Z3495, a LysR-Type Transcriptional Regulator Encoded in O Island 97, Regulates Virulence Gene Expression in Enterohemorrhagic Escherichia coli O157:H7
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Cell Culture
2.2. Bacterial Adherence Assay
2.3. Quantitative Real-Time PCR (qRT-PCR)
2.4. Mouse Colonization Experiments
2.5. Electrophoretic Mobility Shift Assays
2.6. ChIP-seq Analysis
2.7. ChIP-qPCR
2.8. Statistical Analysis
3. Results
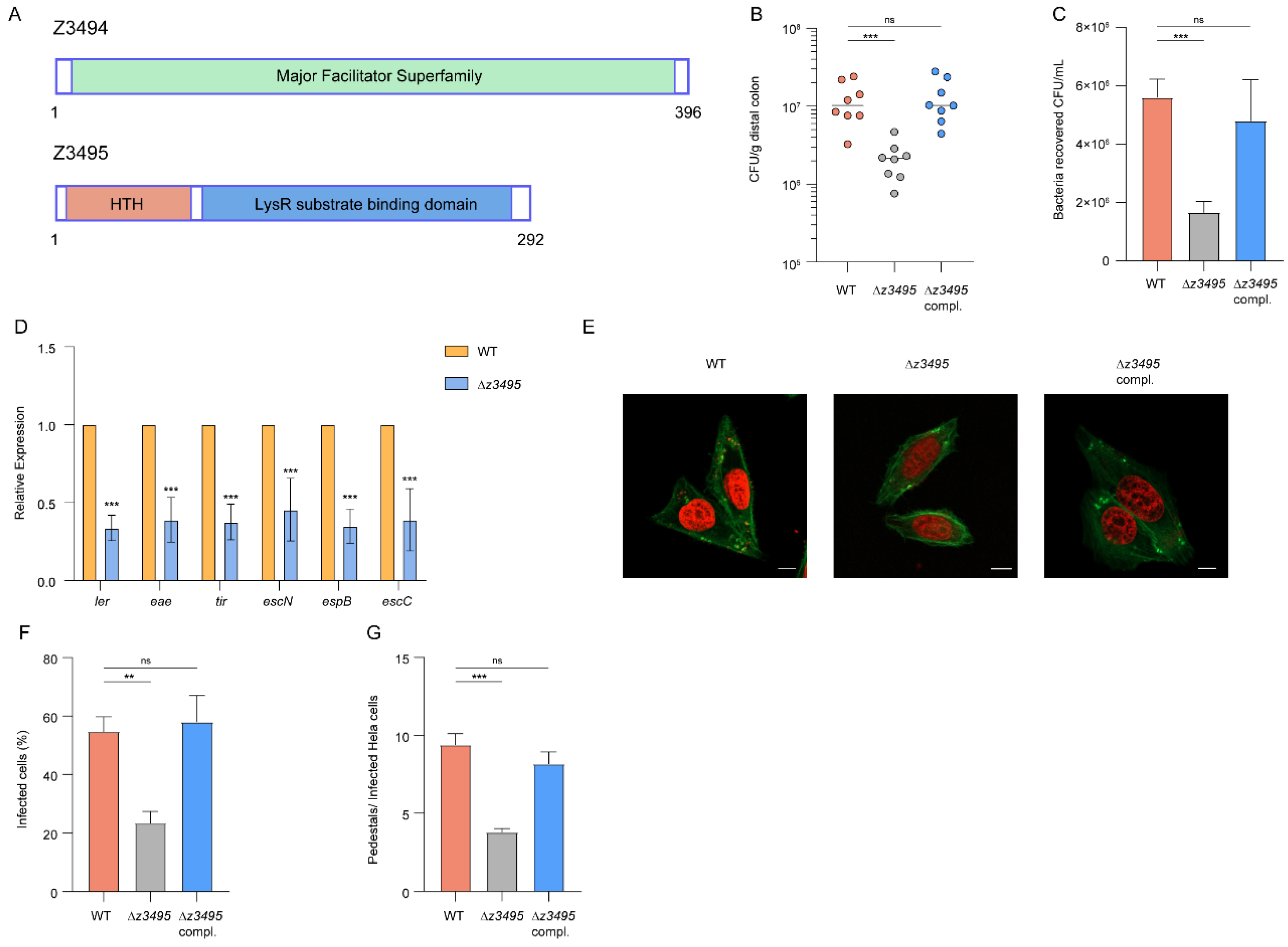
3.1. OI-97 Is Required for O157 Adherence and Colonization
3.2. Z3495 Is Required for O157 Adherence and Colonization
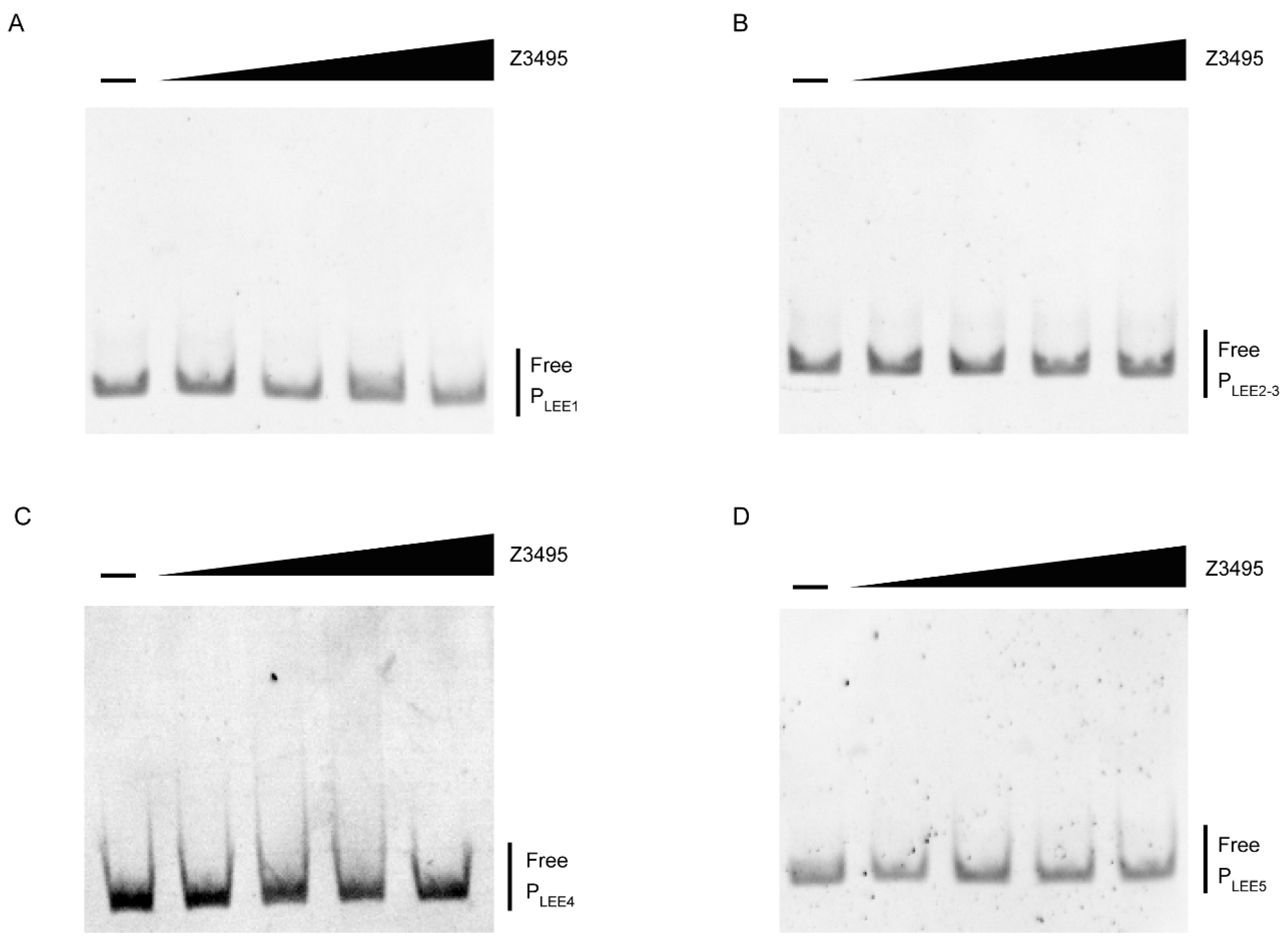
3.3. Z3495 Regulates LEE Expression Indirectly
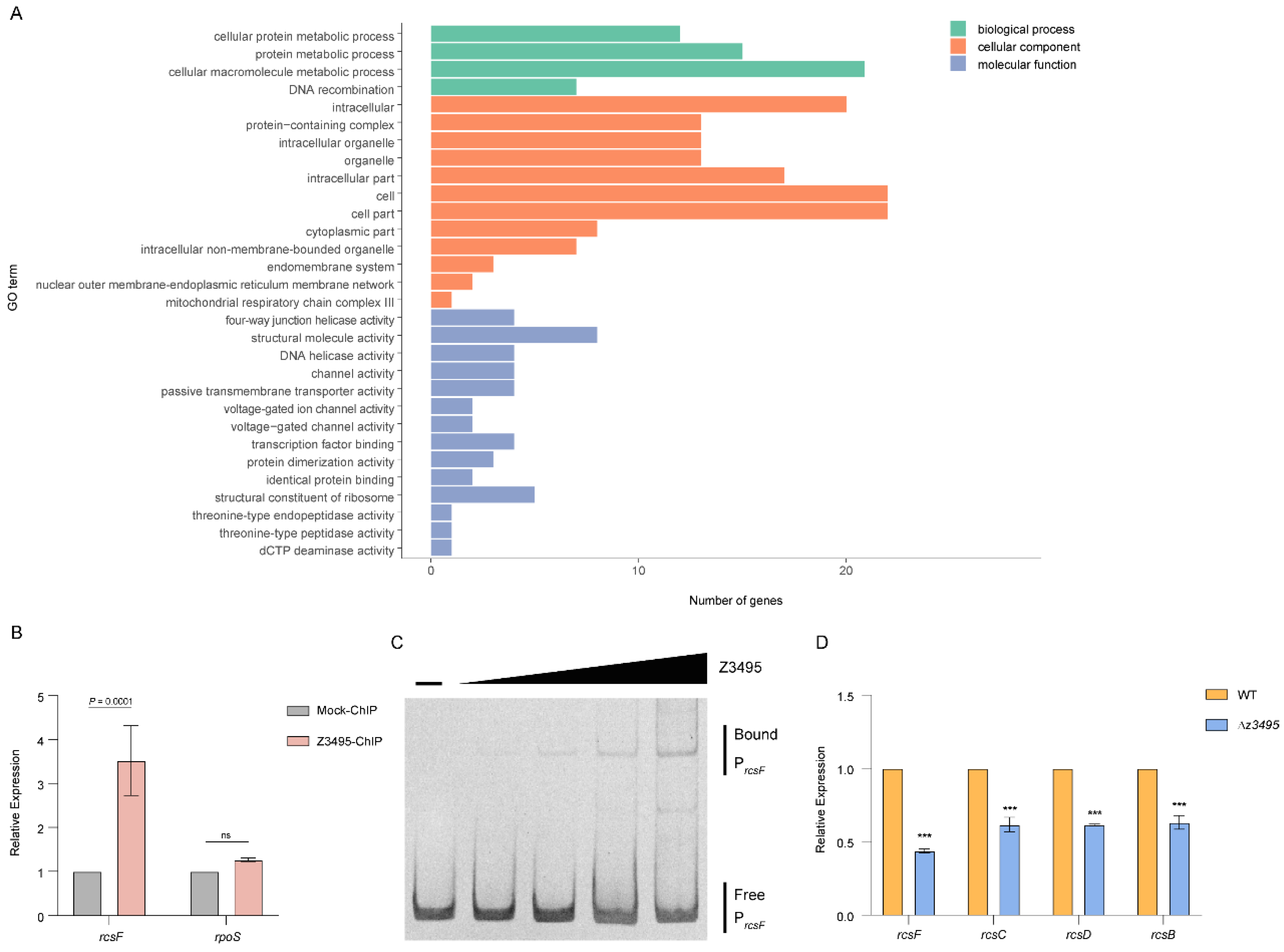
3.4. Z3495 Contributes to O157 Adherence and LEE Expression via rcsF
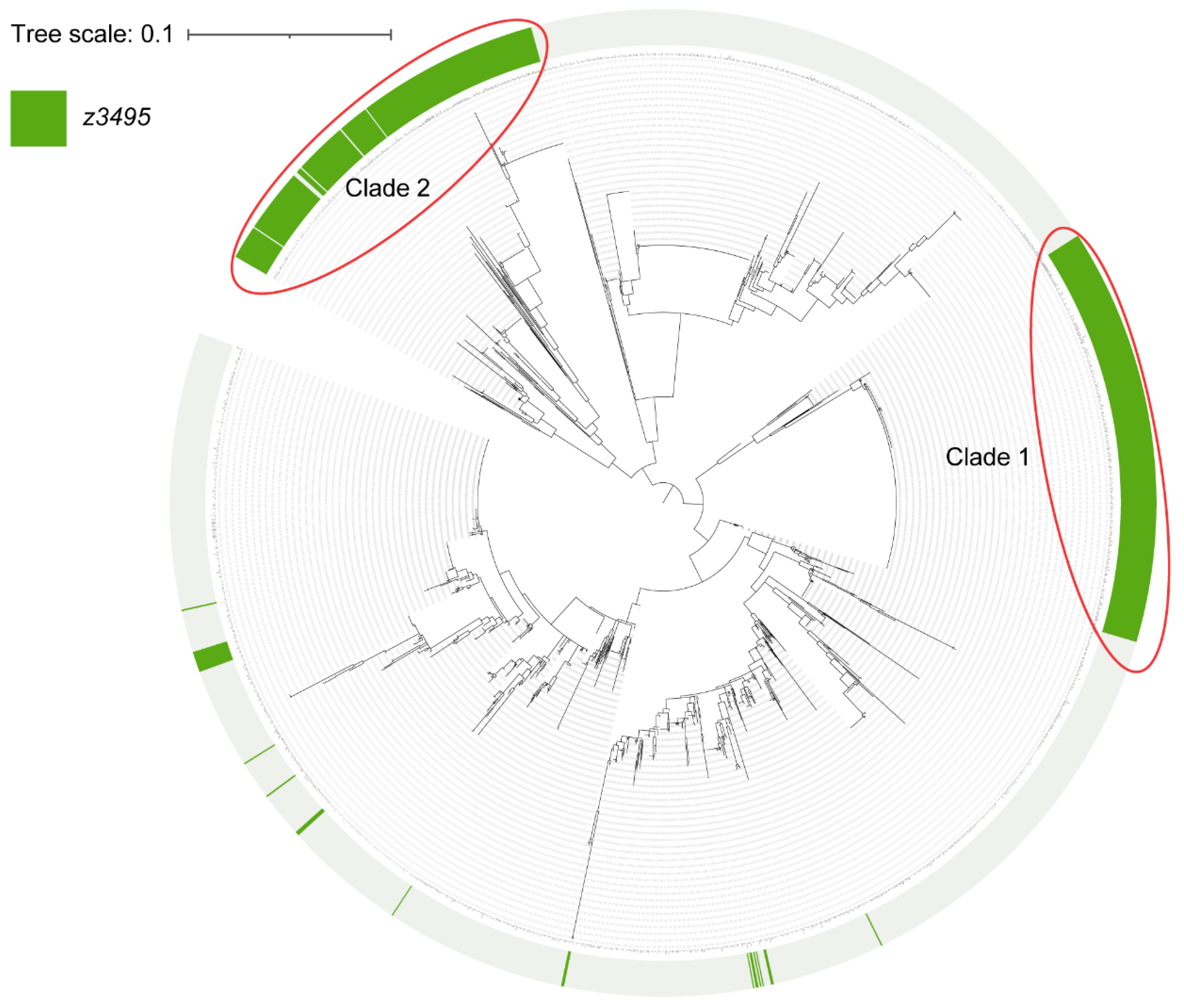
3.5. Z3495 Is a Widespread Regulator of Virulence in Pathogenic Bacteria
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lim, J.Y.; Yoon, J.; Hovde, C.J. A brief overview of Escherichia coli O157:H7 and its plasmid O157. J. Microbiol. Biotechnol. 2010, 20, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Gambushe, S.M.; Zishiri, O.T.; El Zowalaty, M.E. Review of Escherichia coli O157:H7 Prevalence, Pathogenicity, Heavy Metal and Antimicrobial Resistance, African Perspective. Infect. Drug Resist. 2022, 15, 4645–4673. [Google Scholar] [CrossRef]
- Sun, H.; Wang, M.; Liu, Y.; Wu, P.; Yao, T.; Yang, W.; Yan, J.; Yang, B. Regulation of flagellar motility and biosynthesis in enterohemorrhagic Escherichia coli O157:H7. Gut Microbes 2022, 14, 2110822. [Google Scholar] [CrossRef] [PubMed]
- Donnenberg, M.S.; Kaper, J.B.; Finlay, B.B. Interactions between enteropathogenic Escherichia coli and host epithelial cells. Trends Microbiol. 1997, 5, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Ho, N.K.; Henry, A.C.; Johnson-Henry, K.; Sherman, P.M. Pathogenicity, host responses and implications for management of enterohemorrhagic Escherichia coli O157:H7 infection. Can. J. Gastroenterol. 2013, 27, 281–285. [Google Scholar] [CrossRef]
- Elliott, S.J.; Wainwright, L.A.; McDaniel, T.K.; Jarvis, K.G.; Deng, Y.K.; Lai, L.C.; McNamara, B.P.; Donnenberg, M.S.; Kaper, J.B. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 1998, 28, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Sun, H.; Yan, J.; Kang, C.; Wu, J.; Yang, B. Enterohemorrhagic Escherichia coli senses microbiota-derived nicotinamide to increase its virulence and colonization in the large intestine. Cell Rep. 2023, 42, 112638. [Google Scholar] [CrossRef] [PubMed]
- Abe, A.; Heczko, U.; Hegele, R.G.; Brett Finlay, B. Two enteropathogenic Escherichia coli type III secreted proteins, EspA and EspB, are virulence factors. J. Exp. Med. 1998, 188, 1907–1916. [Google Scholar] [CrossRef]
- Holmes, A.; Mühlen, S.; Roe, A.J.; Dean, P. The EspF effector, a bacterial pathogen’s Swiss army knife. Infect. Immun. 2010, 78, 4445–4453. [Google Scholar] [CrossRef]
- Vlisidou, I.; Dziva, F.; La Ragione, R.M.; Best, A.; Garmendia, J.; Hawes, P.; Monaghan, P.; Cawthraw, S.A.; Frankel, G.; Woodward, M.J.; et al. Role of intimin-tir interactions and the tir-cytoskeleton coupling protein in the colonization of calves and lambs by Escherichia coli O157:H7. Infect. Immun. 2006, 74, 758–764. [Google Scholar] [CrossRef]
- Deng, W.; Marshall, N.C.; Rowland, J.L.; McCoy, J.M.; Worrall, L.J.; Santos, A.S.; Strynadka, N.C.J.; Finlay, B.B. Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 2017, 15, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Haack, K.R.; Robinson, C.L.; Miller, K.J.; Fowlkes, J.W.; Mellies, J.L. Interaction of Ler at the LEE5 (tir) operon of enteropathogenic Escherichia coli. Infect. Immun. 2003, 71, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Makino, K.; Ohnishi, M.; Kurokawa, K.; Ishii, K.; Yokoyama, K.; Han, C.G.; Ohtsubo, E.; Nakayama, K.; Murata, T.; et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001, 8, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Boyd, E.F. Bacteriophage-encoded bacterial virulence factors and phage-pathogenicity island interactions. Adv. Virus Res. 2012, 82, 91–118. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Yang, W.; Jiang, X.; Yao, T.; Wang, L.; Yang, B. Virulence-related O islands in enterohemorrhagic Escherichia coli O157:H7. Gut Microbes 2021, 13, 1992237. [Google Scholar] [CrossRef] [PubMed]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Feng, L.; Wang, F.; Wang, L. Enterohemorrhagic Escherichia coli senses low biotin status in the large intestine for colonization and infection. Nat. Commun. 2015, 6, 6592. [Google Scholar] [CrossRef] [PubMed]
- Perna, N.T.; Plunkett, G., 3rd; Burland, V.; Mau, B.; Glasner, J.D.; Rose, D.J.; Mayhew, G.F.; Evans, P.S.; Gregor, J.; Kirkpatrick, H.A.; et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 2001, 409, 529–533. [Google Scholar] [CrossRef]
- Murphy, K.C.; Campellone, K.G. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 2003, 4, 11. [Google Scholar] [CrossRef]
- Dibb-Fuller, M.P.; Best, A.; Stagg, D.A.; Cooley, W.A.; Woodward, M.J. An in-vitro model for studying the interaction of Escherichia coli O157:H7 and other enteropathogens with bovine primary cell cultures. J. Med. Microbiol. 2001, 50, 759–769. [Google Scholar] [CrossRef]
- Liu, B.; Liu, Y.; Yang, B.; Wang, Q.; Liu, X.; Qin, J.; Zhao, K.; Li, F.; Feng, X.; Li, L.; et al. Escherichia coli O157:H7 senses microbiota-produced riboflavin to increase its virulence in the gut. Proc. Natl. Acad. Sci. USA 2022, 119, e2212436119. [Google Scholar] [CrossRef] [PubMed]
- Whetstine, J.R.; Van Rechem, C. A cell-sorting-based protocol for cell cycle small-scale ChIP sequencing. STAR Protoc. 2022, 3, 101243. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, B.; Yang, P.; Wang, T.; Chang, Z.; Wang, J.; Wang, Q.; Li, W.; Wu, J.; Huang, D.; et al. LysR-type transcriptional regulator OvrB encoded in O island 9 drives enterohemorrhagic Escherichia coli O157:H7 virulence. Virulence 2019, 10, 783–792. [Google Scholar] [CrossRef]
- Yan, N. Structural Biology of the Major Facilitator Superfamily Transporters. Annu. Rev. Biophys. 2015, 44, 257–283. [Google Scholar] [CrossRef]
- Zhao, J.; Yu, X.; Zhu, M.; Kang, H.; Ma, J.; Wu, M.; Gan, J.; Deng, X.; Liang, H. Structural and Molecular Mechanism of CdpR Involved in Quorum-Sensing and Bacterial Virulence in Pseudomonas aeruginosa. PLoS Biol. 2016, 14, e1002449. [Google Scholar] [CrossRef]
- Morgan, J.K.; Ortiz, J.A.; Riordan, J.T. The role for TolA in enterohemorrhagic Escherichia coli pathogenesis and virulence gene transcription. Microb. Pathog. 2014, 77, 42–52. [Google Scholar] [CrossRef]
- Haneda, T.; Ishii, Y.; Danbara, H.; Okada, N. Genome-wide identification of novel genomic islands that contribute to Salmonella virulence in mouse systemic infection. FEMS Microbiol. Lett. 2009, 297, 241–249. [Google Scholar] [CrossRef][Green Version]
- Liu, B.; Wang, J.; Wang, L.; Ding, P.; Yang, P.; Yang, B. Transcriptional Activator OvrA Encoded in O Island 19 Modulates Virulence Gene Expression in Enterohemorrhagic Escherichia coli O157:H7. J. Infect. Dis. 2020, 221, 820–829. [Google Scholar] [CrossRef]
- Mellies, J.L.; Barron, A.M.; Carmona, A.M. Enteropathogenic and enterohemorrhagic Escherichia coli virulence gene regulation. Infect. Immun. 2007, 75, 4199–4210. [Google Scholar] [CrossRef]
- Franzin, F.M.; Sircili, M.P. Locus of enterocyte effacement: A pathogenicity island involved in the virulence of enteropathogenic and enterohemorragic Escherichia coli subjected to a complex network of gene regulation. BioMed Res. Int. 2015, 2015, 534738. [Google Scholar] [CrossRef]
- Maddocks, S.E.; Oyston, P.C.F. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 2008, 154 Pt 12, 3609–3623. [Google Scholar] [CrossRef]
- Stec, E.; Witkowska-Zimny, M.; Hryniewicz, M.M.; Neumann, P.; Wilkinson, A.J.; Brzozowski, A.M.; Verma, C.S.; Zaim, J.; Wysocki, S.; Bujacz, G.D. Structural basis of the sulphate starvation response in E. coli: Crystal structure and mutational analysis of the cofactor-binding domain of the Cbl transcriptional regulator. J. Mol. Biol. 2006, 364, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Anantharaman, V.; Balaji, S.; Babu, M.M.; Iyer, L.M. The many faces of the helix-turn-helix domain: Transcription regulation and beyond. FEMS Microbiol. Rev. 2005, 29, 231–262. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Takeuchi, M.; Sato, T. The LysR-type transcriptional regulator YofA controls cell division through the regulation of expression of ftsW in Bacillus subtilis. J. Bacteriol. 2007, 189, 5642–5651. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sperandio, B.; Gautier, C.; McGovern, S.; Ehrlich, D.S.; Renault, P.; Martin-Verstraete, I.; Guédon, E. Control of methionine synthesis and uptake by MetR and homocysteine in Streptococcus mutans. J. Bacteriol. 2007, 189, 7032–7044. [Google Scholar] [CrossRef]
- Cao, H.; Krishnan, G.; Goumnerov, B.; Tsongalis, J.; Tompkins, R.; Rahme, L.G. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc. Natl. Acad. Sci. USA 2001, 98, 14613–14618. [Google Scholar] [CrossRef]
- Lahiri, A.; Das, P.; Chakravortty, D. The LysR-type transcriptional regulator Hrg counteracts phagocyte oxidative burst and imparts survival advantage to Salmonella enterica serovar Typhimurium. Microbiology 2008, 154 Pt 9, 2837–2846. [Google Scholar] [CrossRef][Green Version]
- Axler-Diperte, G.L.; Miller, V.L.; Darwin, A.J. YtxR, a conserved LysR-like regulator that induces expression of genes encoding a putative ADP-ribosyltransferase toxin homologue in Yersinia enterocolitica. J. Bacteriol. 2006, 188, 8033–8043. [Google Scholar] [CrossRef]
- Lahiri, A.; Das, P.; Chakravortty, D. Salmonella Typhimurium: Insight into the multi-faceted role of the LysR-type transcriptional regulators in Salmonella. Int. J. Biochem. Cell Biol. 2009, 41, 2129–2133. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Wei, Y.; Huang, Y.; Qin, J.; Liu, B.; Liu, R.; Chen, X.; Li, D.; Wang, Q.; Li, X.; et al. Z3495, a LysR-Type Transcriptional Regulator Encoded in O Island 97, Regulates Virulence Gene Expression in Enterohemorrhagic Escherichia coli O157:H7. Microorganisms 2024, 12, 140. https://doi.org/10.3390/microorganisms12010140
Wang Q, Wei Y, Huang Y, Qin J, Liu B, Liu R, Chen X, Li D, Wang Q, Li X, et al. Z3495, a LysR-Type Transcriptional Regulator Encoded in O Island 97, Regulates Virulence Gene Expression in Enterohemorrhagic Escherichia coli O157:H7. Microorganisms. 2024; 12(1):140. https://doi.org/10.3390/microorganisms12010140
Chicago/Turabian StyleWang, Qian, Yi Wei, Yu Huang, Jingliang Qin, Bin Liu, Ruiying Liu, Xintong Chen, Dan Li, Qiushi Wang, Xiaoya Li, and et al. 2024. "Z3495, a LysR-Type Transcriptional Regulator Encoded in O Island 97, Regulates Virulence Gene Expression in Enterohemorrhagic Escherichia coli O157:H7" Microorganisms 12, no. 1: 140. https://doi.org/10.3390/microorganisms12010140
APA StyleWang, Q., Wei, Y., Huang, Y., Qin, J., Liu, B., Liu, R., Chen, X., Li, D., Wang, Q., Li, X., Yang, X., Li, Y., & Sun, H. (2024). Z3495, a LysR-Type Transcriptional Regulator Encoded in O Island 97, Regulates Virulence Gene Expression in Enterohemorrhagic Escherichia coli O157:H7. Microorganisms, 12(1), 140. https://doi.org/10.3390/microorganisms12010140





