Methanobrevibacter massiliense and Pyramidobacter piscolens Co-Culture Illustrates Transkingdom Symbiosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical and Regulatory Statement
2.2. Sample Collection
2.3. Methanogen DNA Detection
2.4. Culturing M. massiliense
2.5. Culture Microscopy
2.6. MALDI-TOF Mass Spectrometry Culture
2.7. P. piscolens Antibiotic Susceptibility Testing and M. massiliense Isolation Attempts
2.8. Whole Genome Sequencing and Analysis
3. Results
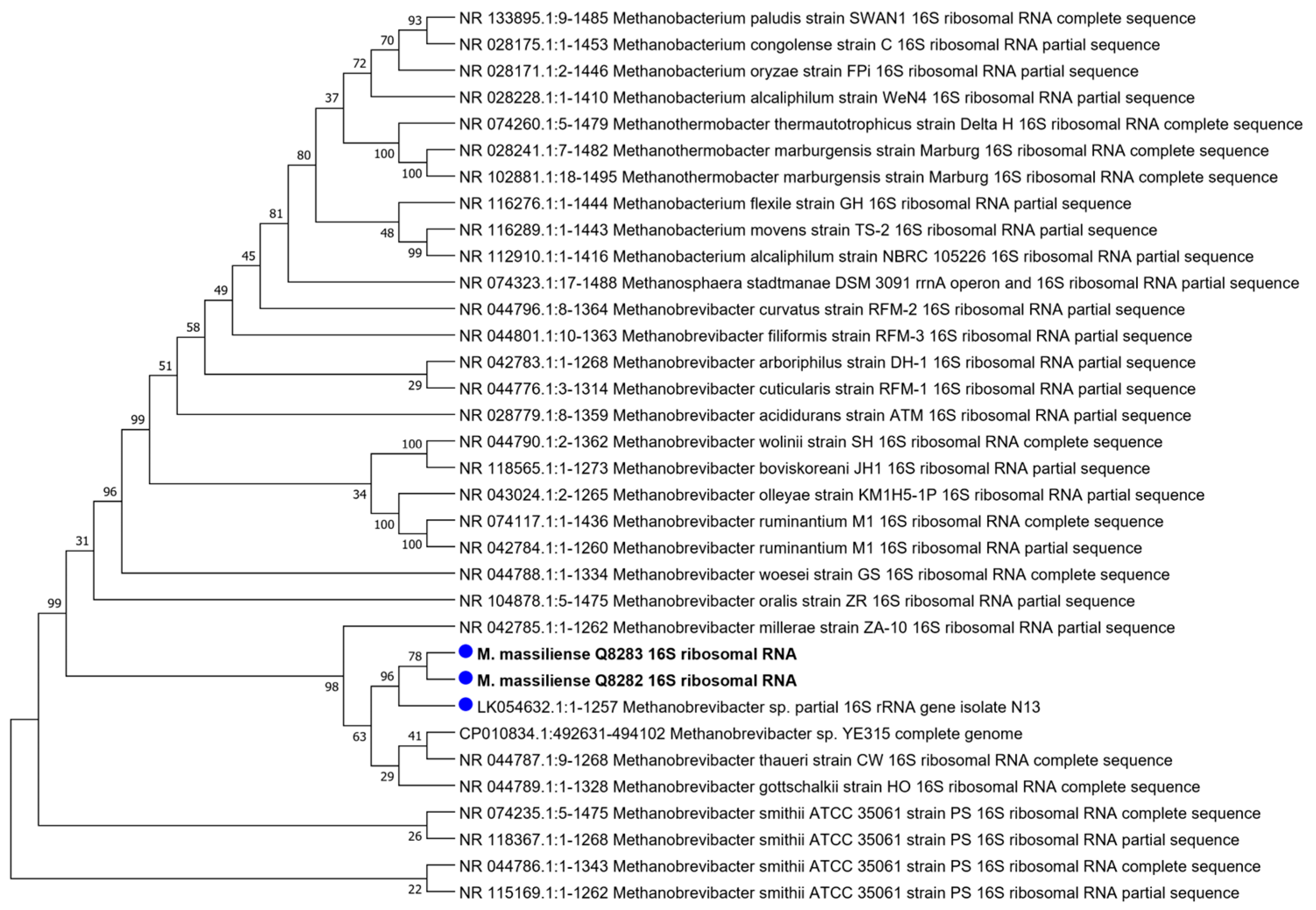
3.1. Molecular Identification
3.2. Co-Culture of M. massiliense with P. piscolens
3.3. Microscopy
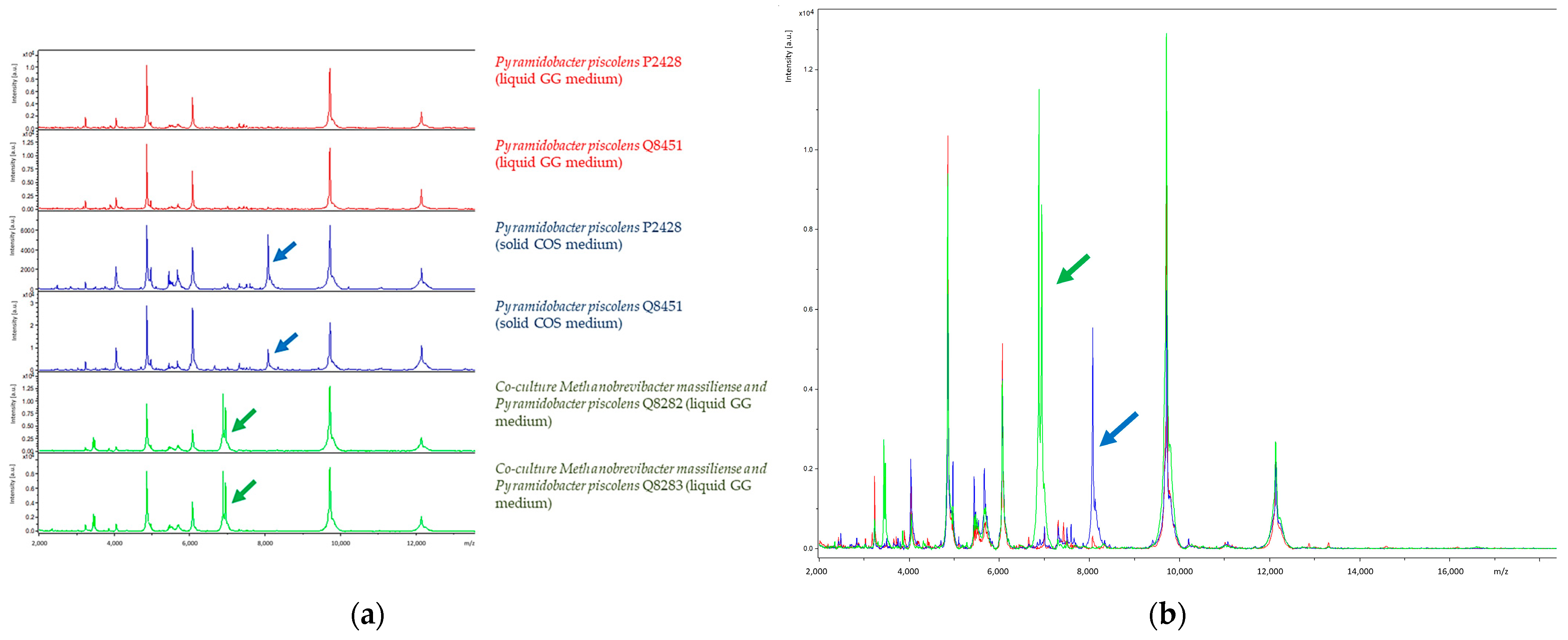
3.4. MALDI-TOF Mass Spectrometry
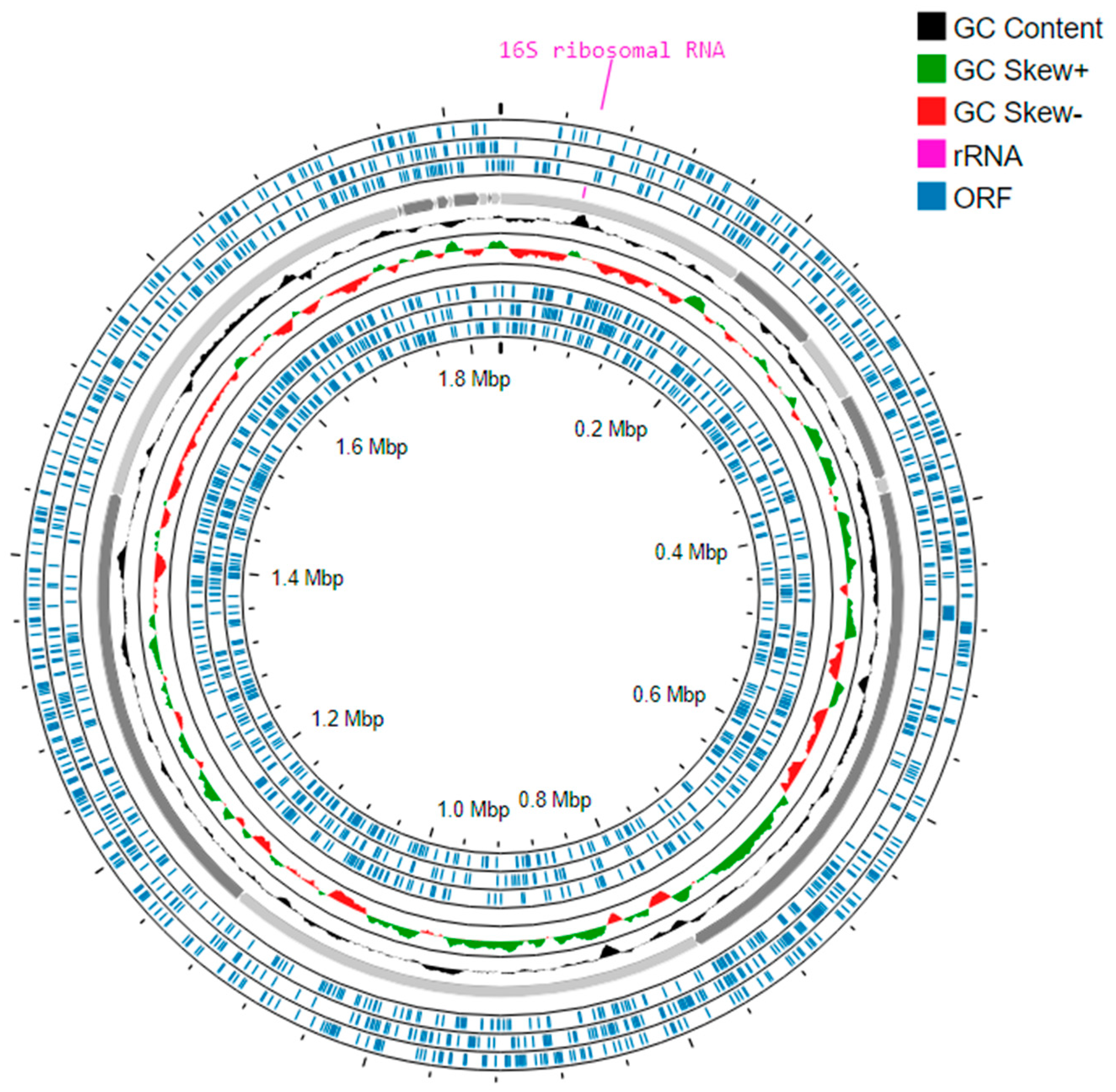
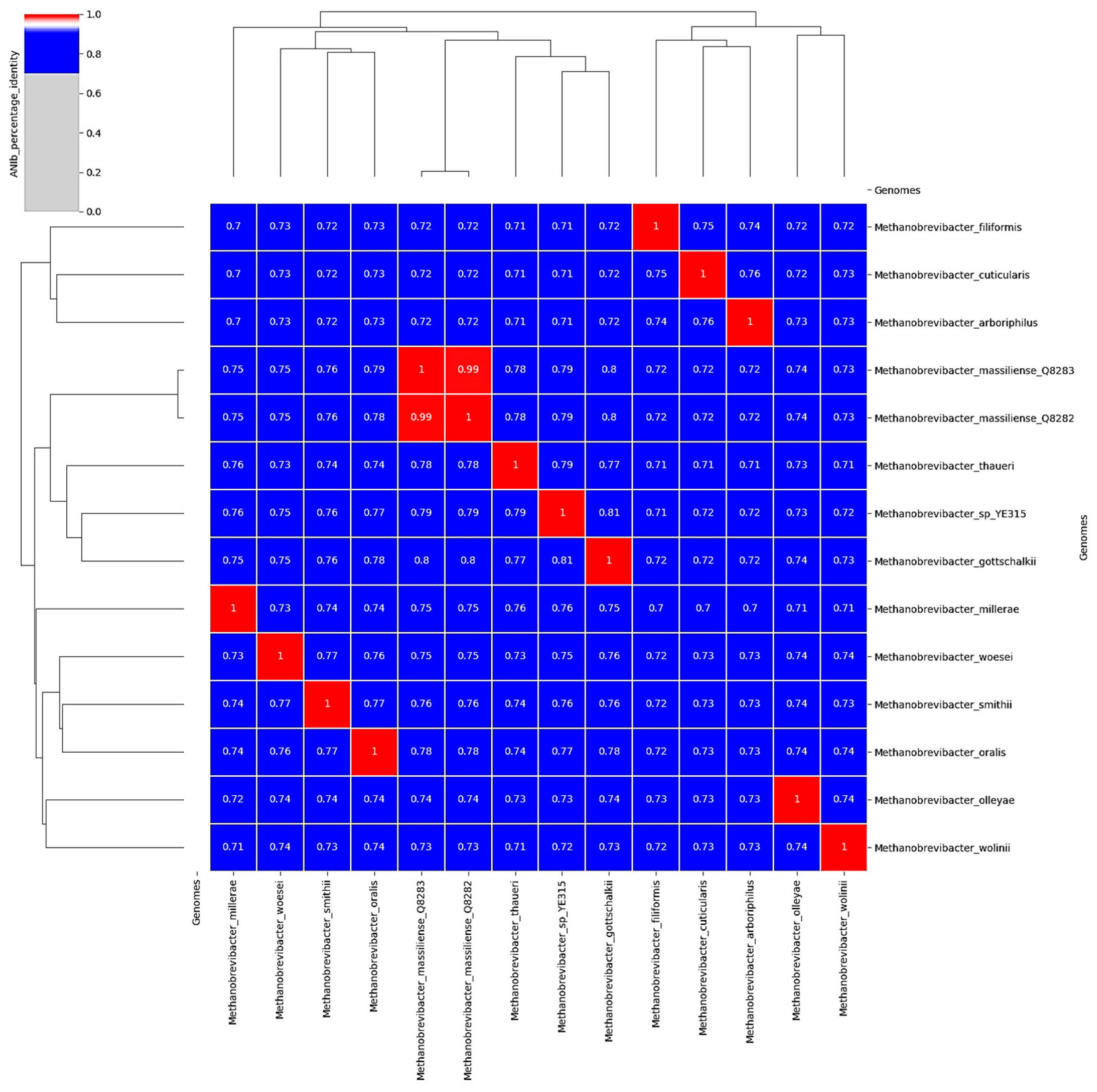
3.5. Whole Genome Sequencing and Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gaci, N.; Borrel, G.; Tottey, W.; O’Toole, P.W.; Brugère, J.-F. Archaea and the Human Gut: New Beginning of an Old Story. World J. Gastroenterol. WJG 2014, 20, 16062–16078. [Google Scholar] [CrossRef] [PubMed]
- Belmok, A.; de Cena, J.A.; Kyaw, C.M.; Damé-Teixeira, N. The Oral Archaeome: A Scoping Review. J. Dent. Res. 2020, 99, 630–643. [Google Scholar] [CrossRef] [PubMed]
- Togo, A.H.; Grine, G.; Khelaifia, S.; des Robert, C.; Brevaut, V.; Caputo, A.; Baptiste, E.; Bonnet, M.; Levasseur, A.; Drancourt, M.; et al. Culture of Methanogenic Archaea from Human Colostrum and Milk. Sci. Rep. 2019, 9, 18653. [Google Scholar] [CrossRef] [PubMed]
- Sereme, Y.; Guindo, C.O.; Filleron, A.; Corbeau, P.; Tran, T.A.; Drancourt, M.; Vitte, J.; Grine, G. Meconial Methanobrevibacter Smithii Suggests Intrauterine Methanogen Colonization in Preterm Neonates. Curr. Res. Microb. Sci. 2021, 2, 100034. [Google Scholar] [CrossRef]
- Grine, G.; Boualam, M.A.; Drancourt, M.M. Methanobrevibacter smithii, a methanogen consistently colonising the newborn stomach. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 2449–2455. [Google Scholar] [CrossRef] [PubMed]
- Guindo, C.O.; Drancourt, M.; Grine, G. Digestive Tract Methanodrome: Physiological Roles of Human Microbiota-Associated Methanogens. Microb. Pathog. 2020, 149, 104425. [Google Scholar] [CrossRef] [PubMed]
- Huynh, H.T.T.; Nkamga, V.D.; Signoli, M.; Tzortzis, S.; Pinguet, R.; Audoly, G.; Aboudharam, G.; Drancourt, M. Restricted Diversity of Dental Calculus Methanogens over Five Centuries, France. Sci. Rep. 2016, 6, 25775. [Google Scholar] [CrossRef]
- Warinner, C. Dental Calculus and the Evolution of the Human Oral Microbiome. J. Calif. Dent. Assoc. 2016, 44, 411–420. [Google Scholar] [CrossRef]
- Weyrich, L.S. The Evolutionary History of the Human Oral Microbiota and Its Implications for Modern Health. Periodontology 2000 2021, 85, 90–100. [Google Scholar] [CrossRef]
- Granehäll, L.; Huang, K.D.; Tett, A.; Manghi, P.; Paladin, A.; O’Sullivan, N.; Rota-Stabelli, O.; Segata, N.; Zink, A.; Maixner, F. Metagenomic Analysis of Ancient Dental Calculus Reveals Unexplored Diversity of Oral Archaeal Methanobrevibacter. Microbiome 2021, 9, 197. [Google Scholar] [CrossRef]
- Grine, G.; Terrer, E.; Boualam, M.A.; Aboudharam, G.; Chaudet, H.; Ruimy, R.; Drancourt, M. Tobacco-Smoking-Related Prevalence of Methanogens in the Oral Fluid Microbiota. Sci. Rep. 2018, 8, 9197. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, J.M.; Campbell, J.H.; Bhandari, A.R.; Jesionowski, A.M.; Vickerman, M.M. Molecular Analysis of 16S rRNA Genes Identifies Potentially Periodontal Pathogenic Bacteria and Archaea in the Plaque of Partially Erupted Third Molars. J. Oral Maxillofac. Surg. 2012, 70, 1507–1514.e6. [Google Scholar] [CrossRef] [PubMed]
- Kulik, E.M.; Sandmeier, H.; Hinni, K.; Meyer, J. Identification of Archaeal rDNA from Subgingival Dental Plaque by PCR Amplification and Sequence Analysis. FEMS Microbiol. Lett. 2001, 196, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Lepp, P.W.; Brinig, M.M.; Ouverney, C.C.; Palm, K.; Armitage, G.C.; Relman, D.A. Methanogenic Archaea and Human Periodontal Disease. Proc. Natl. Acad. Sci. USA 2004, 101, 6176–6181. [Google Scholar] [CrossRef] [PubMed]
- Bringuier, A.; Khelaifia, S.; Richet, H.; Aboudharam, G.; Drancourt, M. Real-Time PCR Quantification of Methanobrevibacter Oralis in Periodontitis. J. Clin. Microbiol. 2013, 51, 993–994. [Google Scholar] [CrossRef] [PubMed]
- Huynh, H.T.T.; Pignoly, M.; Nkamga, V.D.; Drancourt, M.; Aboudharam, G. The Repertoire of Archaea Cultivated from Severe Periodontitis. PLoS ONE 2015, 10, e0121565. [Google Scholar] [CrossRef] [PubMed]
- Belkacemi, S.; Mazel, A.; Tardivo, D.; Tavitian, P.; Stephan, G.; Bianca, G.; Terrer, E.; Drancourt, M.; Aboudharam, G. Peri-Implantitis-Associated Methanogens: A Preliminary Report. Sci. Rep. 2018, 8, 9447. [Google Scholar] [CrossRef] [PubMed]
- Faveri, M.; Gonçalves, L.F.H.; Feres, M.; Figueiredo, L.C.; Gouveia, L.A.; Shibli, J.A.; Mayer, M.P.A. Prevalence and Microbiological Diversity of Archaea in Peri-Implantitis Subjects by 16S Ribosomal RNA Clonal Analysis. J. Periodontal Res. 2011, 46, 338–344. [Google Scholar] [CrossRef]
- Aleksandrowicz, P.; Brzezińska-Błaszczyk, E.; Dudko, A.; Agier, J. Archaea Occurrence in the Subgingival Biofilm in Patients with Peri-Implantitis and Periodontitis. Int. J. Periodontics Restor. Dent. 2020, 40, 677–683. [Google Scholar] [CrossRef]
- Vianna, M.E.; Conrads, G.; Gomes, B.P.F.A.; Horz, H.P. Identification and Quantification of Archaea Involved in Primary Endodontic Infections. J. Clin. Microbiol. 2006, 44, 1274–1282. [Google Scholar] [CrossRef]
- Jiang, Y.T.; Xia, W.W.; Li, C.L.; Jiang, W.; Liang, J.P. Preliminary Study of the Presence and Association of Bacteria and Archaea in Teeth with Apical Periodontitis. Int. Endod. J. 2009, 42, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Vickerman, M.M.; Brossard, K.A.; Funk, D.B.; Jesionowski, A.M.; Gill, S.R.Y. Phylogenetic Analysis of Bacterial and Archaeal Species in Symptomatic and Asymptomatic Endodontic Infections. J. Med. Microbiol. 2007, 56, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Efenberger, M.; Agier, J.; Pawłowska, E.; Brzezińska-Błaszczyk, E. Archaea Prevalence in Inflamed Pulp Tissues. Cent. Eur. J. Immunol. 2015, 40, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Brzezińska-Błaszczyk, E.; Pawłowska, E.; Płoszaj, T.; Witas, H.; Godzik, U.; Agier, J. Presence of Archaea and Selected Bacteria in Infected Root Canal Systems. Can. J. Microbiol. 2018, 64, 317–326. [Google Scholar] [CrossRef]
- Sogodogo, E.; Drancourt, M.; Grine, G. Methanogens as Emerging Pathogens in Anaerobic Abscesses. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 811–818. [Google Scholar] [CrossRef]
- Ito, Y.; Sato, T.; Yamaki, K.; Mayanagi, G.; Hashimoto, K.; Shimauchi, H.; Takahashi, N. Microflora Profiling of Infected Root Canal before and after Treatment Using Culture-Independent Methods. J. Microbiol. Seoul Korea 2012, 50, 58–62. [Google Scholar] [CrossRef]
- Vianna, M.E.; Conrads, G.; Gomes, B.P.F.A.; Horz, H.P. T-RFLP-Based mcrA Gene Analysis of Methanogenic Archaea in Association with Oral Infections and Evidence of a Novel Methanobrevibacter Phylotype. Oral Microbiol. Immunol. 2009, 24, 417–422. [Google Scholar] [CrossRef]
- Huynh, H.T.T.; Pignoly, M.; Drancourt, M.; Aboudharam, G. A New Methanogen “Methanobrevibacter massiliense” Isolated in a Case of Severe Periodontitis. BMC Res. Notes 2017, 10, 657. [Google Scholar] [CrossRef]
- Sogodogo, E.; Doumbo, O.; Aboudharam, G.; Kouriba, B.; Diawara, O.; Koita, H.; Togora, S.; Drancourt, M. First Characterization of Methanogens in Oral Cavity in Malian Patients with Oral Cavity Pathologies. BMC Oral Health 2019, 19, 232. [Google Scholar] [CrossRef]
- Sogodogo, E.; Fellag, M.; Loukil, A.; Nkamga, V.D.; Michel, J.; Dessi, P.; Fournier, P.-E.; Drancourt, M. Nine Cases of Methanogenic Archaea in Refractory Sinusitis, an Emerging Clinical Entity. Front. Public Health 2019, 7, 38. [Google Scholar] [CrossRef]
- Khelaifia, S.; Lagier, J.-C.; Nkamga, V.D.; Guilhot, E.; Drancourt, M.; Raoult, D. Aerobic Culture of Methanogenic Archaea without an External Source of Hydrogen. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 985–991. [Google Scholar] [CrossRef] [PubMed]
- Guindo, C.O.; Davoust, B.; Drancourt, M.; Grine, G. Diversity of Methanogens in Animals’ Gut. Microorganisms 2020, 9, 13. [Google Scholar] [CrossRef] [PubMed]
- Khelaifia, S.; Raoult, D.; Drancourt, M. A Versatile Medium for Cultivating Methanogenic Archaea. PLoS ONE 2013, 8, e61563. [Google Scholar] [CrossRef]
- Pilliol, V.; Guindo, C.O.; Terrer, E.; Aboudharam, G.; Drancourt, M.; Grine, G. Culturing Clinical Methanobrevibacter Smithii Using GG Medium in a Minimal Anaerobe Atmosphere. J. Microbiol. Methods 2023, 207, 106704. [Google Scholar] [CrossRef] [PubMed]
- Lambrecht, J.; Cichocki, N.; Hübschmann, T.; Koch, C.; Harms, H.; Müller, S. Flow Cytometric Quantification, Sorting and Sequencing of Methanogenic Archaea Based on F420 Autofluorescence. Microb. Cell Factories 2017, 16, 180. [Google Scholar] [CrossRef] [PubMed]
- Raskin, L.; Stromley, J.M.; Rittmann, B.E.; Stahl, D.A. Group-Specific 16S rRNA Hybridization Probes to Describe Natural Communities of Methanogens. Appl. Environ. Microbiol. 1994, 60, 1232–1240. [Google Scholar] [CrossRef] [PubMed]
- Amann, R.I.; Binder, B.J.; Olson, R.J.; Chisholm, S.W.; Devereux, R.; Stahl, D.A. Combination of 16S rRNA-Targeted Oligonucleotide Probes with Flow Cytometry for Analyzing Mixed Microbial Populations. Appl. Environ. Microbiol. 1990, 56, 1919–1925. [Google Scholar] [CrossRef]
- Guindo, C.O.; Amir, L.; Couderc, C.; Drancourt, M.; Grine, G. Rapid Identification of Clinically Interesting Methanogens Using an Improved MALDI-TOF-MS Assay. Access Microbiol. 2022, 4, 000372. [Google Scholar] [CrossRef]
- Dridi, B.; Fardeau, M.-L.; Ollivier, B.; Raoult, D.; Drancourt, M. The Antimicrobial Resistance Pattern of Cultured Human Methanogens Reflects the Unique Phylogenetic Position of Archaea. J. Antimicrob. Chemother. 2011, 66, 2038–2044. [Google Scholar] [CrossRef]
- Downes, J.; Vartoukian, S.R.; Dewhirst, F.E.; Izard, J.; Chen, T.; Yu, W.-H.; Sutcliffe, I.C.; Wade, W.G. Pyramidobacter piscolens Gen. Nov., sp. Nov., a Member of the Phylum ‘Synergistetes’ Isolated from the Human Oral Cavity. Int. J. Syst. Evol. Microbiol. 2009, 59, 972–980. [Google Scholar] [CrossRef]
- Khelaifia, S.; Ramonet, P.-Y.; Bedotto Buffet, M.; Drancourt, M. A Semi-Automated Protocol for Archaea DNA Extraction from Stools. BMC Res. Notes 2013, 6, 186. [Google Scholar] [CrossRef] [PubMed]
- Morsli, M.; Kerharo, Q.; Delerce, J.; Roche, P.-H.; Troude, L.; Drancourt, M. Haemophilus Influenzae Meningitis Direct Diagnosis by Metagenomic Next-Generation Sequencing: A Case Report. Pathog. Basel Switz. 2021, 10, 461. [Google Scholar] [CrossRef] [PubMed]
- Morsli, M.; Vincent, J.-J.; Milliere, L.; Colson, P.; Drancourt, M. Direct Next-Generation Sequencing Diagnosis of Echovirus 9 Meningitis, France. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 2037–2039. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and Taxonomy in Diagnostics for Food Security: Soft-Rotting Enterobacterial Plant Pathogens. Anal. Methods 2015, 8, 12–24. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS Is an Automated High-Throughput Platform for State-of-the-Art Genome-Based Taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete Genome Sequence of DSM 30083(T), the Type Strain (U5/41(T)) of Escherichia Coli, and a Proposal for Delineating Subspecies in Microbial Taxonomy. Stand. Genomic Sci. 2014, 9, 2. [Google Scholar] [CrossRef]
- Miller, T.L.; Lin, C. Description of Methanobrevibacter gottschalkii sp. Nov., Methanobrevibacter thaueri sp. Nov., Methanobrevibacter woesei sp. Nov. and Methanobrevibacter wolinii sp. Nov. Int. J. Syst. Evol. Microbiol. 2002, 52, 819–822. [Google Scholar] [CrossRef]
- Li, G.; Yin, P.; Chu, S.; Gao, W.; Cui, S.; Guo, S.; Xu, Y.; Yuan, E.; Zhu, T.; You, J.; et al. Correlation Analysis between GDM and Gut Microbial Composition in Late Pregnancy. J. Diabetes Res. 2021, 2021, 8892849. [Google Scholar] [CrossRef]
- Ma, J.; Sun, J.; Bai, H.; Ma, H.; Wang, K.; Wang, J.; Yu, X.; Pan, Y.; Yao, J. Influence of Flax Seeds on the Gut Microbiota of Elderly Patients with Constipation. J. Multidiscip. Healthc. 2022, 15, 2407–2418. [Google Scholar] [CrossRef]
- Marchandin, H.; Damay, A.; Roudière, L.; Teyssier, C.; Zorgniotti, I.; Dechaud, H.; Jean-Pierre, H.; Jumas-Bilak, E. Phylogeny, Diversity and Host Specialization in the Phylum Synergistetes with Emphasis on Strains and Clones of Human Origin. Res. Microbiol. 2010, 161, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Teng, J.; Wang, X.; Xu, B.; Niu, Y.; Ma, L.; Yan, X. Multi-Omics Analysis Reveals Gut Microbiota-Induced Intramuscular Fat Deposition via Regulating Expression of Lipogenesis-Associated Genes. Anim. Nutr. Zhongguo Xu Mu Shou Yi Xue Hui 2022, 9, 84–99. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Khan, S.; Webb, R.; Denman, S.; McSweeney, C. Characterization and Survey in Cattle of a Rumen pyrimadobacter sp. Which Degrades the Plant Toxin Fluoroacetate. FEMS Microbiol. Ecol. 2020, 96, fiaa077. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Chan, Y.; You, M.; Lacap-Bugler, D.C.; Leung, W.K.; Watt, R.M. In-Depth Snapshot of the Equine Subgingival Microbiome. Microb. Pathog. 2016, 94, 76–89. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, A.; Brusa, T.; Rutili, A.; Canzi, E.; Biavati, B. Isolation and Characterization of Methanobrevibacter oralis sp. Nov. Curr. Microbiol. 1994, 29, 7–12. [Google Scholar] [CrossRef]
- Barnhart, E.P.; McClure, M.A.; Johnson, K.; Cleveland, S.; Hunt, K.A.; Fields, M.W. Potential Role of Acetyl-CoA Synthetase (Acs) and Malate Dehydrogenase (Mae) in the Evolution of the Acetate Switch in Bacteria and Archaea. Sci. Rep. 2015, 5, 12498. [Google Scholar] [CrossRef]
- Ghssein, G.; Brutesco, C.; Ouerdane, L.; Fojcik, C.; Izaute, A.; Wang, S.; Hajjar, C.; Lobinski, R.; Lemaire, D.; Richaud, P.; et al. Biosynthesis of a Broad-Spectrum Nicotianamine-like Metallophore in Staphylococcus aureus. Science 2016, 352, 1105–1109. [Google Scholar] [CrossRef]
- Lyu, Z.; Shao, N.; Akinyemi, T.; Whitman, W.B. Methanogenesis. Curr. Biol. 2018, 28, R727–R732. [Google Scholar] [CrossRef]


| M. massiliense Q8282 | M. massiliense Q8283 | P. piscolens Q8451 | P. piscolens Q8452 | |
|---|---|---|---|---|
| Total sequence length (bp) | 1,834,388 | 2,299,297 | 2,662,074 | 2,638,206 |
| Number of sequences | 19 | 15 | 11 | 4 |
| Longest sequences (bp) | 384,479 | 551,700 | 846,238 | 1,351,082 |
| N50 (bp) | 325,131 | 400,054 | 616,914 | 1,351,082 |
| Gap Ratio (%) | 0 | 0 | 0.001503 | 0.078728 |
| GC content (%) | 31.3 | 30.6 | 59.6 | 59.8 |
| Number of CDSs | 1903 | 2332 | 2512 | 2438 |
| Average protein length | 275.8 | 277.9 | 310.9 | 315.8 |
| Coding ratio (%) | 85.9 | 84.5 | 88 | 87.5 |
| Number of rRNAs | 2 | 4 | 5 | 12 |
| Number of tRNAs | 28 | 32 | 55 | 57 |
| Number of CRISPRs | 3 | 2 | 2 | 3 |
| CheckM completeness (%) | 100 | 97.67 | 100 | 100 |
| CheckM contamination (%) | 0 | 4.72 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pilliol, V.; Beye, M.; Terlier, L.; Balmelle, J.; Kacel, I.; Lan, R.; Aboudharam, G.; Grine, G.; Terrer, E. Methanobrevibacter massiliense and Pyramidobacter piscolens Co-Culture Illustrates Transkingdom Symbiosis. Microorganisms 2024, 12, 215. https://doi.org/10.3390/microorganisms12010215
Pilliol V, Beye M, Terlier L, Balmelle J, Kacel I, Lan R, Aboudharam G, Grine G, Terrer E. Methanobrevibacter massiliense and Pyramidobacter piscolens Co-Culture Illustrates Transkingdom Symbiosis. Microorganisms. 2024; 12(1):215. https://doi.org/10.3390/microorganisms12010215
Chicago/Turabian StylePilliol, Virginie, Mamadou Beye, Laureline Terlier, Julien Balmelle, Idir Kacel, Romain Lan, Gérard Aboudharam, Ghiles Grine, and Elodie Terrer. 2024. "Methanobrevibacter massiliense and Pyramidobacter piscolens Co-Culture Illustrates Transkingdom Symbiosis" Microorganisms 12, no. 1: 215. https://doi.org/10.3390/microorganisms12010215
APA StylePilliol, V., Beye, M., Terlier, L., Balmelle, J., Kacel, I., Lan, R., Aboudharam, G., Grine, G., & Terrer, E. (2024). Methanobrevibacter massiliense and Pyramidobacter piscolens Co-Culture Illustrates Transkingdom Symbiosis. Microorganisms, 12(1), 215. https://doi.org/10.3390/microorganisms12010215






