coetzee, M. The importance of morphological identification of African anopheline mosquitoes (Diptera: Culicidae) for malaria control programmes. Malar. J. 2018, 17, 43. [Google Scholar] [CrossRef]
- Institute of Medicine (US) Committee for the Study on Malaria Prevention and Control (IMUSCSMPC). Malaria: Obstacles and Opportunities. In 7, Vector Biology, Ecology, and Control; Oaks, S.C., Jr., Mitchell, V.S., Pearson, G.W., Carpenter, C.C.J., Eds.; National Academies Press (US): Washington, DC, USA, 1991. Available online: https://www.ncbi.nlm.nih.gov/books/NBK234322/ (accessed on 9 September 2023).[Green Version]
- Kumar, N.P.; Rajavel, A.R.; Natarajan, R.; Jambulingam, P. DNA barcodes can distinguish species of Indian mosquitoes (Diptera: Culicidae). J. Med. Entomol. 2007, 44, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Odero, J.O.; Nambunga, I.H.; Wangrawa, D.W.; Badolo, A.; Weetman, D.; Koekemoer, L.L.; Ferguson, H.M.; Okumu, F.O.; Baldini, F. Advances in the genetic characterization of the malaria vector, Anopheles funestus, and implications for improved surveillance and control. Malar. J. 2023, 22, 230. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation (WHO). Malaria; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Oyegoke, O.O.; Adewumi, T.S.; Aderoju, S.A.; Tsundzukani, N.; Mabunda, E.; Adeleke, M.A.; Maharaj, R.; Okpeku, M. Towards malaria elimination: Analysis of travel history and case forecasting using the SARIMA model in Limpopo Province. Parasitol. Res. 2023, 122, 1775–1785. [Google Scholar] [CrossRef] [PubMed]
- Isaäcson, M. Airport malaria: A review. Bull. World Health Organ. 1989, 67, 737–743. [Google Scholar] [PubMed]
- Piperaki, E. Malaria eradication in the European world: Historical perspective and imminent threats. In Towards Malaria Elimination—A Leap Forward; Manguin, S., Dev, V., Eds.; IntechOpen: London, UK, 2018. [Google Scholar] [CrossRef]
- Van Thiel, P.H. On zoophilism and anthropophilism of Anopheles biotypes and species. Riv. Malariol. 1939, 18, 95–124. [Google Scholar]
- Kuhn, K.G.; Campbell-Lendrum, D.H.; Davies, C.R. A continental risk map for malaria mosquito (Diptera: Culicidae) vectors in Europe. J. Med. Entomol. 2002, 39, 621–630. [Google Scholar] [CrossRef]
- Askling, H.H.; Bruneel, F.; Burchard, G.; Castelli, F.; Chiodini, P.L.; Grobusch, M.P.; Lopez-Vélez, R.; Paul, M.; Petersen, E.; Popescu, C.; et al. Management of imported malaria in Europe. Malar. J. 2012, 11, 328. [Google Scholar] [CrossRef]
- WHO. Regional Office for Europe: Centralized Information System for Infectious Dieases (CISID). 2012. Available online: http://data.euro.who.int/cisid (accessed on 3 March 2022).
- Cibulskis, R.E.; Aregawi, M.; Williams, R.; Otten, M.; Dye, C. Worldwide incidence of malaria in 2009: Estimates, time trends, and a critique of methods. PLoS Med. 2011, 8, e1001142. [Google Scholar] [CrossRef]
- Sabatinelli, G.; Ejov, M.; Joergensen, P. Malaria in the WHO European Region (1971–1999). Eur. Surveill. 2001, 6, 61–65. [Google Scholar] [CrossRef]
- Romi, R.; Sabatinelli, G.; Majori, G. Could Malaria Reappear in Italy? Emerg. Infect. Dis. 2001, 7, 915–919. [Google Scholar] [CrossRef]
- Bertola, M.; Mazzucato, M.; Pombi, M.; Montarsi, F. Updated occurrence and bionomics of potential malaria vectors in Europe: A systematic review (2000–2021). Parasites Vectors 2022, 15, 88. [Google Scholar] [CrossRef] [PubMed]
- WHO. More malaria cases and deaths in 2020 linked to COVID-19 disruptions. In World Malaria Report; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Dye-Braumuller, K.C.; Kanyangarara, M. Malaria in the USA: How Vulnerable Are We to Future Outbreaks? Curr. Trop. Med. Rep. 2021, 8, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Dye-Braumuller, K.; Fredregill, C.; Debboun, M. Mosquitoes, Communities, and Public Health in Texas; Elsevier: Amsterdam, The Netherlands, 2020; pp. 249–278. [Google Scholar]
- Zucker, J.R. Changing patterns of autochthonous malaria transmission in the United States: A review of recent outbreaks. Emerg. Infect. Dis. 1996, 2, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Foster, W.A.; Walker, E.D. Mosquitoes (Culicidae). In Medical and Veterinary Entomology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 261–325. [Google Scholar]
- Darsie, R.F., Jr.; Ward, R.A. Identification and Geographical Distribution of the Mosquitoes of North America, North of Mexico, 2nd ed.; University Press of Florida: Gainesville, FL, USA, 2005; p. 416. ISBN 0-8130-2784-5. [Google Scholar]
- Sinka, M.E.; Rubio-Palis, Y.; Manguin, S.; Patil, A.P.; Temperley, W.H.; Gething, P.W.; Van Boeckel, T.; Kabaria, C.W.; Harbach, R.E.; Hay, S.I. The dominant Anopheles vectors of human malaria in the Americas: Occurrence data, distribution maps and bionomic précis. Parasites Vectors 2010, 3, 72. [Google Scholar] [CrossRef] [PubMed]
- Mace, K.E. Malaria surveillance—United States, 2016. Morb. Mortal. Wkly. Rep. 2019, 68, 1–35. [Google Scholar] [CrossRef] [PubMed]
- Cullen, K.A.; Mace, K.E.; Arguin, P.M. Malaria Surveillance—United States, 2013. MMWR Surveill. Summ. 2016, 65, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Filler, S.J.; MacArthur, J.R.; Parise, M.; Wirtz, R.; Eliades, M.J.; Dasilva, A.; Steketee, R.W. Locally acquired mosquito-transmitted malaria: A guide for investigations in the United States. MMWR Recomm. Rep. 2006, 55, 1–9, Erratum in MMWR Morb. Mortal. Wkly. Rep. 2006, 55, 1075. [Google Scholar]
- Aklilu, E.; Kindu, M.; Gebresilassie, A.; Yared, S.; Tekie, H.; Balkew, M. Environmental Factors Associated with Larval Habitats of Anopheline mosquitoes (Diptera: Culicidae) in Metema District, Northwestern Ethiopia. J. Arthropod Borne Dis. 2020, 14, 153–161. [Google Scholar] [CrossRef]
- Ma, Y.; Qu, F.; Xu, J.; Li, X.; Song, G. Differences in sequences of ribosomal DNA second internal transcribed spacer among three members of Anopheles hyrcanus complex from the Republic of Korea. Entomol. Sin. 2000, 7, 36–40. [Google Scholar]
- World Health Organization. Synopsis of the world malaria situation in 1979. Wkly. Epidemiol. Rec. 1981, 56, 145–149. [Google Scholar]
- Park, J.W.; Yeom, J.S.; Choe, K.W.; Moon, S.H.; Lee, H.C.; Kim, T.S.; Oh, M.D.; Ryu, S.H.; Klein, T.A.; Chai, J.Y.; et al. Vivax malaria: A continuing health threat to the Republic of Korea. Am. J. Trop. Med. Hyg. 2003, 69, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.S.; Yoo, D.-H.; Ju, Y.R.; Lee, W.G.; Roh, J.Y.; Kim, H.-C.; Klein, T.A.; Shin, E.-H. Distribution of malaria vectors and incidence of vivax malaria at Korean army installations near the demilitarized zone. Republic of Korea. Malar. J. 2016, 15, 259. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.J. The malaria situation in the People’s Republic of China. Bull. World Health Organ. 1981, 59, 931–936. [Google Scholar] [PubMed]
- WHO. Global Technical Strategy for Malaria 2016–2030; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- WHO. World Malaria Report 2018; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Zhu, G.; Xia, H.; Zhou, H.; Li, J.; Lu, F.; Liu, Y.; Cao, J.; Gao, Q.; Sattabongkot, J. Susceptibility of Anopheles sinensis to Plasmodium vivax in malarial outbreak areas of central China. Parasites Vectors 2013, 6, 176. [Google Scholar] [CrossRef]
- Zhou, S.S.; Wang, Y.; Tang, L.H. Malaria situation in the People’s Republic of China in 2006. Chin. J. Parasitol. Parasit. Dis. 2007, 25, 439–441. [Google Scholar]
- Lai, S.; Sun, J.; Ruktanonchai, N.W.; Zhou, S.; Yu, J.; Routledge, I.; Wang, L.; Zheng, Y.; Tatem, A.J.; Li, Z. Changing epidemiology and challenges of malaria in China towards elimination. Malar. J. 2019, 18, 107. [Google Scholar] [CrossRef]
- Yin, J.H.; Zhang, L.; Feng, X.Y.; Xia, Z.G. Evolution of anti-malaria policies and measures in P.R. China for achieving and sustaining malaria-free. Front. Public Health 2023, 11, 1094859. [Google Scholar] [CrossRef]
- Sinka, M.E.; Bangs, M.J.; Manguin, S.; Chareonviriyaphap, T.; Patil, A.P.; Temperley, W.H.; Gething, P.W.; Elyazar, I.R.; Kabaria, C.W.; E Harbach, R.; et al. The dominant Anopheles vectors of human malaria in the Asia-Pacific region: Occurrence data, distribution maps and bionomic précis. Parasites Vectors 2011, 4, 89. [Google Scholar] [CrossRef]
- Wiebe, A.; Longbottom, J.; Gleave, K.; Shearer, F.M.; Sinka, M.E.; Massey, N.C.; Cameron, E.; Bhatt, S.; Gething, P.W.; Hemingway, J.; et al. Geographical distributions of African malaria vector sibling species and evidence for insecticide resistance. Malar. J. 2017, 16, 85. [Google Scholar] [CrossRef]
- Battle, K.E.; Gething, P.W.; Elyazar, I.R.F.; Moyes, C.L.; Sinka, M.E.; Howe, R.E.; Guerra, C.A.; Price, R.N.; Baird, J.K.; Hay, S.I. The global public health significance of Plasmodium vivax. Adv. Parasitol. 2012, 80, 1–111. [Google Scholar]
- Torre, A.D.; Tu, Z.; Petrarca, V. On the distribution and genetic differentiation of Anopheles gambiae s.s. molecular forms. Insect Biochem. Mol. Biol. 2005, 35, 755–769. [Google Scholar] [CrossRef] [PubMed]
- Coetzee, M.; Hunt, R.H.; Wilkerson, R.; Torre, A.D.; Coulibaly, M.B.; Besansky, N.J. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa 2013, 3619, 246–274. [Google Scholar] [CrossRef] [PubMed]
- Drake, J.M.; Beier, J.C. Ecological niche and potential distribution of Anopheles arabiensis in Africa in 2050. Malar. J. 2014, 13, 213. [Google Scholar] [CrossRef] [PubMed]
- Moffett, A.; Shackelford, N.; Sarkar, S. Malaria in Africa: Vector species’ niche models and relative risk maps. PLoS ONE 2007, 2, e824. [Google Scholar] [CrossRef] [PubMed]
- Coetzee, M.; Fontenille, D. Advances in the study of Anopheles funestus, a major vector of malaria in Africa. Insect Biochem. Mol. Biol. 2004, 34, 599–605. [Google Scholar] [CrossRef]
- Antonio-Nkondjio, C.; Simard, F. Highlights on Anopheles nili and Anopheles moucheti, malaria vectors in Africa. In Anopheles Mosquitoes—New Insights into Malaria Vectors; InTech: Rijeka, Croatia, 2013. [Google Scholar] [CrossRef]
- Stevenson, J.C.; Norris, D.E. Implicating cryptic and novel anophelines as malaria vectors in Africa. Insects 2017, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Ogunah, J.A.; Lalah, J.O.; Schramm, K.-W. Malaria vector control strategies. What is appropriate towards sustainable global eradication? Sustain. Chem. Pharm. 2020, 18, 100339. [Google Scholar] [CrossRef]
- Kaura, T.; Sylvia Walter, N.; Kaur, U.; Sehgal, R. Different Strategies for Mosquito Control: Challenges and Alternatives; IntechOpen: London, UK, 2023. [Google Scholar] [CrossRef]
- Lanzieri, G. Statistics in Focus: Population and social conditions. Population in Europe 2005: First results. 2006.16. Arch. Eur. Integr. (AEI) 2019, 12, 1024–4352. [Google Scholar]
- Ng’ang’a, P.N.; Aduogo, P.; Mutero, C.M. Long lasting insecticidal mosquito nets (LLINs) ownership, use and coverage following mass distribution campaign in Lake Victoria basin, Western Kenya. BMC Public Health 2021, 21, 1046. [Google Scholar] [CrossRef]
- Black, W.C., 4th; Lanzaro, G.C. Distribution of genetic variation among chromosomal forms of Anopheles gambiae s.s.: Introgressive hybridization, adaptive inversions, or recent reproductive isolation? Insect Mol. Biol. 2001, 10, 3–7. [Google Scholar] [CrossRef]
- Kenea, O.; Balkew, M.; Tekie, H.; Gebre-Michael, T.; Deressa, W.; Loha, E.; Lindtjørn, B.; Overgaard, H.J. Human-biting activities of Anopheles species in south-central Ethiopia. Parasites Vectors 2016, 9, 527. [Google Scholar] [CrossRef]
- Killeen, G.F. Characterizing, controlling and eliminating residual malaria transmission. Malar. J. 2014, 13, 330. [Google Scholar] [CrossRef] [PubMed]
- Giardina, F.; Kasasa, S.; Sié, A.; Utzinger, J.; Tanner, M.; Vounatsou, P. Effects of vector-control interventions on changes in risk of malaria parasitaemia in sub-Saharan Africa: A spatial and temporal analysis. Lancet Glob. Health 2014, 2, e601–e615. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Health. Vector Control Operational Manual for Malaria Elimination in Ethiopia; Ministry of Health: Addis Ababa, Ethiopia, 2017. [Google Scholar]
- Tiono, A.B.; Ouédraogo, A.; Ouattara, D.; Bougouma, E.C.; Coulibaly, S.; Diarra, A.; Faragher, B.; Guelbeogo, M.W.; Grisales, N.; Ouédraogo, I.N.; et al. Efficacy of Olyset Duo, a bednet containing pyriproxyfen and permethrin, versus a permethrin-only net against clinical malaria in an area with highly pyrethroid-resistant vectors in rural Burkina Faso: A cluster-randomised controlled trial. Lancet 2018, 392, 569–580. [Google Scholar] [CrossRef]
- Protopopoff, N.; Mosha, J.F.; Lukole, E.; Charlwood, J.D.; Wright, A.; Mwalimu, C.D.; Manjurano, A.; Mosha, F.W.; Kisinza, W.; Kleinschmidt, I.; et al. Effectiveness of a long-lasting piperonyl butoxide-treated insecticidal net and indoor residual spray interventions, separately and together, against malaria transmitted by pyrethroid-resistant mosquitoes: A cluster, randomised controlled, two-by-two factorial design trial. Lancet 2018, 391, 1577–1588. [Google Scholar] [CrossRef] [PubMed]
- WHO. Use of Indoor Residual Spraying for Scale Up Global Malaria Control and Elimination: WHO Position Statement; World Health Organization: Geneva, Switzerland, 2006. [Google Scholar]
- Kim, D.; Fedak, K.; Kramer, R. Reduction of malaria prevalence by indoor residual spraying: A meta-regression analysis. Am. J. Trop. Med. Hyg. 2012, 87, 117–124. [Google Scholar] [CrossRef] [PubMed]
- WHO. World Health Organization Recommended Insecticides for Indoor Residual Spraying against Malaria Vectors; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- Woyessa, A.; Deressa, W.; Ali, A.; Lindtjørn, B. Malaria risk factors in Butajira area, south-central Ethiopia: A multilevel analysis. Malar. J. 2013, 12, 1731. [Google Scholar] [CrossRef]
- Killeen, G.F.; Tatarsky, A.; Diabate, A.; Chaccour, C.J.; Marshall, J.M.; Okumu, F.O.; Brunner, S.; Newby, G.; Williams, Y.A.; Malone, D.; et al. Developing an expanded vector control toolbox for malaria elimination. BMJ Glob. Public Health 2017, 2, e000211. [Google Scholar] [CrossRef]
- Massebo, F.; Balkew, M.; Gebre-Michael, T.; Lindtjørn, B. Zoophagic behaviour of anopheline mosquitoes in southwest Ethiopia: Opportunity for malaria vector control. Parasites Vectors 2015, 8, 64. [Google Scholar] [CrossRef]
- Franco, O.M.; Gomes, M.G.; Rowland, M.; Coleman, G.P.; Davies, R.C. Controlling malaria using livestock-based interventions: A one health approach. PLoS ONE 2014, 9, e101699. [Google Scholar] [CrossRef]
- WHO. Global Vector Control Response 2017–2030; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- World Health Organization. Ivermectin for Malaria Transmission Control; World Health Organization: Geneva, Switzerland, 2016. [Google Scholar]
- Kobylinski, K.C.; Foy, B.D.; Richardson, J.H. Ivermectin inhibits the sporogony of Plasmodium falciparum in Anopheles gambiae. Malar. J. 2012, 11, 381. [Google Scholar] [CrossRef] [PubMed]
- Kobylinski, K.C.; Ubalee, R.; Ponlawat, A.; Nitatsukprasert, C.; Phasomkulsolsil, S.; Wattanakul, T.; Tarning, J.; Na-Bangchang, K.; McCardle, P.W.; Davidson, S.A.; et al. Ivermectin susceptibility and sporontocidal effect in Greater Mekong Subregion Anopheles. Malar. J. 2017, 16, 280280. [Google Scholar] [CrossRef] [PubMed]
- Smit, M.R.; Ochomo, E.O.; Aljayyoussi, G.; Kwambai, T.K.; Abong’On, B.O.; Chen, T.; Bousema, T.; Slater, H.C.; Waterhouse, D.; Bayoh, N.M.; et al. Safety and mosquitocidal efficacy of high-dose ivermectin when co-administered with dihydroartemisinin–piperaquine in Kenyan adults with uncomplicated malaria (IVERMAL): A randomised, double-blind, placebo-controlled trial. Lancet Infect. Dis. 2018, 18, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Animut, A.; Balkew, M.; Lindtjørn, B. Impact of housing condition on indoor-biting and indoor-resting Anopheles arabiensis density in a highland area, central Ethiopia. Malar. J. 2013, 12, 393. [Google Scholar] [CrossRef] [PubMed]
- Okumu, F.O.; Killeen, G.F.; Ogoma, S.; Biswaro, L.; Smallegange, R.C.; Mbeyela, E.; Titus, E.; Munk, C.; Ngonyani, H.; Takken, W.; et al. Development and field evaluation of a synthetic mosquito lure that is more attractive than humans. PLoS ONE 2009, 5, e8951. [Google Scholar] [CrossRef] [PubMed]
- Okumu, F.O.; Madumla, E.P.; John, A.N.; Lwetoijera, D.W.; Sumaye, R.D. Attracting, trapping and killing disease transmitting mosquitoes using odor-baited stations-the Ifakara Odor-Baited Stations. Parasites Vectors 2010, 3, 12. [Google Scholar] [CrossRef] [PubMed]
- Kaindoa, E.W.; Ngowo, H.S.; Limwagu, A.; Mkandawile, G.; Kihonda, J.; Masalu, J.P.; Bwanary, H.; Diabate, A.; Okumu, F.O. New evidence of mating swarms of the malaria vector, Anopheles arabiensis in Tanzania. Wellcome Open Res. 2017, 2, 88. [Google Scholar] [CrossRef]
- Zahar, A.R. Vector control operations in the African context. Bull. World Health Organ. 1984, 62, 89–100. [Google Scholar] [PubMed]
- WHO. Handbook on Vector Control in Malaria Elimination for the WHO African Region; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- WHO. Malaria Vector Control Commodities Landscape; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- WHO. Vector Control: Methods for Use by Individuals and Communities; World Health Organization: Geneva, Switzerland, 1997. [Google Scholar]
- Singh, N.; Shukla, M.M.; Mishra, A.K.; Singh, M.P.; Paliwal, J.C.; Dash, A.P. Malaria control using indoor residual spraying and larvivorous fish: A case study in Betul, central India. Trop. Med. Int. Health 2006, 11, 1512–1520. [Google Scholar] [CrossRef]
- Alliance for Science. CRISPR may Help Curb Malaria by Altering a Mosquito’s Gut Genes, New Study Suggests; Alliance for Science: New York, NY, USA, 2021. [Google Scholar]
- Hoermann, A.; Tapanelli, S.; Capriotti, P.; Del Corsano, G.; Masters, E.K.; Habtewold, T.; Christophides, G.K.; Windbichler, N. Converting endogenous genes of the malaria mosquito into simple non-autonomous gene drives for population replacement. eLife 2021, 10, e58791. [Google Scholar] [CrossRef]
- Meek, S.R. Vector control in some countries of Southeast Asia: Comparing the vectors and the strategies. Ann. Trop. Med. Parasitol. 1995, 89, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.M. La faune des moustiques du Cambodge. I. Anophelinae (Diptera, Culicidae). Cah. O.R.S.T.O.M. Ser. Entomol. Medicat. Et Parasitol. 1977, 15, 107–122. [Google Scholar]
- Curtis, C.F. Workshop on bednet at the international congress of tropical medicine. Jpn. J. Sanit. Zool. 1993, 44, 65–68. [Google Scholar] [CrossRef]
- Dolan, G.; Kuile, F.O.T.; Jacoutot, V.; White, N.J.; Luxemburger, C.; Malankirii, L. Bed nets for the prevention of malaria and anaemia in pregnancy. Trans. R. Soc. Trop. Med. Hyg. 1993, 87, 620–626. [Google Scholar] [CrossRef] [PubMed]
- Luxemburger, C.; Perea, W.A.; Delmas, G.; Pruja, C.; Pecoul, B.; Moren, A. Permethrin-impregnated bed nets for the prevention of malaria in schoolchildren on the Thai-Burmese border. Trans. R. Soc. Trop. Med. Hyg. 1994, 88, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Kitthawee, S.; Edman, J.D.; Upatham, E.S. Mosquito larvae and associated macroorganisms occurring in gem pits in southern Thai Mai District, Chantaburi Province, Thailand. Southeast Asian J. Trop. Med. Public Health 1993, 24, 143–151. [Google Scholar]
- Williamson, K.B.; Scharff, J.W. Anti-larval sluicing. Malay. Med. J. 1936, 11, 123–151. [Google Scholar]
- Moorhouse, D.E. Some entomological aspects of the malaria eradication pilot project in Malaya. J. Med. Èntomol. 1965, 2, 109–119. [Google Scholar] [CrossRef]
- Kirnowardoyo, S. Status of Anopheles malaria vectors in Indonesia. Southeast Asian J. Trop. Med. Public Health 1985, 16, 129–132. [Google Scholar]
- Center for Disease Control and Prevention (CDC). Larval Control and Other Vector Control Interventions, CDC 24/7: Saving Lives, Protecting People; Center for Disease Control and Prevention (CDC): Atlanta, CA, USA, 2020. [Google Scholar]
- Beaubien, J. Malaria Wiped out in U.S. But Still Plagues U.S. Hospitals, NRP; U.S. Public Health Service: Rockville, MD, USA, 2017. [Google Scholar]
- United State Environmental Protection Agency (EPA). Success in Mosquito Control: An Integrated Approach; United State Environmental Protection Agency (EPA): Washington, DC, USA, 2021. [Google Scholar]
- Cuervo-Parra, J.A.; Cortés, T.R.; Ramirez-Lepe, M. Mosquito-Borne Diseases, Pesticides Used for Mosquito Control, and Development of Resistance to Insecticides. In Insecticides Resistance; IntechOpen: London, UK, 2016. [Google Scholar] [CrossRef]
- Center for Disease Control and Prevention (CDC). Dichlorodiphenyltrichloroethane (DDT) Factsheet, CDC 24/7: Saving Lives, Protecting People; Center for Disease Control and Prevention (CDC): Atlanta, CA, USA, 2021. [Google Scholar]
- Wang, G.H.; Gamez, S.; Raban, R.R.; Marshall, J.M.; Alphey, L.; Li, M.; Rasgon, J.L.; Akbari, O.S. Combating mosquito-borne diseases using genetic control technologies. Nat. Commun. 2021, 12, 4388. [Google Scholar] [CrossRef]
- Beaty, B.J. Genetic manipulation of vectors: A potential novel approach for control of vector-borne diseases. Proc. Natl. Acad. Sci. USA 2000, 97, 10295–10297. [Google Scholar] [CrossRef] [PubMed]
- Wilke, A.B.B.; Marrelli, M.T. Paratransgenesis: A promising new strategy for mosquito vector control. Parasites Vectors 2015, 8, 342. [Google Scholar] [CrossRef] [PubMed]
- Walton, C.; Sharpe, R.G.; Pritchard, S.J.; Thelwell, N.J.; Butlin, R.K. Molecular identification of mosquito species. Biol. J. Linn. Soc. 1999, 68, 241–256. [Google Scholar] [CrossRef]
- Jones, R.T.; Ant, T.H.; Cameron, M.M.; Logan, J.G. 2021.Novel control strategies for mosquito-borne diseases. Philos. Trans. R. Soc. 2021, 376, B3762019080220190802. [Google Scholar] [CrossRef] [PubMed]
- Harbach, R.E. The phylogeny and classification of Anopheles. In Anopheles Mosquitoes—New Insights into Malaria Vectors; Manguin, S., Ed.; InTech: Rijeka, Croatia, 2013; pp. 3–55. [Google Scholar] [CrossRef]
- Christophers, S.R. The male genitalia of Anopheles. Indian J. Med. Res. 1915, 3, 371–394. [Google Scholar]
- Edwards, F.W. A revision of the mosquitos [sic] of the Palaearctic Region. Bull. Entomol. Res. 1921, 12, 263–351. [Google Scholar] [CrossRef]
- Root, F.M. The male genitalia of some American Anopheles mosquitoes. Am. J. Hyg. 1923, 31, 264–279. [Google Scholar] [CrossRef]
- Komp, W.H.W. The species of the subgenus Kerteszia of Anopheles (Diptera, Culicidae). Ann. Entomol. Soc. Am. 1937, 30, 492–529. [Google Scholar] [CrossRef]
- Antunes, P.C.A. A new Anopheles and a new Goeldia from Colombia (Dipt. Culic.). Bull. Entomol. Res. 1937, 28, 69–73. [Google Scholar] [CrossRef]
- Tandina, F.; Doumbo, O.; Yaro, A.S.; Traoré, S.F.; Parola, P.; Robert, V. Mosquitoes (Diptera: Culicidae) and mosquito-borne diseases in Mali, West Africa. Parasite Vectors 2018, 11, 467. [Google Scholar] [CrossRef]
- Harbach, R.E.; Rattanarithikul, R.; Harrison, B.A. Baimaia, a new subgenus for Anopheles kyondawensis Abraham, a unique crabhole-breeding anopheline in southeastern Asia. Proc. Entomol. Soc. Wash. 2005, 107, 750–761. [Google Scholar]
- McKeon, S.N.; Schlichting, C.D.; Povoa, M.M.; Conn, J.E. Ecological suitability and spatial distribution of five Anopheles species in Amazonian Brazil. Am. J. Trop. Med. Hyg. 2013, 88, 1079–1086. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gillies, M.T.; de Meillon, B. The Anophelinae of Africa South of the Sahara (Ethiopian Zoogeographical Region). Publ. South Afr. Inst. Med. Res. 1968, 54, 1–343. [Google Scholar][Green Version]
- Kengne, P.; Awono-Ambene, P.; Antonio Nkondjio, C.; Simard, F.; Fontenille, D. Molecular identification of the Anopheles nili group of African malaria vectors. Med. Vet. Entomol. 2003, 17, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Pedro, P.M.; E Harbach, R.; Somboon, P.; Walton, C.; Butlin, R.K. Mitochondrial DNA variation in the malaria vector Anopheles minimus across China, Thailand, and Vietnam: Evolutionary hypothesis, population structure and population history. Heredity 2011, 106, 241–252. [Google Scholar] [CrossRef]
- Pasteur, N.; Raymond, M. Insecticide resistance genes in mosquitoes: Their mutations, migration, and selection in field populations. J. Hered. 1996, 87, 444–449. [Google Scholar] [CrossRef]
- Faust, C. Why Should We Care about Genetic Diversity in Mosquitoes? BugBitten BMC. 2018. Available online: https://blogs.biomedcentral.com/bugbitten/2018/02/16/care-genetic-diversity-mosquitoes/ (accessed on 9 March 2023).
- Mayr, E.; Bock, W.J. Classifications and other ordering systems. J. Zool. Syst. Evol. Res. 2002, 40, 169–194. [Google Scholar] [CrossRef]
- Edwards, F.W. Genera insectorum, Diptera. Fam. Culicidae Fascicle 1932, 194, 258. [Google Scholar]
- Reid, J.A.; Knight, K.L. Classification within the subgenus Anopheles (Diptera, Culicidae). Ann. Trop. Med. Parasitol. 1961, 55, 474–488. [Google Scholar] [CrossRef]
- Reid, J.A. Anopheline mosquitoes of Malaya and Borneo. Stud. Inst. Med. Res. Malaya 1968, 31, 1–520. [Google Scholar]
- Faran, M.E. Mosquito studies (Diptera, Culicidae) XXXIV. A revision of the Albimanus Section of the subgenus Nyssorhynchus of Anopheles. Contrib. Am. Entomol. Inst. 1980, 15, 1–215. [Google Scholar]
- Linthicum, K.J. A revision of the Argyritarsis Section of the subgenus Nyssorhynchus of Anopheles (Diptera: Culicidae). Mosq. Syst. 1988, 20, 98–271. [Google Scholar]
- Peyton, E.L.; Wilkerson, R.C.; Harbach, R.E. Comparative analysis of the subgenera Kerteszia and Nyssorhynchus of Anopheles (Diptera: Culicidae). Mosq. Syst. 1992, 24, 51–69. [Google Scholar]
- Lobo, N.F.; St Laurent, B.; Sikaala, C.H.; Hamainza, B.; Chanda, J.; Chinula, D.; Krishnankutty, S.M.; Mueller, J.D.; Deason, N.A.; Hoang, Q.T.; et al. Unexpected diversity of Anopheles species in Eastern Zambia: Implications for evaluating vector behavior and interventions using molecular tools. Sci. Rep. 2015, 5, 17952. [Google Scholar] [CrossRef] [PubMed]
- Gillies, M.T.; Coetzee, M. Supplement to the Anophelinae of Africa south of the Sahara. Johannesbg. Publ. South Afr. Inst. Med. Res. 1987, 55, 1–143. [Google Scholar]
- Coetzee, M. Anopheles crypticus, new species from South Africa is distinguished from Anopheles coustani (Diptera: Culicidae). Mosq. Syst. 1994, 26, 125–131. [Google Scholar]
- Lambert, D.M.; Coetzee, M. A dual genetic and taxonomic approach to the resolution of the mosquito taxon Anopheles (Cellia) marshallii (Culicidae). Syst. Entomol. 1982, 7, 321–332. [Google Scholar] [CrossRef]
- Burke, A.; Dandalo, L.; Munhenga, G.; Dahan, Y.; Mbokazi, F.; Coetzee, M.; Ngxongo, S.; Koekemoer, L.; Brooke, B. A new malaria vector mosquito in South Africa. Sci. Rep. 2017, 7, 43779. [Google Scholar] [CrossRef]
- Koekemoer, L.L.; Kamau, L.; Hunt, R.H.; Coetzee, M. A cocktail polymerase chain reaction assay to identify members of the Anopheles funestus (Diptera: Culicidae) group. Am. J. Trop. Med. Hyg. 2002, 66, 804. [Google Scholar] [CrossRef]
- Arnheim, N. Concerted Evolution of Multigene Families. Evolution of Genes and Proteins; Nei, M., Koehm, R.K., Eds.; Sinauer Associates Inc.: Sunderland, MA, USA, 1983; pp. 38–62. [Google Scholar]
- Hackett, B.J.; Gimnig, J.; Guelbeogo, W.; Costantini, C.; Koekemoer, L.L.; Coetzee, M.; Collins, F.H.; Besansky, N.J. Ribosomal DNA internal transcribed spacer (ITS2) sequences differentiate Anopheles funestus and An. rivulorum and uncover a cryptic taxon. Insect Mol. Biol. 2000, 9, 369–374. [Google Scholar] [CrossRef]
- Weeraratne, T.C.; Surendran, S.N.; Karunaratne, S.H.P.P. DNA barcoding of morphologically characterized mosquitoes belonging to the subfamily Culicinae from Sri Lanka. Parasites Vectors 2018, 11, 266. [Google Scholar] [CrossRef] [PubMed]
- Batovska, J.; Blacket, M.J.; Brown, K.; Lynch, S.E. Molecular identification of mosquitoes (Diptera: Culicidae) in southeastern Australia. Ecol. Evol. 2016, 6, 3001–3011. [Google Scholar] [CrossRef] [PubMed]
- WHO. Mosquitoes of the Genus Anopheles in Countries of the WHO European Region; WHO: Geneva, Switzerland, 2008. [Google Scholar]
- Madeira, S.; Duarte, A.; Boinas, F.; Osório, H.C. A DNA barcode reference library of Portuguese mosquitoes. Zoonoses Public Health 2021, 68, 926–936. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Sharma, G.; Das, M.K.; Pande, V.; Singh, O.P. Intragenomic sequence variations in the second internal transcribed spacer (ITS2) ribosomal DNA of the malaria vector Anopheles stephensi. PLoS ONE 2021, 16, e0253173. [Google Scholar] [CrossRef] [PubMed]
- Iyiola, O.A.; Shaibu, R.D.; Shaibu, R.D.; Shittu, O.; Adelaja, O.J.; Kamaldeen-Ibrahim, A.T.; Fadipe, T.O.; Alaba, A.E.; Adejuwon, S.F.; Oyinlola, B.O. Genetic diversity, and molecular characterization of mosquitoes (Diptera: Culicidae) in North-Central Nigeria using ribosomal DNA ITS2 and mitochondrial 16S-DNA Sequences. Iraqi J. Vet. Med. 2020, 44, 78–91. [Google Scholar] [CrossRef]
- Davidson, J.R.; Wahid, I.; Sudirman, R.; Small, S.T.; Hendershot, A.L.; Baskin, R.N.; Burton, T.A.; Makuru, V.; Xiao, H.; Yu, X.; et al. Molecular analysis reveals a high diversity of Anopheles species in Karama, West Sulawesi, Indonesia. Parasites Vectors 2020, 13, 379. [Google Scholar] [CrossRef] [PubMed]
- St Laurent, B.; Cooke, M.; Krishnankutty, S.M.; Asih, P.; Mueller, J.D.; Kahindi, S.; Thumloup, J.; Oriango, R.M.; Stevenson, J.C.; Ayoma, E.; et al. Molecular characterization reveals diverse and unknown malaria vectors in the Western Kenyan highlands. Am. J. Trop. Med. Hyg. 2016, 94, 327–335. [Google Scholar] [CrossRef]
- Van Bortel, W.; Trung, H.D.; Roelants, P.; Harbach, R.E.; Backeljau, T.; Coosemans, M. Molecular identification of Anopheles minimus s.l. beyond distinguishing the members of the species complex. Insect Mol. Biol. 2000, 9, 335–340. [Google Scholar] [CrossRef]
- Gao, Q.; Beebe, N.W.; Cooper, R.D. Molecular Identification of the Malaria Vectors Anopheles anthropophagus and Anopheles sinensis (Diptera: Culicidae) in Central China Using Polymerase Chain Reaction and Appraisal of Their Position Within the Hyrcanus Group. J. Med. Entomol. 2004, 41, 5–11. [Google Scholar] [CrossRef]
- Fumagalli, M.; Vieira, F.G.; Linderoth, T.; Nielsen, R. NgsTools: Methods for population genetics analyses from next-generation sequencing data. Bioinformatics 2014, 30, 1486–1487. [Google Scholar] [CrossRef]
- Holliday, J.A.; Hallerman, E.M.; Haak, D.C. Genotyping and sequencing technologies in population genetics and genomics. In Population Genomics. Population Genomics; Rajora, O., Ed.; Springer: Cham, Switzerland, 2018. [Google Scholar] [CrossRef]
- Harrison, B.A. A new interpretation of affinities within the Anopheles hyrcanus complex of southeast Asia. Mosq. Syst. 1972, 4, 73–83. [Google Scholar]
- Hill, S.M.; Urwin, R.; Crampton, J. Synthetic DNA probes for the identification of sibling species within the Anopheles gambiae complex. Med. Vet. Entomol. 1992, 5, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Hill, S.M.; Urwin, R.; Crampton, J. A simplified, non-radioactive DNA probe protocol for the field identification of insect vector specimens. Trans. R. Soc. Trop. Med. Hyg. 1992, 86, 213–215. [Google Scholar] [CrossRef] [PubMed]
- Hill, S.M.; Crampton, J.M. Synthetic DNA probes to identify members of the Anopheles gambiae complex and to distinguish the two major vector of malaria within the complex An. gambiae s.s. and An. arabiensis. Am. J. Trop. Med. Hyg. 1994, 50, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.G.K.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar] [CrossRef] [PubMed]
- Ari, S.; Arıkan, M. Next-Generation Sequencing: Advantages, disadvantages, and future. In Plant Omics-Trends and Applications; Springer International Publishing: Cham, Switzerland, 2016; pp. 109–136. [Google Scholar] [CrossRef]
- Hert, D.G.; Fredlake, C.P.; Barron, A.E. Advantages and limitations of next generation sequencing technologies: A comparison of electrophoresis and non-electrophoresis methods. Electrophoresis 2008, 29, 4618–4626. [Google Scholar] [CrossRef] [PubMed]
- Shendure, J.; Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 2008, 26, 1135–1145. [Google Scholar] [CrossRef]
- König, K.; Peifer, M.; Fassunke, J.; Ihle, M.A.; Künstlinger, H.; Heydt, C.; Stamm, K.; Ueckeroth, F.; Vollbrecht, C.; Bos, M.; et al. Implementation of amplicon parallel sequencing leads to improvement of diagnosis and therapy of lung cancer patients. J. Thorac. Oncol. 2015, 10, 1049–1057. [Google Scholar] [CrossRef]
- Rivas, M.A.; Beaudoin, M.; Gardet, A.; Stevens, C.; Sharma, Y.; Zhang, C.K.; Boucher, G.; Ripke, S.; Ellinghaus, D.; Burtt, N.; et al. Deep resequencing of GWAS loci identifies independent low-frequency variants associated with inflammatory bowel disease. Nat. Genet. 2011, 43, 1066–1073. [Google Scholar] [CrossRef]
- Schuster, S.C. Next-generation sequencing transforms today’s biology. Nat. Methods 2008, 5, 16–18. [Google Scholar] [CrossRef]
- Gkazi, A. An Overview of Next-Generation Sequencing. Technol. Netw. Genom. Res. 2021. Available online: https://www.technologynetworks.com/genomics/articles/an-overview-of-next-generation-sequencing-346532 (accessed on 16 July 2023).
- Alzu’bi, A.A.; Zhou, L.; Watzlaf, V. Personal genomic information management and personalized medicine: Challenges, current solutions, and roles of HIM professionals. Perspect. Health Inf. Manag. 2014, 11, 1c. [Google Scholar] [PubMed]
- Gullapalli, R.R.; Desai, K.V.; Santana-Santos, L.; Kant, J.A.; Becich, M.J. Next generation sequencing in clinical medicine: Challenges and lessons for pathology and biomedical informatics. J. Pathol. Inform. 2012, 3, 40. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Yuan, Y.; Wang, H.; Yu, D.; Liu, Y.; Zhang, A.; Gowda, M.; Nair, S.K.; Hao, Z.; Lu, Y.; et al. Applications of genotyping-by-sequencing (GBS) in maize genetics and breeding. Sci. Rep. 2020, 10, 16308. [Google Scholar] [CrossRef] [PubMed]
- Wickland, D.P.; Battu, G.; Hudson, K.A.; Diers, B.W.; Hudson, M.E. A comparison of genotyping-by-sequencing analysis methods on low-coverage crop datasets shows advantages of a new workflow. GB-eaSy. BMC Bioinform. 2017, 18, 586. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.K. Chapter 13—Bioassay-guided isolation and evaluation of herbal drugs. In Evaluating Natural Products and Traditional Medicine; Elsevier: Amsterdam, The Netherlands, 2019; pp. 515–537. [Google Scholar]
- Bhatia, D.; Wing, R.A.; Singh, K. Genotyping by sequencing, its implications, and benefits. Crop Improv. 2013, 40, 101–111. [Google Scholar]
- Mishra, A.; Singh, P.K.; Bhandawat, A.; Sharma, V.; Sharma, V.; Singh, P.; Roy, J.; Sharma, H. Chapter 8—Analysis of SSR and SNP markers. Bioinformatics Methods and Applications; Academic Press: Cambridge, MA, USA, 2021; pp. 131–144. [Google Scholar]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Peterson, B.K.; Weber, J.N.; Kay, E.H.; Fisher, H.S.; Hoekstra, H.E. Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 2012, 7, e37135. [Google Scholar] [CrossRef]
- Rheindt, F.E.; Fujita, M.K.; Wilton, P.R.; Edwards, S.V. Introgression and phenotypic assimilation in Zimmerius flycatchers (Tyrannidae): Population genetic and phylogenetic inferences from genome-wide SNPs. Syst. Biol. 2012, 63, 134–152. [Google Scholar] [CrossRef]
- Davey, J.W.; Blaxter, M.L.; Davey, J.W.; Blaxter, M.L. RADSeq: Next-generation population genetics. Brief. Funct. Genom. 2010, 9, 416–423, Erratum in Brief. Funct. Genom. 2011, 10, 108. [Google Scholar] [CrossRef]
- Phelps, M.P.; Seeb, L.W.; Seeb, J.E. Transforming ecology and conservation biology through genome editing. Conserv. Biol. 2020, 34, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, R.J. RNA-Seq: Basics, applications and protocol. Technol. Netw. Genom. Res. 2018. Available online: https://www.technologynetworks.com/genomics/articles/rna-seq-basics-applications-and-protocol-299461 (accessed on 15 August 2023).
- Ozsolak, F.; Milos, P.M. RNA sequencing: Advances, challenges, and opportunities. Nat. Rev. Genet. 2011, 12, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.A.M.; Dehler, C.E.; Krol, E. Transcriptomic responses in the fish intestine. Dev. Comp. Immunol. 2016, 64, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Metzker, M.L. Sequencing technologies—The next generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Koboldt, D.C.; Ding, L.; Mardis, E.R.; Wilson, R.K. Challenges of sequencing human genomes. Brief. Bioinform. 2010, 11, 484–498. [Google Scholar] [CrossRef] [PubMed]
- Chavshin, A.R.; Oshaghi, M.A.; Vatandoost, H.; Hanafi-Bojd, A.A.; Raeisi, A.; Nikpoor, F. Molecular characterization, biological forms and sporozoite rate of Anopheles stephensi in southern Iran. Asian Pac. J. Trop. Biomed. 2014, 4, 47–51. [Google Scholar] [CrossRef]
- Makanda, M.; Kemunto, G.; Wamuyu, L.; Bargul, J.; Muema, J.; Mutunga, J. Diversity and Molecular Characterization of Mosquitoes (Diptera: Culicidae) in Selected Ecological Regions in Kenya. F1000Research 2019, 8, 262. [Google Scholar] [CrossRef]
- Beebe, N.W.; Foley, D.H.; Cooper, R.D.; Bryan, J.H.; Saul, A. DNA probes for the Anopheles punctulatus complex. Am. J. Trop. Med. Hyg. 1996, 54, 395–398. [Google Scholar] [CrossRef]
- Cornel, A.J.; Lee, Y.; Almeida, A.P.G.; Johnson, T.; Mouatcho, J.; Venter, M.; de Jager, C.; Braack, L. Mosquito community composition in South Africa and some neighboring countries. Parasites Vectors 2018, 11, 331. [Google Scholar] [CrossRef]
- Akeju, A.V.; Olusi, T.A.; Simon-Oke, I.A. Molecular identification and wing variations among malaria vectors in Akure North Local Government Area, Nigeria. Sci. Rep. 2022, 12, 7674. [Google Scholar] [CrossRef] [PubMed]
- Badmos, A.O.; Alaran, A.J.; Adebisi, Y.A.; Bouaddi, O.; Onibon, Z.; Dada, A.; Lin, X.; Lucero-Prisno, D.E. What sub-Saharan African countries can learn from malaria elimination in China. Trop. Med. Health 2021, 49, 86. [Google Scholar] [CrossRef] [PubMed]
- Lukindu, M. Molecular Tools and Genetic Approaches for Studying the Genetic Structure of Major Malaria Vectors in Sub-Saharan Africa; University of Notre Dame: Notre Dame, IN, USA, 2023. [Google Scholar] [CrossRef]
- Abdelwhab, O.F.; Elaagip, A.; Albsheer, M.M.; Ahmed, A.; Paganotti, G.M.; Hamid, M.M.A. Molecular and morphological identification of suspected Plasmodium vivax vectors in Central and Eastern Sudan. Malar. J. 2021, 20, 132. [Google Scholar] [CrossRef] [PubMed]
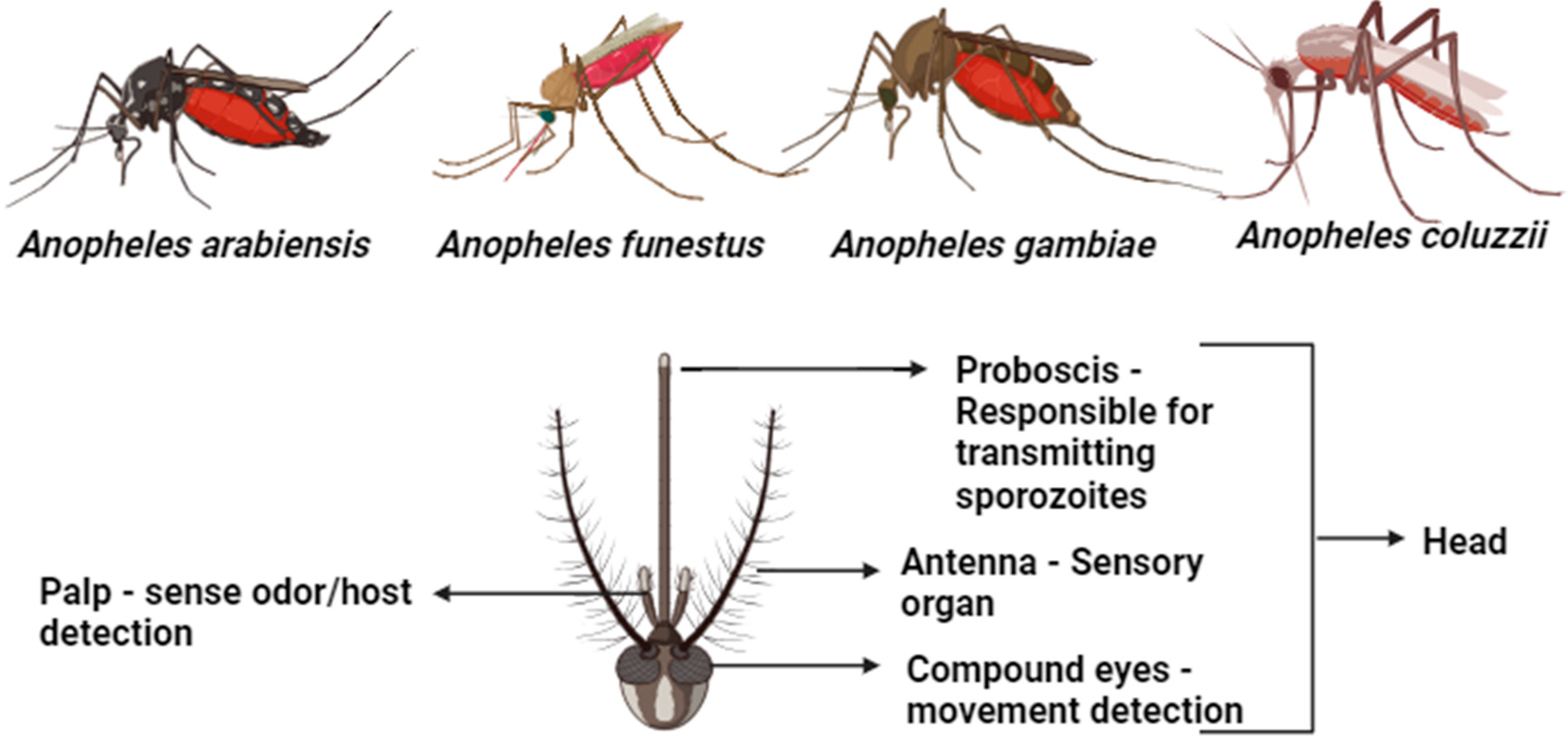
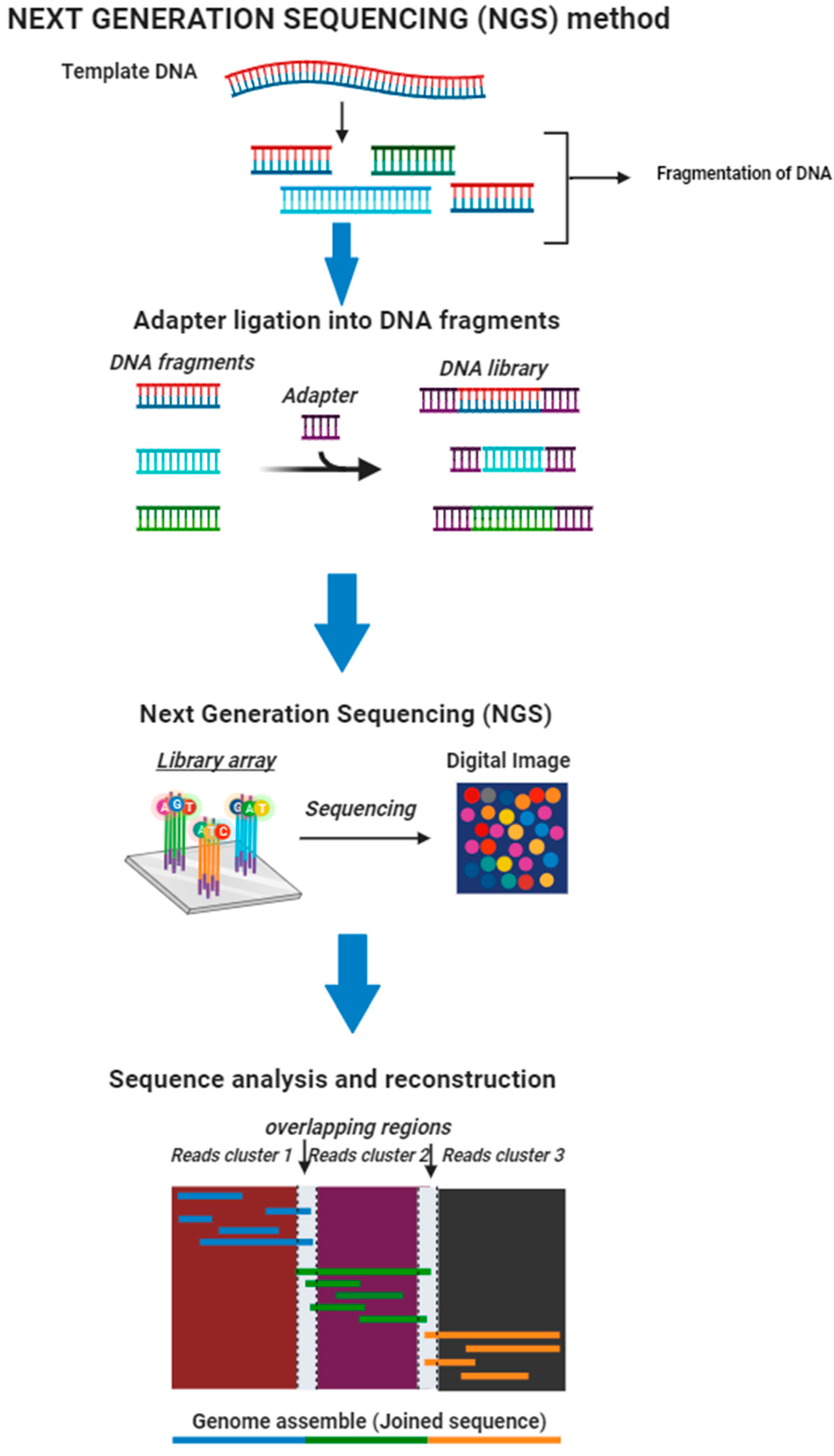
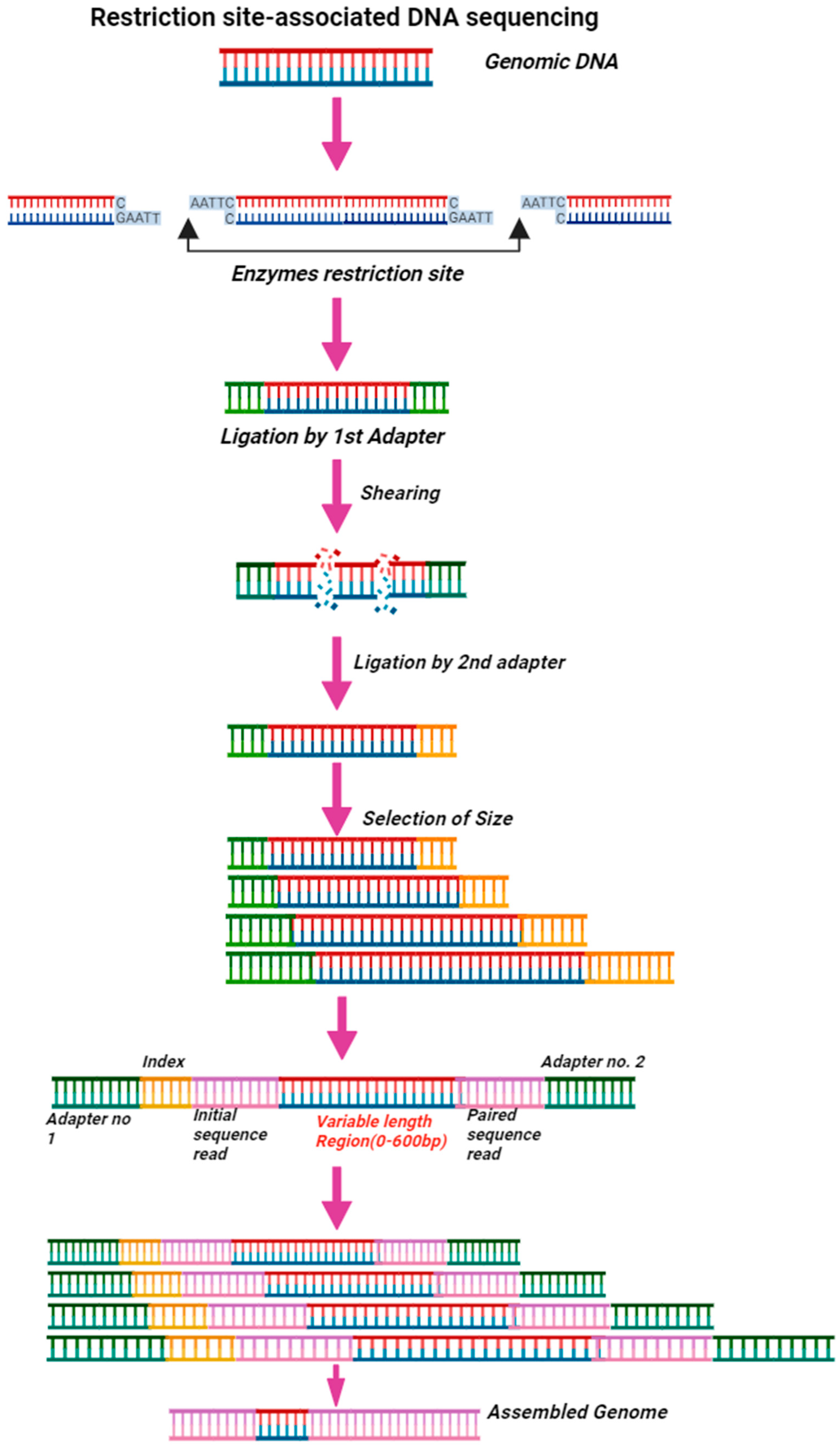
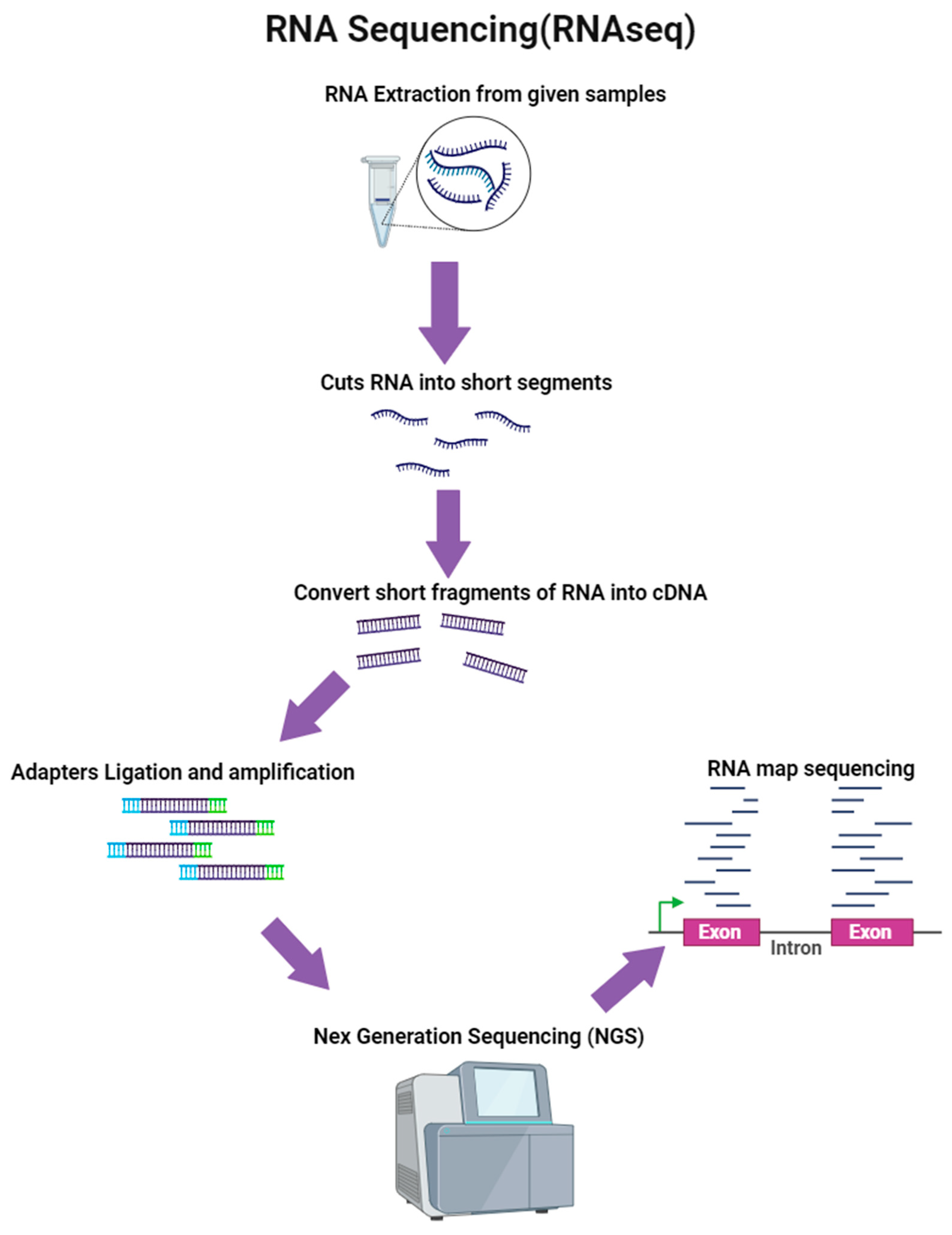
| Markers | Countries/Regions | Anopheles Complex Identified | Reference | |
|---|---|---|---|---|
| 1. | COI ITS2 | Sri Lanka | An. culicinae complex | Weeraratne et al. [143] |
| 2. | COI | Australia (Victoria State) | An. culicidae complex | Batovska et al. [144] |
| 3. | ITS2 COI | Middle Asia and Kazakhstan | An. Maculipennis complex | WHO, [145] |
| 4. | COI ITS2 | Portugal | An. maculipennis complex, An. claviger complex, and Aedes detritus complex | Madeira et al. [146] |
| 5. | ITS2 5.8S 28S | India, i.e., Gurugram, Nuh, Alwar, and New Delhi from northern India, Ranchi, Raipur, and Gadhchiroli from central India, Goa, Bangalore, Mangalore, Chennai, and Mysuru from southern India | An. stephensi | Mishra et al. [147] |
| 6. | ITS2 16S-rDNA | North-central Nigeria | An. culicidae | Iyiola et al. [148] |
| 7. | ITS2 COI | Karama, west Sulawesi, and Indonesia | An. aconitus; An. barbirostris; An. karwari; An. peditaeniatus; An. tessellatus; An. vagus; An. kochi; An. flavirostris; An. nigerrimus; and An. maculatus | Davidson et al. [149] |
| 8. | ITS2 COI | Kenyan highlands (Nyanza Province) | An. gambiae and An. funestus | St Laurent et al. [150] |
| 9. | ITS2 D3 28SDomain | Cameroon, Burkina Faso, Ivory Coast. and Senegal | An. nili | Kengne et al. [124] |
| 10. | ITS2 | South-east Asia (Hanoi suburbs; Hoa Binh; Ninh Binh; Khanh Hoa; Dak Lak; Binh Thuan; Vientiane; Kanchanaburi; Rattanakiry) | An. minimus | Van Bortel et al. [151] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hadebe, M.T.; Malgwi, S.A.; Okpeku, M. Revolutionizing Malaria Vector Control: The Importance of Accurate Species Identification through Enhanced Molecular Capacity. Microorganisms 2024, 12, 82. https://doi.org/10.3390/microorganisms12010082
Hadebe MT, Malgwi SA, Okpeku M. Revolutionizing Malaria Vector Control: The Importance of Accurate Species Identification through Enhanced Molecular Capacity. Microorganisms. 2024; 12(1):82. https://doi.org/10.3390/microorganisms12010082
Chicago/Turabian StyleHadebe, Mzwandile Thabani, Samson Anjikwi Malgwi, and Moses Okpeku. 2024. "Revolutionizing Malaria Vector Control: The Importance of Accurate Species Identification through Enhanced Molecular Capacity" Microorganisms 12, no. 1: 82. https://doi.org/10.3390/microorganisms12010082
APA StyleHadebe, M. T., Malgwi, S. A., & Okpeku, M. (2024). Revolutionizing Malaria Vector Control: The Importance of Accurate Species Identification through Enhanced Molecular Capacity. Microorganisms, 12(1), 82. https://doi.org/10.3390/microorganisms12010082






