The Natural and Clinical History of Plague: From the Ancient Pandemics to Modern Insights
Abstract
1. Introduction
1.1. The First Pandemic—The Plague of Justinian
1.2. The Second Pandemic—Black Death
1.3. The Third Plague Pandemic
2. Ancient DNA (aDNA) Studies
3. Evolutionary Origins
4. Microorganism Profile
4.1. Discovery of the Bubonic Plague Agent
4.2. Pathogenesis and Virulence Factors
4.3. Adaptations of Yersinia pestis
5. The Epidemiology of Plague
5.1. The Urban and Sylvatic Plagues
5.2. Transmission of Plague
6. Diagnosis, Treatment, and Prevention of Plague
6.1. The Clinical Diagnosis of Plague
6.1.1. Clinical Presentations
6.1.2. Laboratory Testing
6.2. Treatment of Plague
6.3. Prevention
7. Vaccine
8. Things to Do in Plague Research
9. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Asensi, V.; Fierer, J. Of Rats and Men: Poussin’s Plague at Ashdod. Emerg. Infect. Dis. 2018, 24, 186–187. [Google Scholar] [CrossRef]
- Little, L.K. Plague and the End of Antiquity: The Pandemic of 541–750; Little, L.K., Ed.; Cambridge University Press: New York, NY, USA, 2007. [Google Scholar]
- Barbieri, R.; Signoli, M.; Chevé, D.; Costedoat, C.; Tzortzis, S.; Aboudharam, G.; Raoult, D.; Drancourt, M. Yersinia pestis: The Natural History of Plague. Clin. Microbiol. Rev. 2020, 34, 1–44. [Google Scholar] [CrossRef]
- Mordechai, L.; Eisenberg, M.; Newfield, T.P.; Izdebski, A.; Kay, J.E.; Poinar, H. The Justinianic Plague: An inconsequential pandemic? Proc. Natl. Acad. Sci. USA 2019, 116, 25546–25554. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.R.; Rosenthal, K.S.; Pfaller, M.A. Medical Microbiology, 8th ed.; Elsevier: Philadelphia, PA, USA, 2016. [Google Scholar]
- WHO. WHO Expert Committee on Plague, Fourth Report; World Health Organization Technical Report Series; WTO: Geneva, Switzerland, 1970; Volume 447, pp. 1–25. [Google Scholar]
- Glatter, K.A.; Finkelman, P. History of the Plague: An Ancient Pandemic for the Age of COVID-19. Am. J. Med. 2021, 134, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Spyrou, M.A.; Musralina, L.; Gnecchi Ruscone, G.A.; Kocher, A.; Borbone, P.-G.; Khartanovich, V.I.; Buzhilova, A.; Djansugurova, L.; Bos, K.I.; Kühnert, D.; et al. The source of the Black Death in fourteenth-century central Eurasia. Nature 2022, 606, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Bramanti, B.; Stenseth, N.C.; Walløe, L.; Lei, X. Plague: A Disease Which Changed the Path of Human Civilization. Adv. Exp. Med. Biol. 2016, 918, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Raoult, D.; Mouffok, N.; Bitam, I.; Piarroux, R.; Drancourt, M. Plague: History and contemporary analysis. J. Infect. 2013, 66, 18–26. [Google Scholar] [CrossRef]
- Riedel, S. Plague: From natural disease to bioterrorism. Proceedings 2005, 18, 116–124. [Google Scholar] [CrossRef]
- Spyrou, M.A.; Keller, M.; Tukhbatova, R.I.; Scheib, C.L.; Nelson, E.A.; Andrades Valtuena, A.; Neumann, G.U.; Walker, D.; Alterauge, A.; Carty, N.; et al. Phylogeography of the second plague pandemic revealed through analysis of historical Yersinia pestis genomes. Nat. Commun. 2019, 10, 4470. [Google Scholar] [CrossRef]
- Karlsson, G. Plague without rats: The case of fifteenth-century Iceland. J. Mediev. Hist. 1996, 22, 263–284. [Google Scholar] [CrossRef]
- Perry, R.D.; Fetherston, J.D. Yersinia pestis—Etiologic agent of plague. Clin. Microbiol. Rev. 1997, 10, 35–66. [Google Scholar] [CrossRef] [PubMed]
- Benedictow, O.J. The Complete History of the Black Death; Boydell & Brewer: Martlesham, UK, 2021. [Google Scholar]
- Cohn, S.K., Jr. Epidemiology of the Black Death and successive waves of plague. Med. Hist. 2008, 52, 74–100. [Google Scholar] [CrossRef]
- Guellil, M.; Kersten, O.; Namouchi, A.; Luciani, S.; Marota, I.; Arcini, C.A.; Iregren, E.; Lindemann, R.A.; Warfvinge, G.; Bakanidze, L.; et al. A genomic and historical synthesis of plague in 18th century Eurasia. Proc. Natl. Acad. Sci. USA 2020, 117, 28328–28335. [Google Scholar] [CrossRef] [PubMed]
- Alfani, G. Plague in seventeenth-century Europe and the decline of Italy: An epidemiological hypothesis. Eur. Rev. Econ. Hist. 2013, 17, 408–430. [Google Scholar] [CrossRef]
- Eckert, E.A. The retreat of plague from Central Europe, 1640–1720: A geomedical approach. Bull. Hist. Med. 2000, 74, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Bonnici, W. Inspector of Hospitals Ralph Green and the plague in Malta of 1813. J. R. Army Med. Corps 1998, 144, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Moll, I.; Salas Vives, P.; Pujadas-Mora, J.M. Vers une nouvelle modernité sanitaire: L’épidémie de peste de Majorque en 1820. Ann. Démogr. Hist. 2017, 134, 125–149. [Google Scholar] [CrossRef]
- Contreras Mas, A. Legends and documents on the way of the contagion in the epidemic of plague of the majorcan east in 1820. Acad. J. Health Sci. 2021, 36, 174–178. [Google Scholar]
- Bramanti, B.; Dean, K.R.; Walløe, L.; Chr Stenseth, N. The Third Plague Pandemic in Europe. Proc. Biol. Sci. 2019, 286, 20182429. [Google Scholar] [CrossRef]
- Yersin, A. La peste bubonique à Hong-Kong. Ann. Inst. Pasteur Paris 1894, 8, 662–667. [Google Scholar]
- Lynteris, C. In search of lost fleas: Reconsidering Paul-Louis Simond’s contribution to the study of the propagation of plague. Med. Hist. 2022, 66, 242–263. [Google Scholar] [CrossRef]
- Ogata, M. Über die Pestepidemie in Formosa. Centralbl. Bukt. Parasitenkd. Infect. Krankh. 1897, 21, 769–777. [Google Scholar]
- Simond, P.-L. La propagation de la peste. Ann. Inst. Pasteur 1898, 12, 625–687. [Google Scholar]
- Ansari, I.; Grier, G.; Byers, M. Deliberate release: Plague—A review. J. Biosaf. Biosecur. 2020, 2, 10–22. [Google Scholar] [CrossRef]
- Danforth, M.; Novak, M.; Petersen, J.; Mead, P.; Kingry, L.; Weinburke, M.; Buttke, D.; Hacker, G.; Tucker, J.; Niemela, M.; et al. Investigation of and Response to 2 Plague Cases, Yosemite National Park, California, USA, 2015. Emerg. Infect. Dis. 2016, 22, 2045–2053. [Google Scholar] [CrossRef] [PubMed]
- Guy, J.; Gansukh, B. Teenage Boy Dies from Bubonic Plague after Eating Marmot. Available online: https://edition.cnn.com/2020/07/15/asia/mongolia-plague-death-scli-intl/index.html# (accessed on 19 December 2023).
- Cui, Y.; Yu, C.; Yan, Y.; Li, D.; Li, Y.; Jombart, T.; Weinert, L.A.; Wang, Z.; Guo, Z.; Xu, L.; et al. Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis. Proc. Natl. Acad. Sci. USA 2013, 110, 577–582. [Google Scholar] [CrossRef]
- Morelli, G.; Song, Y.; Mazzoni, C.J.; Eppinger, M.; Roumagnac, P.; Wagner, D.M.; Feldkamp, M.; Kusecek, B.; Vogler, A.J.; Li, Y.; et al. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat. Genet. 2010, 42, 1140–1143. [Google Scholar] [CrossRef]
- Bos, K.I.; Herbig, A.; Sahl, J.; Waglechner, N.; Fourment, M.; Forrest, S.A.; Klunk, J.; Schuenemann, V.J.; Poinar, D.; Kuch, M.; et al. Eighteenth century Yersinia pestis genomes reveal the long-term persistence of an historical plague focus. eLife 2016, 5, e12994. [Google Scholar] [CrossRef]
- Keller, M.; Spyrou, M.A.; Scheib, C.L.; Neumann, G.U.; Kropelin, A.; Haas-Gebhard, B.; Paffgen, B.; Haberstroh, J.; Ribera, I.L.A.; Raynaud, C.; et al. Ancient Yersinia pestis genomes from across Western Europe reveal early diversification during the First Pandemic (541–750). Proc. Natl. Acad. Sci. USA 2019, 116, 12363–12372. [Google Scholar] [CrossRef]
- Rasmussen, S.; Allentoft, M.E.; Nielsen, K.; Orlando, L.; Sikora, M.; Sjogren, K.G.; Pedersen, A.G.; Schubert, M.; Van Dam, A.; Kapel, C.M.; et al. Early divergent strains of Yersinia pestis in Eurasia 5000 years ago. Cell 2015, 163, 571–582. [Google Scholar] [CrossRef]
- Wagner, D.M.; Klunk, J.; Harbeck, M.; Devault, A.; Waglechner, N.; Sahl, J.W.; Enk, J.; Birdsell, D.N.; Kuch, M.; Lumibao, C.; et al. Yersinia pestis and the Plague of Justinian 541–543 AD: A genomic analysis. Lancet Infect. Dis. 2014, 14, 319–326. [Google Scholar] [CrossRef]
- Clavel, P.; Louis, L.; Sarkissian, C.; Thèves, C.; Gillet, C.; Chauvey, L.; Tressières, G.; Schiavinato, S.; Calvière-Tonasso, L.; Telmon, N.; et al. Improving the extraction of ancient Yersinia pestis genomes from the dental pulp. iScience 2023, 26, 106787. [Google Scholar] [CrossRef]
- Drancourt, M.; Aboudharam, G.; Signoli, M.; Dutour, O.; Raoult, D. Detection of 400-year-old Yersinia pestis DNA in human dental pulp: An approach to the diagnosis of ancient septicemia. Proc. Natl. Acad. Sci. USA 1998, 95, 12637–12640. [Google Scholar] [CrossRef]
- Seifert, L.; Harbeck, M.; Thomas, A.; Hoke, N.; Zöller, L.; Wiechmann, I.; Grupe, G.; Scholz, H.C.; Riehm, J.M. Strategy for sensitive and specific detection of Yersinia pestis in skeletons of the black death pandemic. PLoS ONE 2013, 8, e75742. [Google Scholar] [CrossRef]
- Schuenemann, V.J.; Bos, K.; DeWitte, S.; Schmedes, S.; Jamieson, J.; Mittnik, A.; Forrest, S.; Coombes, B.K.; Wood, J.W.; Earn, D.J.; et al. Targeted enrichment of ancient pathogens yielding the pPCP1 plasmid of Yersinia pestis from victims of the Black Death. Proc. Natl. Acad. Sci. USA 2011, 108, E746–E752. [Google Scholar] [CrossRef]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef] [PubMed]
- Klunk, J.; Vilgalys, T.P.; Demeure, C.E.; Cheng, X.; Shiratori, M.; Madej, J.; Beau, R.; Elli, D.; Patino, M.I.; Redfern, R.; et al. Evolution of immune genes is associated with the Black Death. Nature 2022, 611, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Pal, V.; Kumar, M.; Tripathi, N.K.; Goel, A.K. Development of a PCR-lateral flow assay for rapid detection of Yersinia pestis, the causative agent of plague. Acta Trop. 2021, 220, 105958. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Schubert, M.; Ermini, L.; Der Sarkissian, C.; Jónsson, H.; Ginolhac, A.; Schaefer, R.; Martin, M.D.; Fernández, R.; Kircher, M.; McCue, M.; et al. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat. Protoc. 2014, 9, 1056–1082. [Google Scholar] [CrossRef] [PubMed]
- Peltzer, A.; Jager, G.; Herbig, A.; Seitz, A.; Kniep, C.; Krause, J.; Nieselt, K. EAGER: Efficient ancient genome reconstruction. Genome Biol. 2016, 17, 60. [Google Scholar] [CrossRef]
- Hubler, R.; Key, F.M.; Warinner, C.; Bos, K.I.; Krause, J.; Herbig, A. HOPS: Automated detection and authentication of pathogen DNA in archaeological remains. Genome Biol. 2019, 20, 280. [Google Scholar] [CrossRef]
- Achtman, M.; Morelli, G.; Zhu, P.; Wirth, T.; Diehl, I.; Kusecek, B.; Vogler, A.J.; Wagner, D.M.; Allender, C.J.; Easterday, W.R.; et al. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl. Acad. Sci. USA 2004, 101, 17837–17842. [Google Scholar] [CrossRef]
- Achtman, M.; Zurth, K.; Morelli, G.; Torrea, G.; Guiyoule, A.; Carniel, E. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 1999, 96, 14043–14048. [Google Scholar] [CrossRef]
- Chain, P.S.; Carniel, E.; Larimer, F.W.; Lamerdin, J.; Stoutland, P.O.; Regala, W.M.; Georgescu, A.M.; Vergez, L.M.; Land, M.L.; Motin, V.L.; et al. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 2004, 101, 13826–13831. [Google Scholar] [CrossRef]
- Chouikha, I.; Hinnebusch, B.J. Silencing urease: A key evolutionary step that facilitated the adaptation of Yersinia pestis to the flea-borne transmission route. Proc. Natl. Acad. Sci. USA 2014, 111, 18709–18714. [Google Scholar] [CrossRef]
- Susat, J.; Lübke, H.; Immel, A.; Brinker, U.; Macāne, A.; Meadows, J.; Steer, B.; Tholey, A.; Zagorska, I.; Gerhards, G.; et al. A 5000-year-old hunter-gatherer already plagued by Yersinia pestis. Cell Rep. 2021, 35, 109278. [Google Scholar] [CrossRef]
- McNally, A.; Thomson, N.R.; Reuter, S.; Wren, B.W. ‘Add, stir and reduce’: Yersinia spp. as model bacteria for pathogen evolution. Nat. Rev. Microbiol. 2016, 14, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Atkinson, S.; Chen, Z.; Cui, Y.; Du, Z.; Han, Y.; Sebbane, F.; Slavin, P.; Song, Y.; Yan, Y.; et al. Yersinia pestis and Plague: Some knowns and unknowns. Zoonoses 2023, 3. [Google Scholar] [CrossRef] [PubMed]
- Worsham, P.L.; Roy, C. Pestoides F, a Yersinia pestis Strain Lacking Plasminogen Activator, is Virulent by the Aerosol Route. In The Genus Yersinia: Entering the Functional Genomic Era; Skurnik, M., Bengoechea, J.A., Granfors, K., Eds.; Springer: Boston, MA, USA, 2003; pp. 129–131. [Google Scholar] [CrossRef]
- Samoilova, S.V.; Samoilova, L.V.; Yezhov, I.N.; Drozdov, I.G.; Anisimov, A.P. Virulence of pPst+ and pPst− strains of Yersinia pestis for guinea-pigs. J. Med. Microbiol. 1996, 45, 440–444. [Google Scholar] [CrossRef] [PubMed]
- Demeure, C.; Dussurget, O.; Fiol, G.M.; Le Guern, A.S.; Savin, C.; Pizarro-Cerda, J. Yersinia pestis and plague: An updated view on evolution, virulence determinants, immune subversion, vaccination and diagnostics. Microbes Infect. 2019, 21, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Wren, B.W. The Yersiniae—A model genus to study the rapid evolution of bacterial pathogens. Nat. Rev. Microbiol. 2003, 1, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.Y.; Tan, I.K.; Tan, M.F.; Dutta, A.; Choo, S.W. Evolutionary study of Yersinia genomes deciphers emergence of human pathogenic species. Sci. Rep. 2016, 6, 36116. [Google Scholar] [CrossRef]
- Rascovan, N.; Sjogren, K.G.; Kristiansen, K.; Nielsen, R.; Willerslev, E.; Desnues, C.; Rasmussen, S. Emergence and Spread of Basal Lineages of Yersinia pestis during the Neolithic Decline. Cell 2019, 176, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Sebbane, F.; Uversky, V.N.; Anisimov, A.P. Yersinia pestis Plasminogen Activator. Biomolecules 2020, 10, 1554. [Google Scholar] [CrossRef]
- Namouchi, A.; Guellil, M.; Kersten, O.; Hänsch, S.; Ottoni, C.; Schmid, B.V.; Pacciani, E.; Quaglia, L.; Vermunt, M.; Bauer, E.L.; et al. Integrative approach using Yersinia pestis genomes to revisit the historical landscape of plague during the Medieval Period. Proc. Natl. Acad. Sci. USA 2018, 115, E11790–E11797. [Google Scholar] [CrossRef]
- Kutyrev, V.V.; Eroshenko, G.A.; Motin, V.L.; Nosov, N.Y.; Krasnov, J.M.; Kukleva, L.M.; Nikiforov, K.A.; Al’khova, Z.V.; Oglodin, E.G.; Guseva, N.P. Phylogeny and Classification of Yersinia pestis through the Lens of Strains from the Plague Foci of Commonwealth of Independent States. Front. Microbiol. 2018, 9, 1106. [Google Scholar] [CrossRef]
- Bos, K.I.; Schuenemann, V.J.; Golding, G.B.; Burbano, H.A.; Waglechner, N.; Coombes, B.K.; McPhee, J.B.; DeWitte, S.N.; Meyer, M.; Schmedes, S.; et al. A draft genome of Yersinia pestis from victims of the Black Death. Nature 2011, 478, 506–510. [Google Scholar] [CrossRef]
- Yang, R. Plague: Recognition, Treatment, and Prevention. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef]
- Zhou, J.; Bi, Y.; Xu, X.; Qiu, Y.; Wang, Q.; Feng, N.; Cui, Y.; Yan, Y.; Zhou, L.; Tan, Y.; et al. Bioluminescent tracking of colonization and clearance dynamics of plasmid-deficient Yersinia pestis strains in a mouse model of septicemic plague. Microbes Infect. 2014, 16, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Chen, Z.; Yang, R. Yersinia pestis: Mechanisms of entry into and resistance to the host cell. Front. Cell. Infect. Microbiol. 2013, 3. [Google Scholar] [CrossRef]
- Bland, D.M.; Miarinjara, A.; Bosio, C.F.; Calarco, J.; Hinnebusch, B.J. Acquisition of yersinia murine toxin enabled Yersinia pestis to expand the range of mammalian hosts that sustain flea-borne plague. PLoS Pathog. 2021, 17, e1009995. [Google Scholar] [CrossRef]
- Hinnebusch, B.J.; Rudolph, A.E.; Cherepanov, P.; Dixon, J.E.; Schwan, T.G.; Forsberg, Å. Role of Yersinia Murine Toxin in Survival of Yersinia pestis in the Midgut of the Flea Vector. Science 2002, 296, 733–735. [Google Scholar] [CrossRef] [PubMed]
- Anisimov, A.P.; Lindler, L.E.; Pier, G.B. Intraspecific Diversity of Yersinia pestis. Clin. Microbiol. Rev. 2004, 17, 434–464. [Google Scholar] [CrossRef] [PubMed]
- Butler, T. Plague history: Yersin’s discovery of the causative bacterium in 1894 enabled, in the subsequent century, scientific progress in understanding the disease and the development of treatments and vaccines. Clin. Microbiol. Infect. 2014, 20, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Drancourt, M.; Signoli, M.; Dang, L.V.; Bizot, B.; Roux, V.; Tzortzis, S.; Raoult, D. Yersinia pestis Orientalis in remains of ancient plague patients. Emerg. Infect. Dis. 2007, 13, 332–333. [Google Scholar] [CrossRef]
- Devignat, R. Varieties of Pasteurella pestis; new hypothesis. Bull. World Health Organ. 1951, 4, 247–263. [Google Scholar]
- Drancourt, M. Finally, plague is plague. Clin. Microbiol. Infect. 2012, 18, 105–106. [Google Scholar] [CrossRef]
- Drancourt, M.; Raoult, D. Genotyping Yersinia pestis in historical plague. Lancet Infect. Dis. 2011, 11, 894–895. [Google Scholar] [CrossRef] [PubMed]
- Drancourt, M.; Roux, V.; Dang, L.V.; Tran-Hung, L.; Castex, D.; Chenal-Francisque, V.; Ogata, H.; Fournier, P.E.; Crubézy, E.; Raoult, D. Genotyping, Orientalis-like Yersinia pestis, and plague pandemics. Emerg. Infect. Dis. 2004, 10, 1585–1592. [Google Scholar] [CrossRef]
- Seifert, L.; Wiechmann, I.; Harbeck, M.; Thomas, A.; Grupe, G.; Projahn, M.; Scholz, H.C.; Riehm, J.M. Genotyping Yersinia pestis in Historical Plague: Evidence for Long-Term Persistence of Y. pestis in Europe from the 14th to the 17th Century. PLoS ONE 2016, 11, e0145194. [Google Scholar] [CrossRef] [PubMed]
- Motin, V.L.; Georgescu, A.M.; Elliott, J.M.; Hu, P.; Worsham, P.L.; Ott, L.L.; Slezak, T.R.; Sokhansanj, B.A.; Regala, W.M.; Brubaker, R.R.; et al. Genetic variability of Yersinia pestis isolates as predicted by PCR-based IS100 genotyping and analysis of structural genes encoding glycerol-3-phosphate dehydrogenase (glpD). J. Bacteriol. 2002, 184, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Tong, Z.; Song, Y.; Han, Y.; Pei, D.; Pang, X.; Zhai, J.; Li, M.; Cui, B.; Qi, Z.; et al. Genetics of metabolic variations between Yersinia pestis biovars and the proposal of a new biovar, microtus. J. Bacteriol. 2004, 186, 5147–5152. [Google Scholar] [CrossRef]
- Bendiner, E. Alexandre Yersin: Pursuer of plague. Hosp. Pract. (Off. Ed.) 1989, 24, 121–128, 131–132, 135–138. [Google Scholar]
- Butler, T. Plague and Other Yersinia Infections; Plenum Press: New York, NY, USA, 1983. [Google Scholar]
- Adeolu, M.; Alnajar, S.; Naushad, S.; Gupta, R. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5575–5599. [Google Scholar] [CrossRef]
- Parte, A.C.; Sarda Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Goker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Sun, Y.C.; Jarrett, C.O.; Bosio, C.F.; Hinnebusch, B.J. Retracing the evolutionary path that led to flea-borne transmission of Yersinia pestis. Cell Host Microbe 2014, 15, 578–586. [Google Scholar] [CrossRef]
- Zimbler, D.L.; Schroeder, J.A.; Eddy, J.L.; Lathem, W.W. Early emergence of Yersinia pestis as a severe respiratory pathogen. Nat. Commun. 2015, 6, 7487. [Google Scholar] [CrossRef]
- Pullen, J.K.; Anderson, G.W., Jr.; Welkos, S.L.; Friedlander, A.M. Analysis of the Yersinia pestis V protein for the presence of linear antibody epitopes. Infect. Immun. 1998, 66, 521–527. [Google Scholar] [CrossRef]
- Do, Y.; Koh, H.; Park, C.G.; Dudziak, D.; Seo, P.; Mehandru, S.; Choi, J.H.; Cheong, C.; Park, S.; Perlin, D.S.; et al. Targeting of LcrV virulence protein from Yersinia pestis to dendritic cells protects mice against pneumonic plague. Eur. J. Immunol. 2010, 40, 2791–2796. [Google Scholar] [CrossRef]
- Kingston, R.; Burke, F.; Robinson, J.H.; Bedford, P.A.; Jones, S.M.; Knight, S.C.; Williamson, E.D. The fraction 1 and V protein antigens of Yersinia pestis activate dendritic cells to induce primary T cell responses. Clin. Exp. Immunol. 2007, 149, 561–569. [Google Scholar] [CrossRef]
- Hu, P.; Elliott, J.; McCready, P.; Skowronski, E.; Garnes, J.; Kobayashi, A.; Brubaker, R.R.; Garcia, E. Structural organization of virulence-associated plasmids of Yersinia pestis. J. Bacteriol. 1998, 180, 5192–5202. [Google Scholar] [CrossRef]
- Spinner, J.L.; Winfree, S.; Starr, T.; Shannon, J.G.; Nair, V.; Steele-Mortimer, O.; Hinnebusch, B.J. Yersinia pestis survival and replication within human neutrophil phagosomes and uptake of infected neutrophils by macrophages. J. Leukoc. Biol. 2014, 95, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Cavanaugh, D.C.; Randall, R. The role of multiplication of Pasteurella pestis in mononuclear phagocytes in the pathogenesis of flea-borne plague. J. Immunol. 1959, 83, 348–363. [Google Scholar] [CrossRef]
- Straley, S.C.; Skrzypek, E.; Plano, G.V.; Bliska, J.B. Yops of Yersinia spp. pathogenic for humans. Infect. Immun. 1993, 61, 3105–3110. [Google Scholar] [CrossRef]
- Leung, K.Y.; Reisner, B.S.; Straley, S.C. YopM inhibits platelet aggregation and is necessary for virulence of Yersinia pestis in mice. Infect. Immun. 1990, 58, 3262–3271. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, S.; Williams, P. Yersinia virulence factors—A sophisticated arsenal for combating host defences. F1000Research 2016, 5, 1370. [Google Scholar] [CrossRef]
- Portnoy, D.A.; Falkow, S. Virulence-associated plasmids from Yersinia enterocolitica and Yersinia pestis. J. Bacteriol. 1981, 148, 877–883. [Google Scholar] [CrossRef]
- Reuter, S.; Connor, T.R.; Barquist, L.; Walker, D.; Feltwell, T.; Harris, S.R.; Fookes, M.; Hall, M.E.; Petty, N.K.; Fuchs, T.M.; et al. Parallel independent evolution of pathogenicity within the genus Yersinia. Proc. Natl. Acad. Sci. USA 2014, 111, 6768–6773. [Google Scholar] [CrossRef]
- Huang, X.Z.; Lindler, L.E. The pH 6 antigen is an antiphagocytic factor produced by Yersinia pestis independent of Yersinia outer proteins and capsule antigen. Infect. Immun. 2004, 72, 7212–7219. [Google Scholar] [CrossRef] [PubMed]
- Lindler, L.E.; Tall, B.D. Yersinia pestis pH 6 antigen forms fimbriae and is induced by intracellular association with macrophages. Mol. Microbiol. 1993, 8, 311–324. [Google Scholar] [CrossRef]
- Hinnebusch, B.J.; Jarrett, C.O.; Bland, D.M. “Fleaing” the Plague: Adaptations of Yersinia pestis to Its Insect Vector That Lead to Transmission. Annu. Rev. Microbiol. 2017, 71, 215–232. [Google Scholar] [CrossRef] [PubMed]
- Hinnebusch, B.J.; Erickson, D.L. Yersinia pestis biofilm in the flea vector and its role in the transmission of plague. Curr. Top. Microbiol. Immunol. 2008, 322, 229–248. [Google Scholar] [CrossRef]
- Hitchen, P.G.; Prior, J.L.; Oyston, P.C.; Panico, M.; Wren, B.W.; Titball, R.W.; Morris, H.R.; Dell, A. Structural characterization of lipo-oligosaccharide (LOS) from Yersinia pestis: Regulation of LOS structure by the PhoPQ system. Mol. Microbiol. 2002, 44, 1637–1650. [Google Scholar] [CrossRef] [PubMed]
- Oyston, P.C.; Dorrell, N.; Williams, K.; Li, S.R.; Green, M.; Titball, R.W.; Wren, B.W. The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect. Immun. 2000, 68, 3419–3425. [Google Scholar] [CrossRef]
- Hinnebusch, B.J.; Chouikha, I.; Sun, Y.C. Ecological Opportunity, Evolution, and the Emergence of Flea-Borne Plague. Infect. Immun. 2016, 84, 1932–1940. [Google Scholar] [CrossRef]
- Perry, R.D.; Fetherston, J.D. Yersiniabactin iron uptake: Mechanisms and role in Yersinia pestis pathogenesis. Microbes Infect. 2011, 13, 808–817. [Google Scholar] [CrossRef]
- Perry, R.D.; Pendrak, M.L.; Schuetze, P. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J. Bacteriol. 1990, 172, 5929–5937. [Google Scholar] [CrossRef]
- Gao, H.; Zhou, D.; Li, Y.; Guo, Z.; Han, Y.; Song, Y.; Zhai, J.; Du, Z.; Wang, X.; Lu, J.; et al. The iron-responsive Fur regulon in Yersinia pestis. J. Bacteriol. 2008, 190, 3063–3075. [Google Scholar] [CrossRef] [PubMed]
- Malek, M.A.; Bitam, I.; Levasseur, A.; Terras, J.; Gaudart, J.; Azza, S.; Flaudrops, C.; Robert, C.; Raoult, D.; Drancourt, M. Yersinia pestis halotolerance illuminates plague reservoirs. Sci. Rep. 2017, 7, 40022. [Google Scholar] [CrossRef]
- Zhou, D.; Yang, R. Molecular Darwinian evolution of virulence in Yersinia pestis. Infect. Immun. 2009, 77, 2242–2250. [Google Scholar] [CrossRef]
- Cavanaugh, D.C. K. F. Meyer’s Work on Plague. J. Infect. Dis. 1974, 129, S10–S12. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.F.; Eddie, B. Persistence of Sylvatic Plague. Proc. Soc. Exp. Biol. Med. 1938, 38, 333–334. [Google Scholar] [CrossRef]
- Biggins, D.E.; Kosoy, M.Y. Influences of introduced plague on North American mammals: Implications from ecology of plague in Asia. J. Mammal. 2001, 82, 906–916. [Google Scholar] [CrossRef]
- Mahmoudi, A.; Kryštufek, B.; Sludsky, A.; Schmid, B.V.; AMP, D.E.A.; Lei, X.; Ramasindrazana, B.; Bertherat, E.; Yeszhanov, A.; Stenseth, N.C.; et al. Plague reservoir species throughout the world. Integr. Zool. 2021, 16, 820–833. [Google Scholar] [CrossRef]
- Abbott, R.C.; Rocke, T.E. Plague: U.S. Geological Survey Circular 1372. 2012; 79p, Plus Appendix. Available online: https://pubs.usgs.gov/circ/1372/pdf/C1372_Plague.pdf (accessed on 2 January 2024).
- Elbroch, L.M.; Quigley, H.B.; Vickers, T.W. Plague, pumas and potential zoonotic exposure in the Greater Yellowstone Ecosystem. Environ. Conserv. 2020, 47, 75–78. [Google Scholar] [CrossRef]
- Ayyadurai, S.; Houhamdi, L.; Lepidi, H.; Nappez, C.; Raoult, D.; Drancourt, M. Long-term persistence of virulent Yersinia pestis in soil. Microbiology 2008, 154, 2865–2871. [Google Scholar] [CrossRef]
- Nikiforov, V.V.; Gao, H.; Zhou, L.; Anisimov, A. Plague: Clinics, Diagnosis and Treatment. Adv. Exp. Med. Biol. 2016, 918, 293–312. [Google Scholar] [CrossRef]
- Chanteau, S.; Rahalison, L.; Ralafiarisoa, L.; Foulon, J.; Ratsitorahina, M.; Ratsifasoamanana, L.; Carniel, E.; Nato, F. Development and testing of a rapid diagnostic test for bubonic and pneumonic plague. Lancet 2003, 361, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Dennis, D.T.; Chu, M.C. A major new test for plague. Lancet 2003, 361, 191–192. [Google Scholar] [CrossRef] [PubMed]
- Galy, A.; Loubet, P.; Peiffer-Smadja, N.; Yazdanpanah, Y. La peste: Mise au point et actualités. Rev. Med. Interne 2018, 39, 863–868. [Google Scholar] [CrossRef] [PubMed]
- Pechous, R.D.; Sivaraman, V.; Stasulli, N.M.; Goldman, W.E. Pneumonic Plague: The Darker Side of Yersinia pestis. Trends Microbiol. 2016, 24, 190–197. [Google Scholar] [CrossRef]
- Randremanana, R.; Andrianaivoarimanana, V.; Nikolay, B.; Ramasindrazana, B.; Paireau, J.; Ten Bosch, Q.A.; Rakotondramanga, J.M.; Rahajandraibe, S.; Rahelinirina, S.; Rakotomanana, F.; et al. Epidemiological characteristics of an urban plague epidemic in Madagascar, August-November, 2017: An outbreak report. Lancet Infect. Dis. 2019, 19, 537–545. [Google Scholar] [CrossRef]
- Nelson, C.A.; Meaney-Delman, D.; Fleck-Derderian, S.; Cooley, K.M.; Yu, P.A.; Mead, P.S. Antimicrobial Treatment and Prophylaxis of Plague: Recommendations for Naturally Acquired Infections and Bioterrorism Response. MMWR Recomm. Rep. 2021, 70, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Tourdjman, M.; Ibraheem, M.; Brett, M.; Debess, E.; Progulske, B.; Ettestad, P.; McGivern, T.; Petersen, J.; Mead, P. Misidentification of Yersinia pestis by automated systems, resulting in delayed diagnoses of human plague infections--Oregon and New Mexico, 2010–2011. Clin Infect Dis 2012, 55, e58–e60. [Google Scholar] [CrossRef]
- Prentice, M.B.; Rahalison, L. Plague. Lancet 2007, 369, 1196–1207. [Google Scholar] [CrossRef]
- Rollins, S.E.; Rollins, S.M.; Ryan, E.T. Yersinia pestis and the plague. Am. J. Clin. Pathol. 2003, 119 (Suppl. S1), S78–S85. [Google Scholar] [CrossRef]
- Qu, S.; Shi, Q.; Zhou, L.; Guo, Z.; Zhou, D.; Zhai, J.; Yang, R. Ambient stable quantitative PCR reagents for the detection of Yersinia pestis. PLoS Negl. Trop. Dis. 2010, 4, e629. [Google Scholar] [CrossRef]
- Mölsä, M.; Hemmilä, H.; Katz, A.; Niemimaa, J.; Forbes, K.M.; Huitu, O.; Stuart, P.; Henttonen, H.; Nikkari, S. Monitoring biothreat agents (Francisella tularensis, Bacillus anthracis and Yersinia pestis) with a portable real-time PCR instrument. J. Microbiol. Methods 2015, 115, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Green, M.S.; LeDuc, J.; Cohen, D.; Franz, D.R. Confronting the threat of bioterrorism: Realities, challenges, and defensive strategies. Lancet Infect. Dis. 2019, 19, e2–e13. [Google Scholar] [CrossRef] [PubMed]
- Galimand, M.; Guiyoule, A.; Gerbaud, G.; Rasoamanana, B.; Chanteau, S.; Carniel, E.; Courvalin, P. Multidrug Resistance in Yersinia pestis Mediated by a Transferable Plasmid. N. Engl. J. Med. 1997, 337, 677–681. [Google Scholar] [CrossRef]
- Salalli, R.; Dange, J.R.; Dhiman, S.; Sharma, T. Vaccines development in India: Advances, regulation, and challenges. Clin. Exp. Vaccine Res. 2023, 12, 193–208. [Google Scholar] [CrossRef]
- Andrianaivoarimanana, V.; Wagner, D.M.; Birdsell, D.N.; Nikolay, B.; Rakotoarimanana, F.; Randriantseheno, L.N.; Vogler, A.J.; Sahl, J.W.; Hall, C.M.; Somprasong, N.; et al. Transmission of Antimicrobial Resistant Yersinia pestis During a Pneumonic Plague Outbreak. Clin. Infect. Dis. 2022, 74, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Gupta, G.; Khan, A.A.; Rao, D.N. Cell-mediated immune response and Th/Th cytokine profile of B-T constructs of F1 and V antigen of Yersinia pestis. Scand. J. Immunol. 2010, 71, 186–198. [Google Scholar] [CrossRef]
- Cui, Y.; Yang, X.; Xiao, X.; Anisimov, A.P.; Li, D.; Yan, Y.; Zhou, D.; Rajerison, M.; Carniel, E.; Achtman, M.; et al. Genetic variations of live attenuated plague vaccine strains (Yersinia pestis EV76 lineage) during laboratory passages in different countries. Infect. Genet. Evol. 2014, 26, 172–179. [Google Scholar] [CrossRef]
- Feng, J.; Deng, Y.; Fu, M.; Hu, X.; Luo, W.; Lu, Z.; Dai, L.; Yang, H.; Zhao, X.; Du, Z.; et al. Construction of a Live-Attenuated Vaccine Strain of Yersinia pestis EV76-B-SHUΔpla and Evaluation of Its Protection Efficacy in a Mouse Model by Aerosolized Intratracheal Inoculation. Front. Cell Infect. Microbiol. 2020, 10, 473. [Google Scholar] [CrossRef]
- FDA Grants Orphan Drug Designation for Plague Vaccine. Available online: https://globalbiodefense.com/2017/03/10/fda-grants-orphan-drug-designation-plague-vaccine/ (accessed on 2 January 2024).

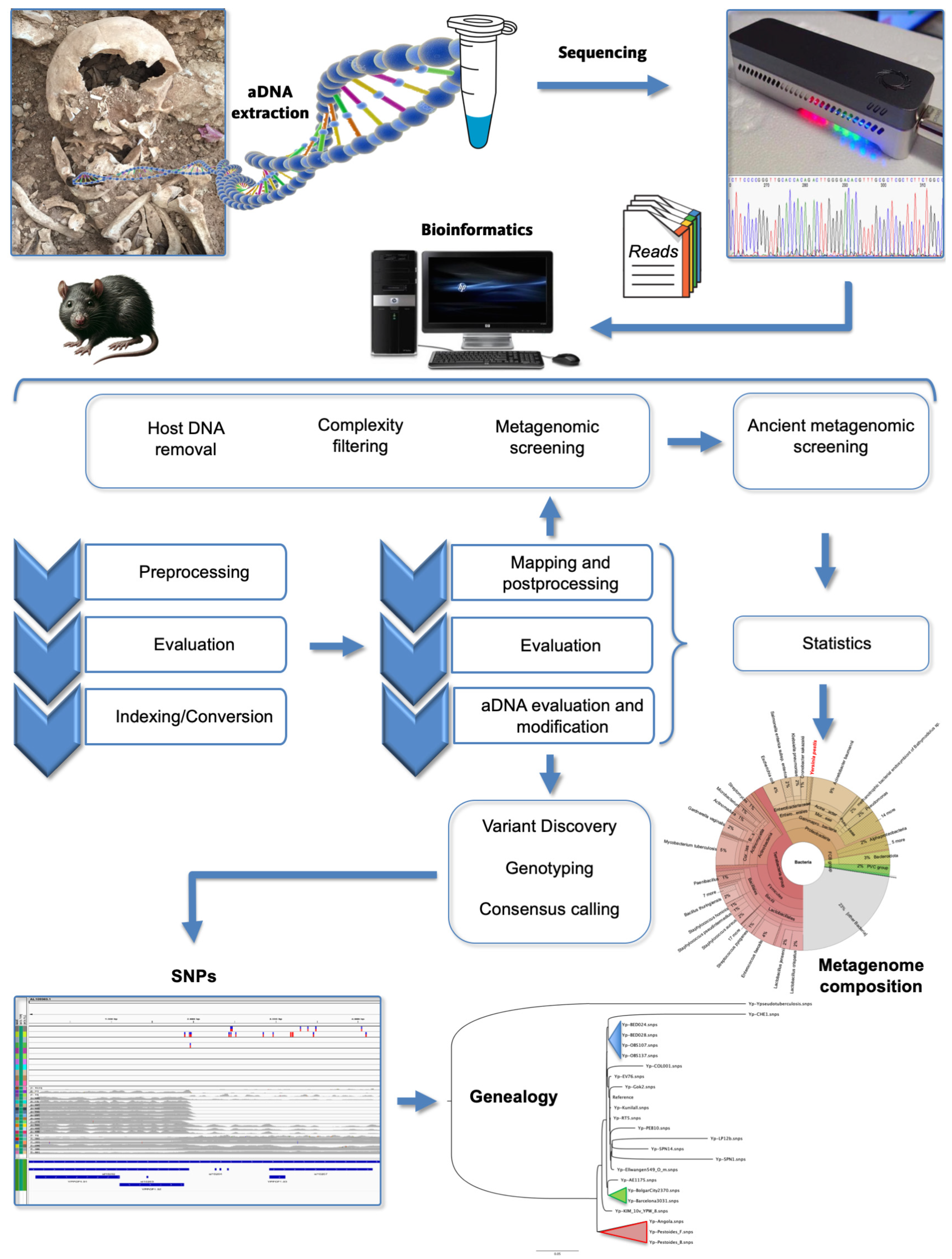
 ) Branch 0, (
) Branch 0, ( ) Branch 1, (
) Branch 1, ( ) Branch 2, (
) Branch 2, ( ) Branch 3, and (
) Branch 3, and ( ) Branch 4. Branch length does not represent the evolutionary time. * The genome of Y. pestis, strain CO92 (including plasmids) is usually used as reference for reads mapping from aDNA [64,66].
) Branch 4. Branch length does not represent the evolutionary time. * The genome of Y. pestis, strain CO92 (including plasmids) is usually used as reference for reads mapping from aDNA [64,66].
 ) Branch 0, (
) Branch 0, ( ) Branch 1, (
) Branch 1, ( ) Branch 2, (
) Branch 2, ( ) Branch 3, and (
) Branch 3, and ( ) Branch 4. Branch length does not represent the evolutionary time. * The genome of Y. pestis, strain CO92 (including plasmids) is usually used as reference for reads mapping from aDNA [64,66].
) Branch 4. Branch length does not represent the evolutionary time. * The genome of Y. pestis, strain CO92 (including plasmids) is usually used as reference for reads mapping from aDNA [64,66].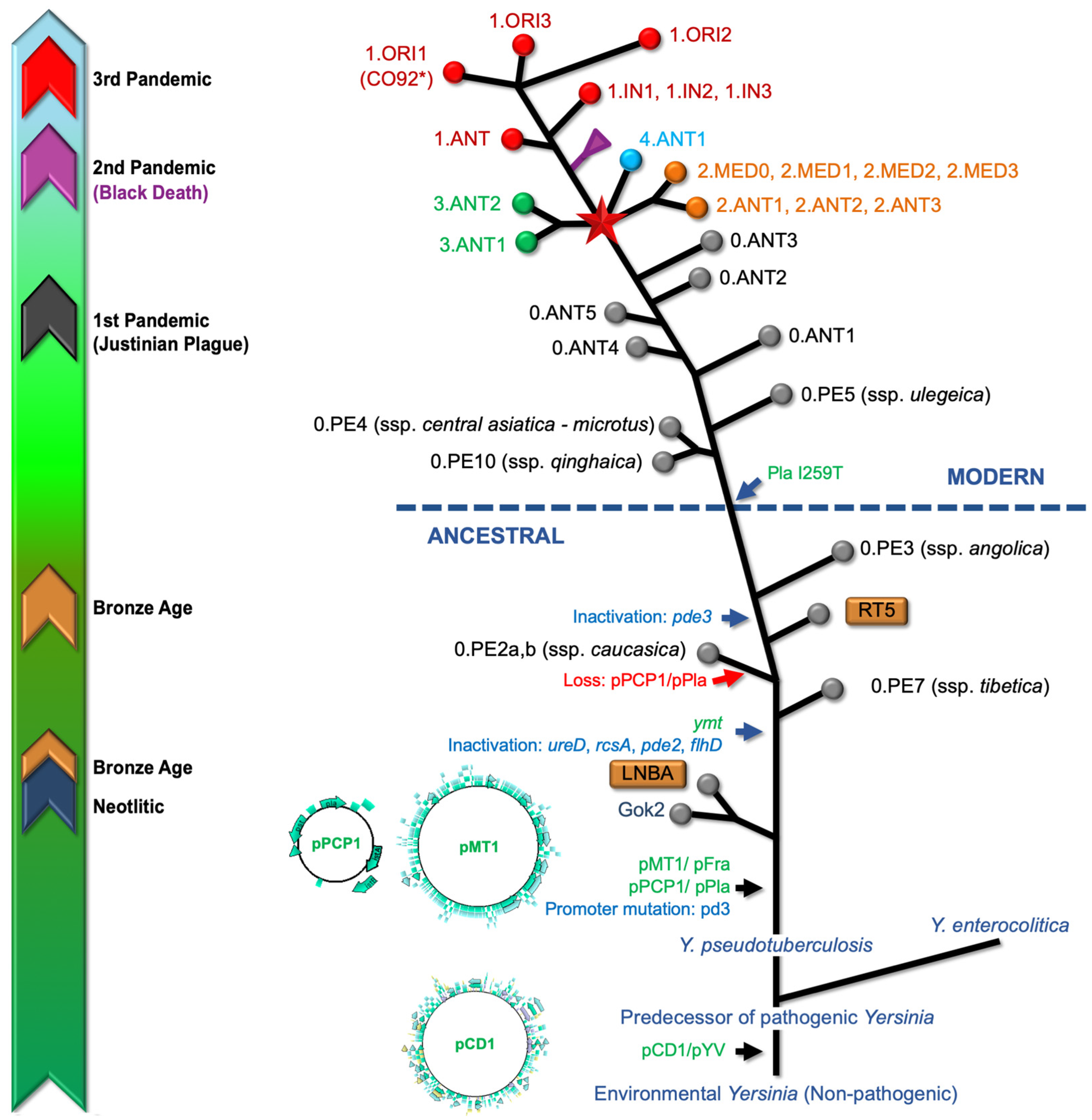
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bennasar-Figueras, A. The Natural and Clinical History of Plague: From the Ancient Pandemics to Modern Insights. Microorganisms 2024, 12, 146. https://doi.org/10.3390/microorganisms12010146
Bennasar-Figueras A. The Natural and Clinical History of Plague: From the Ancient Pandemics to Modern Insights. Microorganisms. 2024; 12(1):146. https://doi.org/10.3390/microorganisms12010146
Chicago/Turabian StyleBennasar-Figueras, Antoni. 2024. "The Natural and Clinical History of Plague: From the Ancient Pandemics to Modern Insights" Microorganisms 12, no. 1: 146. https://doi.org/10.3390/microorganisms12010146
APA StyleBennasar-Figueras, A. (2024). The Natural and Clinical History of Plague: From the Ancient Pandemics to Modern Insights. Microorganisms, 12(1), 146. https://doi.org/10.3390/microorganisms12010146




