Metagenome and Resistome Analysis of Beta-Lactam-Resistant Bacteria Isolated from River Waters in Surabaya, Indonesia
Abstract
1. Introduction
2. Materials and Methods
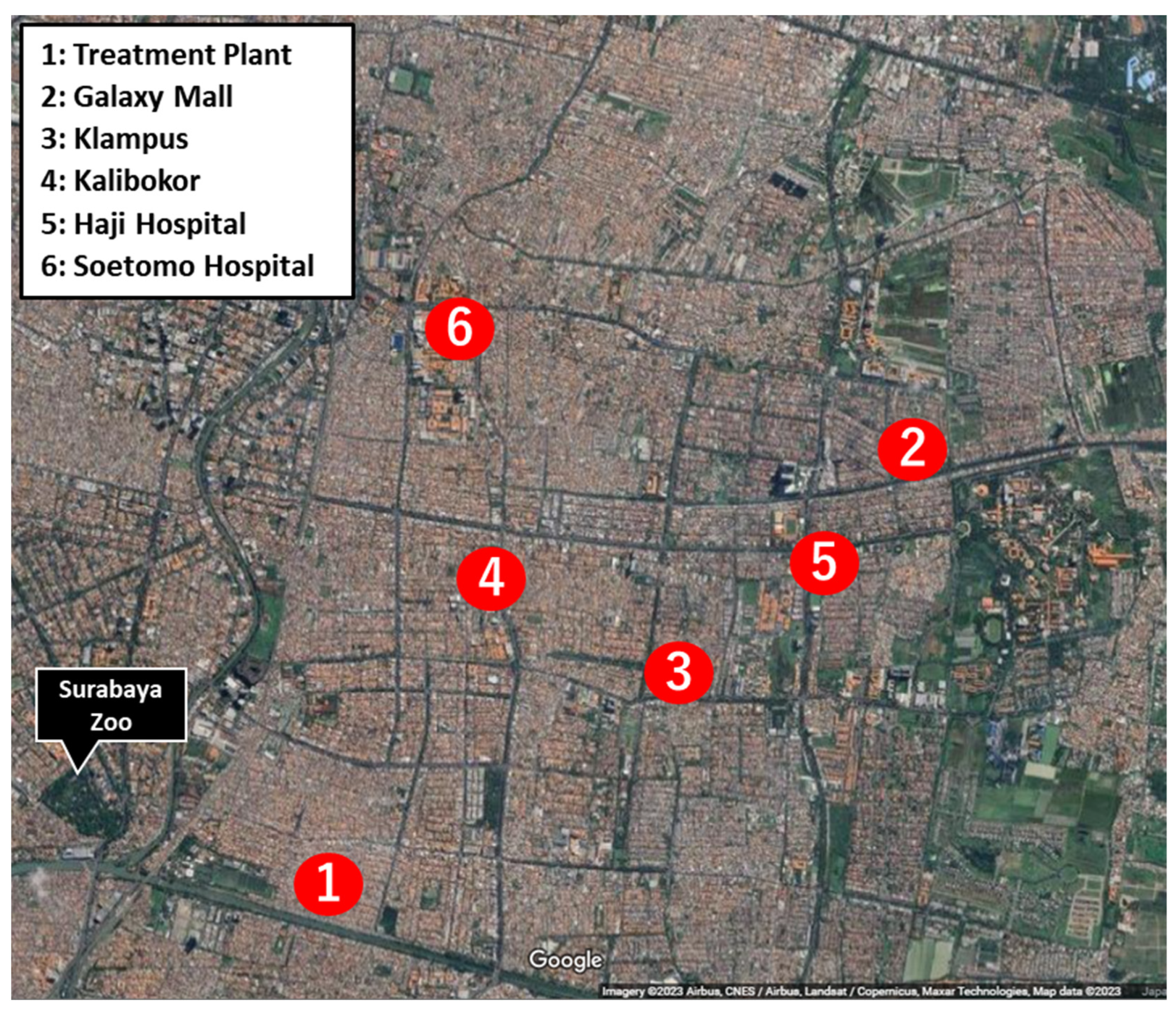
2.1. Sample Collection and Culture Condition
2.2. Metagenomic DNA-Seq Analysis of Extended Spectrum Beta-Lactamase (ESBL)-Producing Bacteria
2.3. Resistome Analysis
3. Results
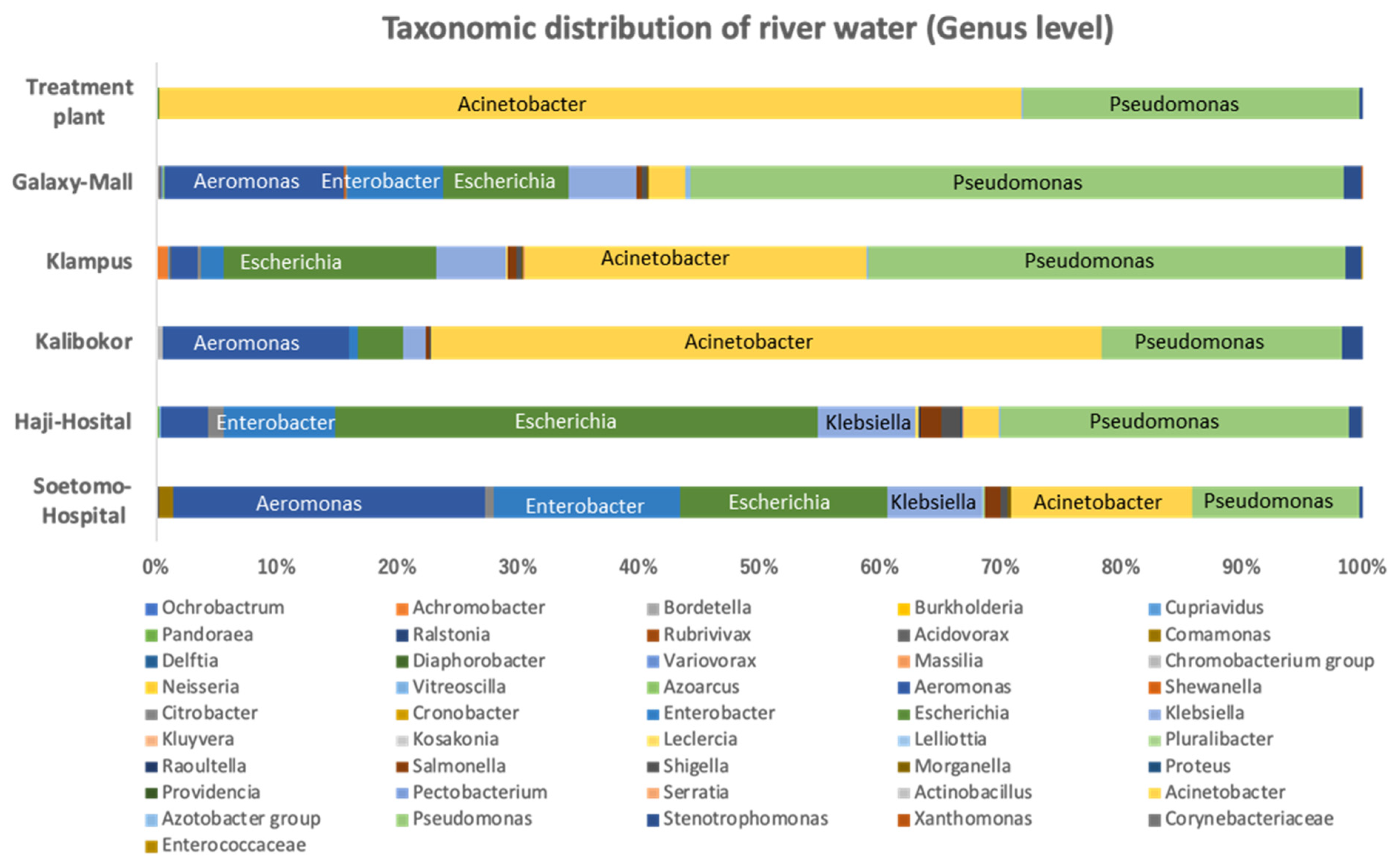
3.1. Bacterial Proportion of Strains Growing on CHROMagar ESBL Plate from Indonesian River Water
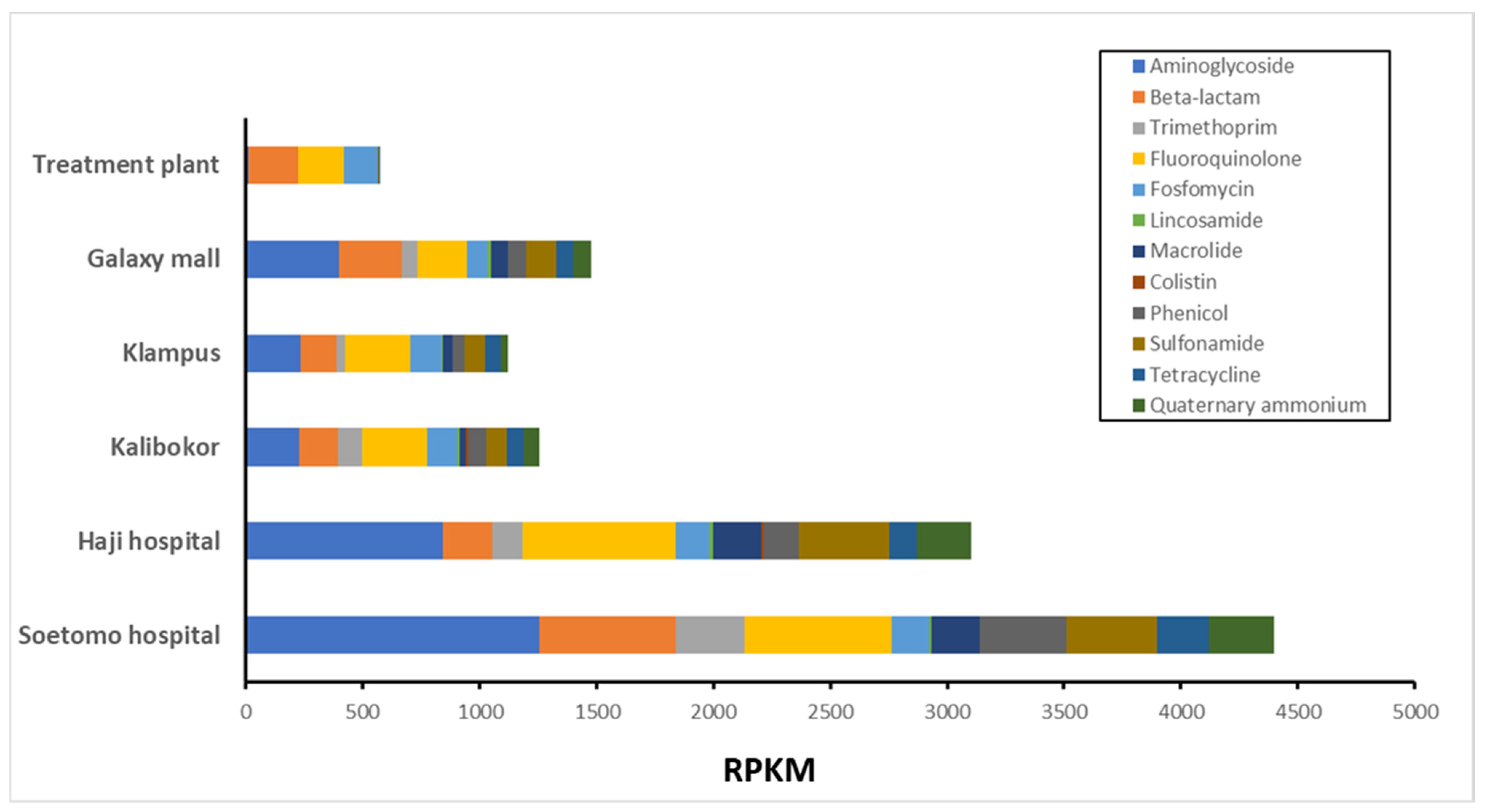
3.2. ARG Proportion of Growing on CHROMagar ESBL Plate from Indonesian River Water
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- O’Neill, J. Review on Antimicrobial Resistance. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; Welcome Trust: London, UK, 2014; pp. 1–16. [Google Scholar]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- McEwen, S.A.; Collignon, P.J. Antimicrobial resistance: A one health perspective. Microbiol. Spectr. 2018, 6, 521–547. [Google Scholar] [CrossRef]
- Aslam, B.; Khurshid, M.; Arshad, M.I.; Muzammil, S.; Rasool, M.; Yasmeen, N.; Shah, T.; Chaudhry, T.H.; Rasool, M.H.; Shahid, A.; et al. Antibiotic resistance: One health one world outlook. Front. Cell. Infect. Microbiol. 2021, 11, 771510. [Google Scholar] [CrossRef]
- Mackenzie, J.S.; Jeggo, M. The one health approach—Why is it so important? Trop. Med. Infect. Dis. 2019, 4, 88. [Google Scholar] [CrossRef]
- Su, M.; Satola, S.W.; Read, T.D. Genome-based prediction of bacterial antibiotic resistance. J. Clin. Microbiol. 2019, 57, e01405–e01418. [Google Scholar] [CrossRef] [PubMed]
- de Abreu, V.A.C.; Perdigão, J.; Almeida, S. Metagenomic approaches to analyze antimicrobial resistance: An overview. Front. Genet. 2021, 11, 575592. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Pachter, L. Bioinformatics for whole-genome shotgun sequencing of microbial communities. PLoS Comput. Biol. 2005, 1, 106–112. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Takeuchi, F.; Sekizuka, T.; Yamashita, A.; Ogasawara, Y.; Mizuta, K.; Kuroda, M. MePIC, metagenomic pathogen identification for clinical specimens. Jpn. J. Infect. Dis. 2014, 67, 62–65. [Google Scholar] [CrossRef]
- Huson, D.H.; Auch, A.F.; Qi, J.; Schuster, S.C. MEGAN analysis of metagenomic data. Genome Res. 2007, 17, 377–386. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Gillings, M.R. Integrons: Past, present, and future. Microbiol. Mol. Biol. Rev. 2014, 78, 257–277. [Google Scholar] [CrossRef] [PubMed]
- Muurinen, J.; Muziasari, W.I.; Hultman, J.; Pärnänen, K.; Narita, V.; Lyra, C.; Fadlillah, L.N.; Rizki, L.P.; Nurmi, W.; Tiedje, J.M.; et al. Antibiotic resistomes and microbiomes in the surface water along the code river in Indonesia reflect drainage basin anthropogenic activities. Environ. Sci. Technol. 2022, 56, 14994–15006. [Google Scholar] [CrossRef]
- Girlich, D.; Grosperrin, V.; Naas, T.; Dortet, L. CHROMagar™ ESBL/mSuperCARBA bi-plate medium for detection of ESBL- and carbapenemase-producing Enterobacteriaceae from spiked stools. Diagn. Microbiol. Infect. Dis. 2019, 95, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Vurayai, M.; Strysko, J.; Kgomanyane, K.; Bayani, O.; Mokomane, M.; Machiya, T.; Arscott-Mills, T.; Goldfarb, D.M.; Steenhoff, A.P.; McGann, C.; et al. Characterizing the bioburden of ESBL-producing organisms in a neonatal unit using chromogenic culture media: A feasible and efficient environmental sampling method. Antimicrob. Resist. Infect. Control 2022, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Carvalheira, A.; Silva, J.; Teixeira, P. Acinetobacter spp. In food and drinking water—A review. Food Microbiol. 2021, 95, 103675. [Google Scholar] [CrossRef]
- Mena, K.D.; Gerba, C.P. Risk assessment of Pseudomonas aeruginosa in water. Rev. Environ. Contam. Toxicol. 2009, 201, 71–115. [Google Scholar] [CrossRef]
- Puspandari, N.; Sunarno, S.; Febrianti, T.; Febriyana, D.; Saraswati, R.D.; Rooslamiati, I.; Amalia, N.; Nursofiah, S.; Hartoyo, Y.; Herna, H.; et al. Extended spectrum beta-lactamase-producing Escherichia coli surveillance in the human, food chain, and environment sectors: Tricycle project (pilot) in Indonesia. One Health 2021, 13, 100331. [Google Scholar] [CrossRef]
- Luo, X.; Mu, K.; Zhao, Y.; Zhang, J.; Qu, Y.; Hu, D.; Jia, Y.; Dai, P.; Weng, J.; Wang, D.; et al. Emergence of blaNDM-1-carrying Aeromonas caviae K433 isolated from patient with community-acquired pneumonia. Front. Microbiol. 2022, 13, 825389. [Google Scholar] [CrossRef]
- Klein, E.Y.; Van Boeckel, T.P.; Martinez, E.M.; Pant, S.; Gandra, S.; Levin, S.A.; Goossens, H.; Laxminarayan, R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 2018, 115, E3463–E3470. [Google Scholar] [CrossRef]
- Ayukekbong, J.A.; Ntemgwa, M.; Atabe, A.N. The threat of antimicrobial resistance in developing countries: Causes and control strategies. Antimicrob. Resist. Infect. Control 2017, 6, 47. [Google Scholar] [CrossRef] [PubMed]
- Aarestrup, F.M. The livestock reservoir for antimicrobial resistance: A personal view on changing patterns of risks, effects of interventions and the way forward. Philos. Trans. R. Soc. B 2015, 370, 20140085. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nomoto, R.; Osawa, K.; Kinoshita, S.; Kitagawa, K.; Nakanishi, N.; Sarassari, R.; Raharjo, D.; Fujisawa, M.; Kuntaman, K.; Shirakawa, T. Metagenome and Resistome Analysis of Beta-Lactam-Resistant Bacteria Isolated from River Waters in Surabaya, Indonesia. Microorganisms 2024, 12, 199. https://doi.org/10.3390/microorganisms12010199
Nomoto R, Osawa K, Kinoshita S, Kitagawa K, Nakanishi N, Sarassari R, Raharjo D, Fujisawa M, Kuntaman K, Shirakawa T. Metagenome and Resistome Analysis of Beta-Lactam-Resistant Bacteria Isolated from River Waters in Surabaya, Indonesia. Microorganisms. 2024; 12(1):199. https://doi.org/10.3390/microorganisms12010199
Chicago/Turabian StyleNomoto, Ryohei, Kayo Osawa, Shohiro Kinoshita, Koichi Kitagawa, Noriko Nakanishi, Rosantia Sarassari, Dadik Raharjo, Masato Fujisawa, Kuntaman Kuntaman, and Toshiro Shirakawa. 2024. "Metagenome and Resistome Analysis of Beta-Lactam-Resistant Bacteria Isolated from River Waters in Surabaya, Indonesia" Microorganisms 12, no. 1: 199. https://doi.org/10.3390/microorganisms12010199
APA StyleNomoto, R., Osawa, K., Kinoshita, S., Kitagawa, K., Nakanishi, N., Sarassari, R., Raharjo, D., Fujisawa, M., Kuntaman, K., & Shirakawa, T. (2024). Metagenome and Resistome Analysis of Beta-Lactam-Resistant Bacteria Isolated from River Waters in Surabaya, Indonesia. Microorganisms, 12(1), 199. https://doi.org/10.3390/microorganisms12010199




