Gut Microbiome Disruption Following SARS-CoV-2: A Review
Abstract
1. Introduction
2. Methods
2.1. Systematic Review
2.2. Longitudinal Study Methodology
Bioinformatics and Statistical Analysis
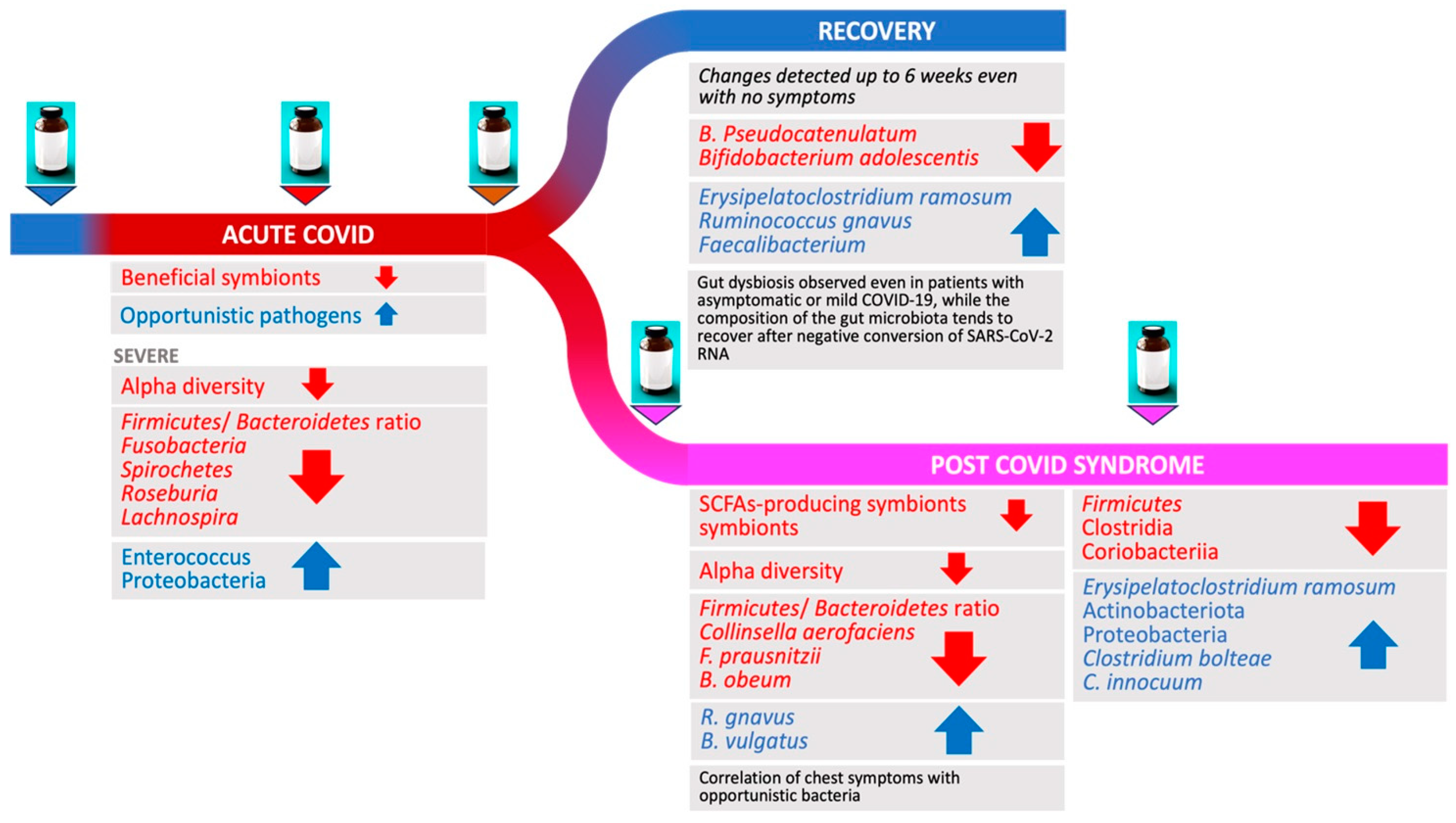
3. Changes in Gut Microbiome during and Following Acute COVID-19
3.1. Acute COVID-19
3.2. COVID-19 Severity and Gut Microbiome Changes
3.3. Gut Microbiome in Patients Recovering from COVID-19
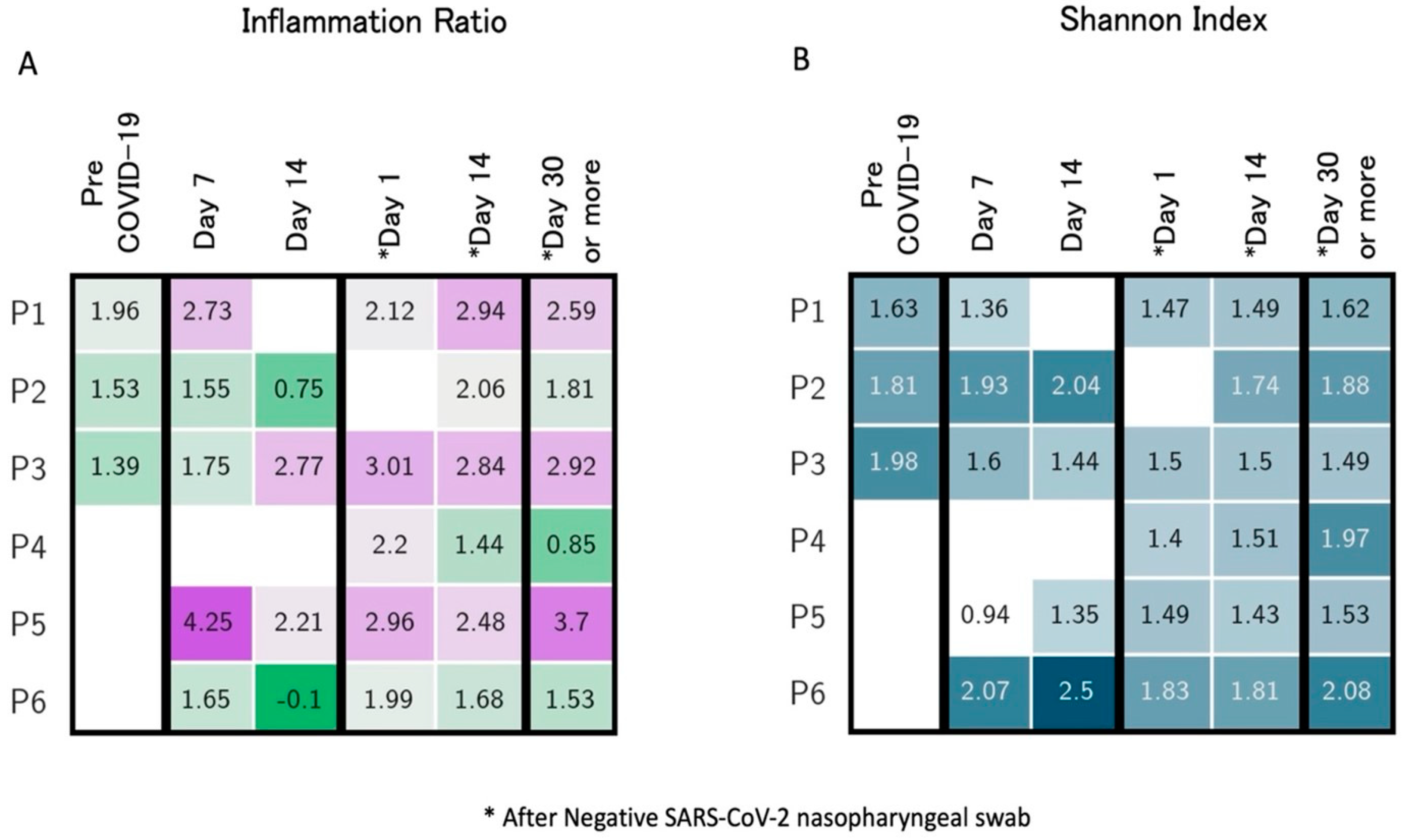
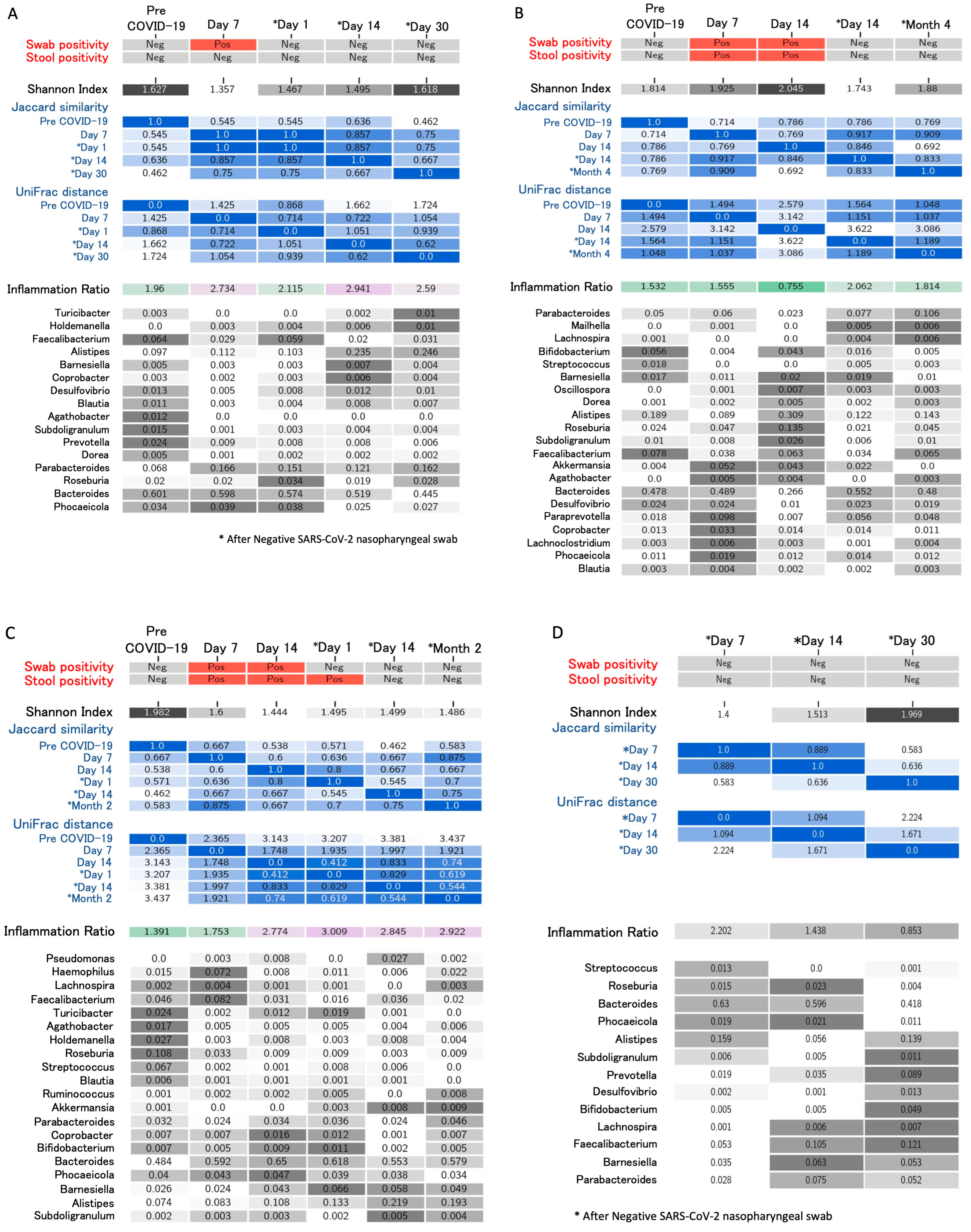
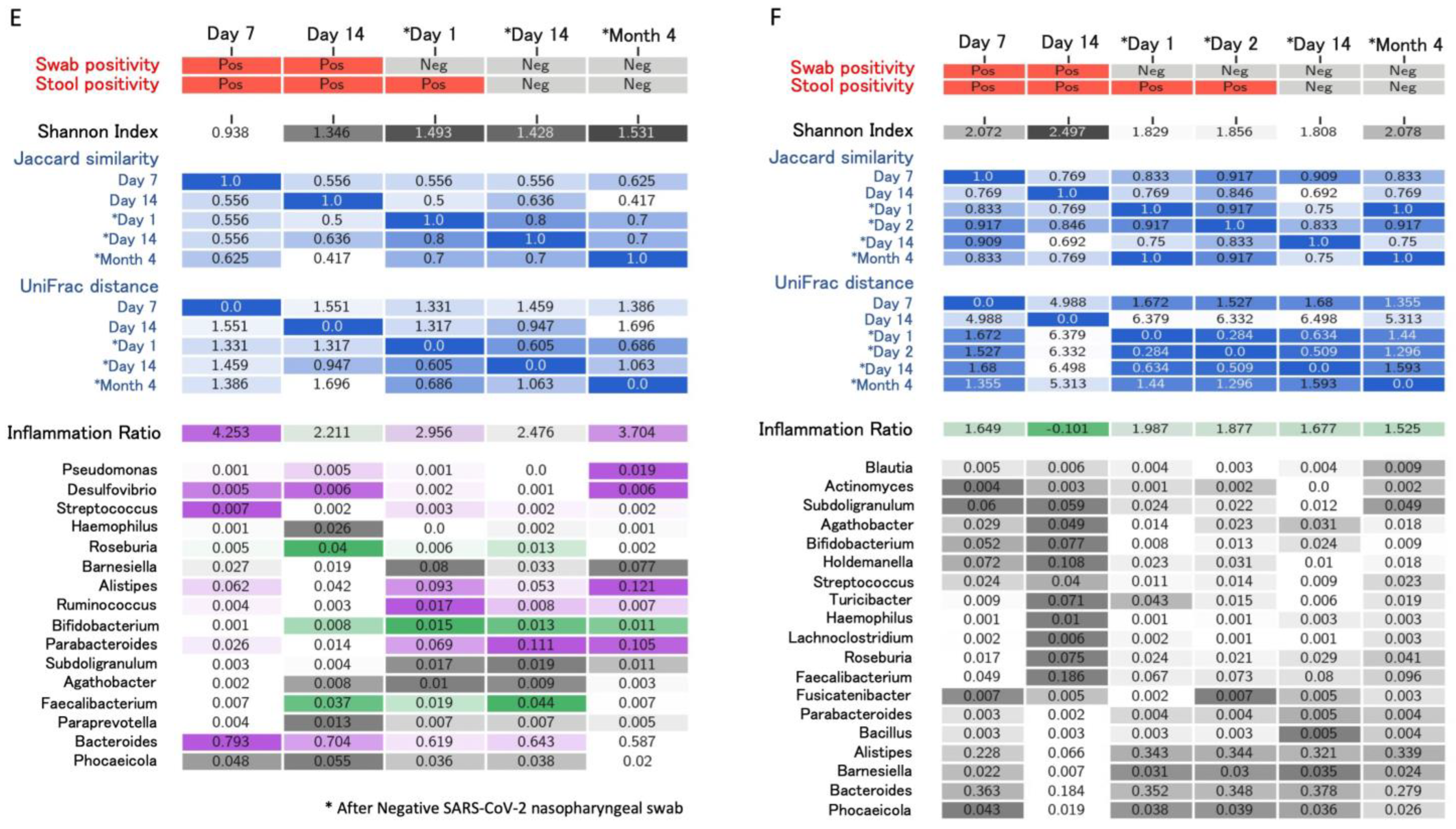
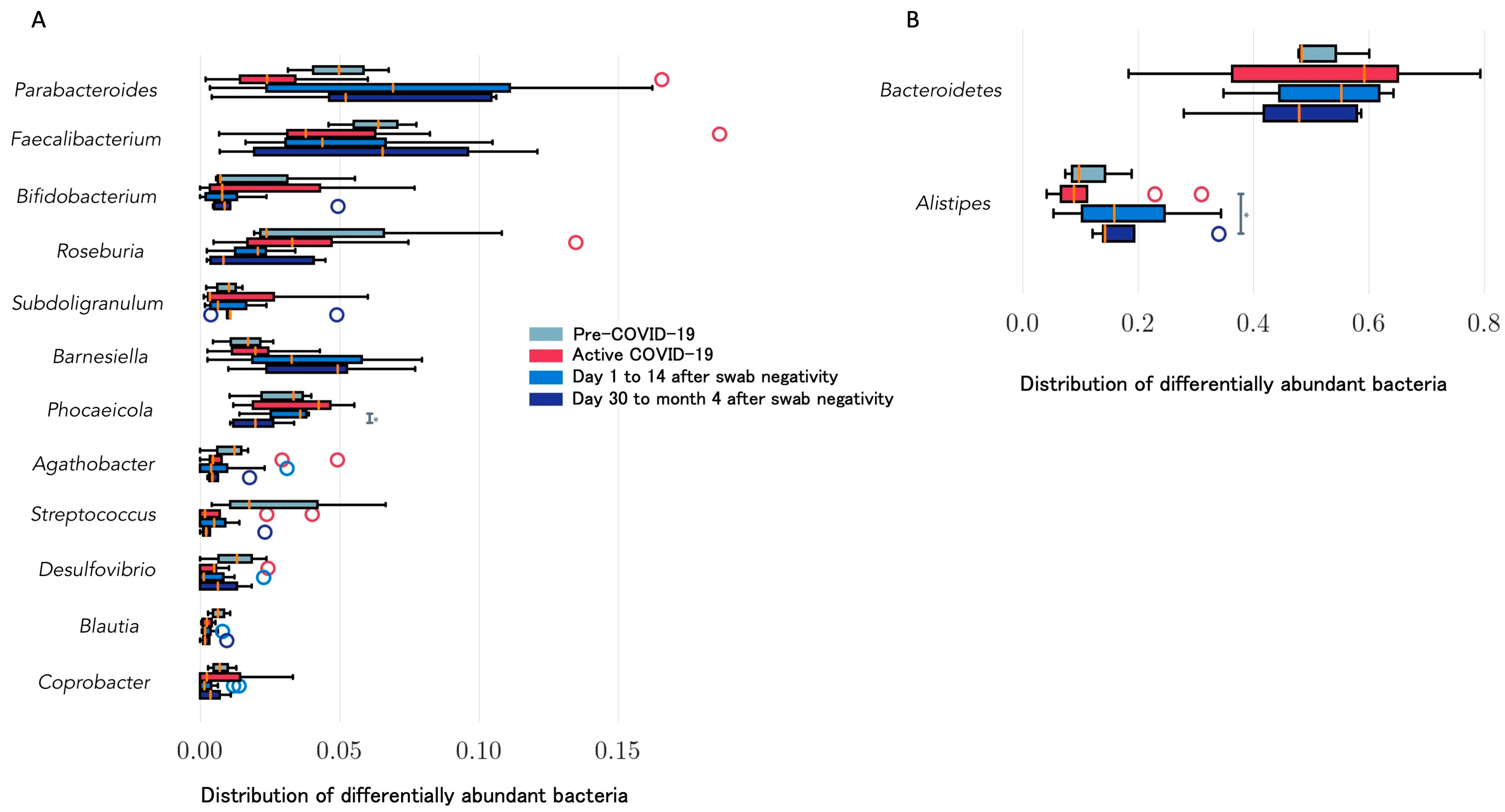
4. Longitudinal Study of Microbiome Changes: A Case Series
5. Microbiome Changes and PCS
6. Microbiome Manipulation and Treatment Potential
7. Discussion and Conclusions
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Zuo, T.; Zhan, H.; Zhang, F.; Liu, Q.; Tso, E.Y.K.; Lui, G.C.Y.; Chen, N.; Li, A.; Lu, W.; Francis, K.; et al. Alterations in Fecal Fungal Microbiome of Patients With COVID-19 During Time of Hospitalization until Discharge. Gastroenterology 2020, 159, 1302–1310.e5. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Pang, X.; Wu, J.; Liu, T.; Wang, B.; Cao, H. Gut microbiota in COVID-19: New insights from inside. Gut Microbes 2023, 15, 2201157. [Google Scholar] [CrossRef]
- Zhang, F.; Lau, R.I.; Liu, Q.; Su, Q.; Chan, F.K.L.; Ng, S.C. Gut microbiota in COVID-19: Key microbial changes, potential mechanisms and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Righi, E.; Lambertenghi, L.; Gorska, A.; Sciammarella, C.; Ivaldi, F.; Mirandola, M. Assunta Sartor, Evelina Tacconelli. Impact of COVID-19 and Antibiotic Treatments on Gut Microbiome: A Role for Enterococcus spp. Biomedicines 2022, 10, 2786. [Google Scholar] [CrossRef] [PubMed]
- Post COVID-19 Condition (Long COVID) [Internet]. Available online: https://www.who.int/europe/news-room/fact-sheets/item/post-covid-19-condition (accessed on 3 October 2023).
- Liu, Q.; Mak, J.W.Y.; Su, Q.; Yeoh, Y.K.; Lui, G.C.Y.; Ng, S.S.S.; Zhang, F.; Li, A.Y.L.; Lu, W.; Hui, D.S.; et al. Gut microbiota dynamics in a prospective cohort of patients with post-acute COVID-19 syndrome. Gut 2022, 71, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Lau, R.I.; Liu, Q.; Chan, F.K.L.; Ng, S.C. Post-acute COVID-19 syndrome and gut dysbiosis linger beyond 1 year after SARS-CoV-2 clearance. Gut 2023, 72, 1230–1232. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Huang, L.; Wang, Y.; Li, X.; Ren, L.; Gu, X.; Kang, L.; Guo, L.; Liu, M.; Zhou, X.; et al. 6-month consequences of COVID-19 in patients discharged from hospital: A cohort study. Lancet Lond. Engl. 2021, 397, 220–232. [Google Scholar] [CrossRef]
- Righi, E.; Mirandola, M.; Mazzaferri, F.; Razzaboni, E.; Zaffagnini, A.; Erbogasto, A.; Dalla Vecchia, I.; Auerbach, N.; Ivaldi, F.; Mongardi, M.; et al. Long-Term Patient-Centred Follow-up in a Prospective Cohort of Patients with COVID-19. Infect. Dis. Ther. 2021, 10, 1579–1590. [Google Scholar] [CrossRef]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Huson, D.H.; Beier, S.; Flade, I.; Górska, A.; El-Hadidi, M.; Mitra, S.; Ruscheweyh, H.J.; Tappu, R. MEGAN Community Edition—Interactive Exploration and Analysis of Large-Scale Microbiome Sequencing Data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef] [PubMed]
- Hunter, J.D. MATPLOTLIB: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Jones, E.; Oliphant, T.; Peterson, P. SciPy: Open Source Scientific Tools for Python. 2001. Available online: https://scipy.org/ (accessed on 23 June 2023).
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- scikit-bio: A Bioinformatics Library for Data Scientists, Students, and Developers. 2020. Available online: https://scikit.bio (accessed on 20 April 2023).
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Gaibani, P.; D’Amico, F.; Bartoletti, M.; Lombardo, D.; Rampelli, S.; Fornaro, G.; Coladonato, S.; Siniscalchi, A.; Re, M.C.; Viale, P.; et al. The Gut Microbiota of Critically Ill Patients With COVID-19. Front. Cell Infect. Microbiol. 2021, 11, 670424. [Google Scholar] [CrossRef]
- Mazzarelli, A.; Giancola, M.L.; Farina, A.; Marchioni, L.; Rueca, M.; Gruber, C.E.M.; Bartolini, B.; Ascoli Bartoli, T.; Maffongelli, G.; Capobianchi, M.R.; et al. 16S rRNA gene sequencing of rectal swab in patients affected by COVID-19. PLoS ONE 2021, 16, e0247041. [Google Scholar] [CrossRef]
- Moreira-Rosário, A.; Marques, C.; Pinheiro, H.; Araújo, J.R.; Ribeiro, P.; Rocha, R.; Mota, I.; Pestana, D.; Ribeiro, R.; Pereira, A.; et al. Gut Microbiota Diversity and C-Reactive Protein Are Predictors of Disease Severity in COVID-19 Patients. Front. Microbiol. 2021, 12, 705020. [Google Scholar] [CrossRef]
- Khan, M.; Mathew, B.J.; Gupta, P.; Garg, G.; Khadanga, S.; Vyas, A.K.; Singh, A.K. Gut dysbiosis and il-21 response in patients with severe covid-19. Microorganisms 2021, 9, 1292. [Google Scholar] [CrossRef]
- Reinold, J.; Farahpour, F.; Fehring, C.; Dolff, S.; Konik, M.; Korth, J.; van Baal, L.; Hoffmann, D.; Buer, J.; Witzke, O.; et al. A Pro-Inflammatory Gut Microbiome Characterizes SARS-CoV-2 Infected Patients and a Reduction in the Connectivity of an Anti-Inflammatory Bacterial Network Associates With Severe COVID-19. Front. Cell Infect. Microbiol. 2021, 11, 747816. [Google Scholar] [CrossRef] [PubMed]
- Newsome, R.C.; Gauthier, J.; Hernandez, M.C.; Abraham, G.E.; Robinson, T.O.; Williams, H.B.; Sloan, M.; Owing, A.; Laird, H.; Christian, T.; et al. The gut microbiome of COVID-19 recovered patients returns to uninfected status in a minority-dominated United States cohort. Gut Microbes 2021, 13, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.N.; Joo, E.J.; Lee, C.W.; Ahn, K.S.; Kim, H.L.; Park, D.I.; Park, S.K. Reversion of Gut Microbiota during the Recovery Phase in Patients with Asymptomatic or Mild COVID-19: Longitudinal Study. Microorganisms 2021, 9, 1237. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Zhang, Y.; Li, Y.; Wu, Q.; Wu, J.; Park, S.K.; Guo, C.; Lu, J. Meta-analysis of 16S rRNA microbial data identified alterations of the gut microbiota in COVID-19 patients during the acute and recovery phases. BMC Microbiol. 2022, 22, 274. [Google Scholar] [CrossRef]
- de Nies, L.; Galata, V.; Martin-Gallausiaux, C.; Despotovic, M.; Busi, S.B.; Snoeck, C.J.; Delacour, L.; Budagavi, D.P.; Laczny, C.C.; Habier, J.; et al. Altered infective competence of the human gut microbiome in COVID-19. Microbiome 2023, 11, 46. [Google Scholar] [CrossRef] [PubMed]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.M.; Rodriguez-Palacios, A. The Genus Alistipes: Gut Bacteria With Emerging Implications to Inflammation, Cancer, and Mental Health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef] [PubMed]
- Gentilotti, E.; Górska, A.; Tami, A.; Gusinow, R.; Mirandola, M.; Rodríguez Baño, J.; Palacios Baena, Z.R.; Rossi, E.; Hasenauer, J.; Lopes-Rafegas, I.; et al. Clinical phenotypes and quality of life to define post-COVID-19 syndrome: A cluster analysis of the multinational, prospective ORCHESTRA cohort. EClinicalMedicine 2023, 62, 102107. [Google Scholar] [CrossRef] [PubMed]
- Righi, E.; Mirandola, M.; Mazzaferri, F.; Dossi, G.; Razzaboni, E.; Zaffagnini, A.; Ivaldi, F.; Visentin, A.; Lambertenghi, L.; Arena, C.; et al. Determinants of persistence of symptoms and impact on physical and mental wellbeing in Long COVID: A prospective cohort study. J. Infect. 2022, 84, 566–572. [Google Scholar] [CrossRef]
- Chen, Y.; Gu, S.; Chen, Y.; Lu, H.; Shi, D.; Guo, J.; Wu, W.R.; Yang, Y.; Li, Y.; Xu, K.Y.; et al. Six-month follow-up of gut microbiota richness in patients with COVID-19. Gut 2022, 71, 222–225. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, J.; Zhang, D.; Ma, W.L.; Wang, X. Linking the gut microbiota to persistent symptoms in survivors of COVID-19 after discharge. J. Microbiol. 2021, 59, 941–948. [Google Scholar] [CrossRef]
- Vestad, B.; Ueland, T.; Lerum, T.V.; Dahl, T.B.; Holm, K.; Barratt-Due, A.; Kåsine, T.; Dyrhol-Riise, A.M.; Stiksrud, B.; Tonby, K. Respiratory dysfunction three months after severe COVID-19 is associated with gut microbiota alterations. J. Intern. Med. 2022, 291, 801–812. [Google Scholar] [CrossRef]
- Liu, Q.; Su, Q.; Zhang, F.; Tun, H.M.; Mak, J.W.Y.; Lui, G.C.Y.; Ng, S.S.S.; Ching, J.Y.L.; Li, A.; Lu, W.; et al. Multi-kingdom gut microbiota analyses define COVID-19 severity and post-acute COVID-19 syndrome. Nat. Commun. 2022, 13, 6806. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, V.L.; Littlefield, K.M.; Watson, R.; Palmer, B.E.; Lozupone, C. Inflammation-associated gut microbiome in postacute sequelae of SARS-CoV-2 points towards new therapeutic targets. Gut 2023, 73, 376–378. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhou, Y.; Ma, Y.; Chen, P.; Tang, J.; Yang, B.; Li, H.; Liang, M.; Xue, Y.; Liu, J.; et al. Gut Microbiota Dysbiosis Correlates With Long COVID-19 at One-Year After Discharge. J. Korean Med. Sci. 2023, 38, e120. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Berni Canani, R.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Davani-Davari, D.; Negahdaripour, M.; Karimzadeh, I.; Seifan, M.; Mohkam, M.; Masoumi, S.J.; Berenjian, A.; Ghasemi, Y. Prebiotics: Definition, Types, Sources, Mechanisms, and Clinical Applications. Foods 2019, 8, 92. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, K.; Kim, W. Gut microbiota restoration through fecal microbiota transplantation: A new atopic dermatitis therapy. Exp. Mol. Med. 2021, 53, 907–916. [Google Scholar] [CrossRef]
- Alenazy, M.F.; Aljohar, H.I.; Alruwaili, A.R.; Daghestani, M.H.; Alonazi, M.A.; Labban, R.S.; El-Ansary, A.K.; Balto, H.A. Gut Microbiota Dynamics in Relation to Long-COVID-19 Syndrome: Role of Probiotics to Combat Psychiatric Complications. Metabolites 2022, 12, 912. [Google Scholar] [CrossRef]
- Yeoh, Y.K.; Zuo, T.; Lui, G.C.Y.; Zhang, F.; Liu, Q.; Li, A.Y.; Chung, A.C.; Cheung, C.P.; Tso, E.Y.; Fung, K.S.; et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 2021, 70, 698–706. [Google Scholar] [CrossRef]
- Hegazy, M.; Ahmed Ashoush, O.; Tharwat Hegazy, M.; Wahba, M.; Lithy, R.M.; Abdel-Hamid, H.M.; Abd Elshafy, S.A.; Abdelfatah, D.; El-Din Ibrahim, M.H.; Abdelghani, A. Beyond probiotic legend: ESSAP gut microbiota health score to delineate SARS-COV-2 infection severity. Br. J. Nutr. 2022, 127, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhang, L.; Lin, W.; Tang, W.; Chan, F.K.L.; Ng, S.C. Review article: Probiotics, prebiotics and dietary approaches during COVID-19 pandemic. Trends Food Sci. Technol. 2021, 108, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Oliva, S.; Di Nardo, G.; Ferrari, F.; Mallardo, S.; Rossi, P.; Patrizi, G.; Cucchiara, S.; Stronati, L. Randomised clinical trial: The effectiveness of Lactobacillus reuteri ATCC 55730 rectal enema in children with active distal ulcerative colitis. Aliment. Pharmacol. Ther. 2012, 35, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Baud, D.; Dimopoulou Agri, V.; Gibson, G.R.; Reid, G.; Giannoni, E. Using probiotics to flatten the curve of coronavirus disease COVID-2019 pandemic. Front. Public Health 2020, 8, 186. [Google Scholar] [CrossRef]
- Din, A.U.; Mazhar, M.; Waseem, M.; Ahmad, W.; Bibi, A.; Hassan, A.; Ali, N.; Gang, W.; Qian, G.; Ullah, R.; et al. SARS-CoV-2 microbiome dysbiosis linked disorders and possible probiotics role. Biomed Pharmacother. Biomed. Pharmacother. 2021, 133, 110947. [Google Scholar] [CrossRef] [PubMed]
- McAleer, J.P.; Kolls, J.K. Contributions of the intestinal microbiome in lung immunity. Eur. J. Immunol. 2018, 48, 39–49. [Google Scholar] [CrossRef]
- Allie, S.R.; Bradley, J.E.; Mudunuru, U.; Schultz, M.D.; Graf, B.A.; Lund, F.E.; Randall, T.D. The establishment of resident memory B cells in the lung requires local antigen encounter. Nat. Immunol. 2019, 20, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.C.S.; Zhang, L.; Ching, J.Y.L.; Mak, J.W.Y.; Huang, J.; Wang, S.; Mok, C.K.P.; Wong, A.; Chiu, Q.L.; Fung, J.T.; et al. Effects of Gut Microbiome Modulation on Reducing Adverse Health Outcomes among Elderly and Diabetes Patients during the COVID-19 Pandemic: A Randomised, Double-Blind, Placebo-Controlled Trial (IMPACT Study). Nutrients 2023, 15, 1982. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xu, Z.; Mak, J.W.Y.; Chow, K.M.; Lui, G.; Li, T.C.M.; Wong, C.K.; Chan, P.K.S.; Ching, J.Y.L.; Fujiwara, J.; et al. Gut microbiota-derived synbiotic formula (SIM01) as a novel adjuvant therapy for COVID-19: An open-label pilot study. J. Gastroenterol. Hepatol. 2022, 37, 823–831. [Google Scholar] [CrossRef]
- Wischmeyer, P.; Tang, H.; Ren, Y.; Bohannon, L.; Ramirez, Z.; Andermann, T.; Messina, J.A.; Sung, J.A.; Jensen, D.; Jung, S.; et al. Efficacy of Probiotic Treatment as Post-Exposure Prophylaxis for COVID-19: A Double-Blind, Placebo Controlled Randomized Tria [Internet]. 2022. Available online: https://europepmc.org/article/PPR/PPR545983 (accessed on 3 October 2023).
- Johnson, B.A.; Hage, A.; Kalveram, B.; Mears, M.; Plante, J.A.; Rodriguez, S.E.; Ding, Z.; Luo, X.; Bente, D.; Bradrick, S.S.; et al. Peptidoglycan-Associated Cyclic Lipopeptide Disrupts Viral Infectivity. J. Virol. 2019, 93, e01282-19. [Google Scholar] [CrossRef]
- Gutiérrez-Castrellón, P.; Gandara-Martí, T.; Abreu YAbreu, A.T.; Nieto-Rufino, C.D.; López-Orduña, E.; Jiménez-Escobar, I.; Jiménez-Gutiérrez, C.; López-Velazquez, G.; Espadaler-Mazo, J. Probiotic improves symptomatic and viral clearance in Covid19 outpatients: A randomized, quadruple-blinded, placebo-controlled trial. Gut Microbes 2022, 14, 2018899. [Google Scholar] [CrossRef]
- Ceccarelli, G.; Borrazzo, C.; Pinacchio, C.; Santinelli, L.; Innocenti, G.P.; Cavallari, E.N.; Celani, L.; Marazzato, M.; Alessandri, F.; Ruberto, F.; et al. Oral Bacteriotherapy in Patients With COVID-19: A Retrospective Cohort Study. Front. Nutr. 2020, 7, 613928. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Han, H.; Li, X.; Chen, C.; Xie, X.; Su, G.; Ye, S.; Wang, C.; He, Q.; Wang, F.; et al. Probiotics use is associated with improved clinical outcomes among hospitalized patients with COVID-19. Ther. Adv. Gastroenterol. 2021, 14, 17562848211035670. [Google Scholar] [CrossRef] [PubMed]
- Piscotta, F.J.; Hoffmann, H.H.; Choi, Y.J.; Small, G.I.; Ashbrook, A.W.; Koirala, B.; Campbell, E.A.; Darst, S.A.; Rice, C.M.; Brady, S.F. Metabolites with SARS-CoV-2 Inhibitory Activity Identified from Human Microbiome Commensals. Msphere 2021, 6, e0071121. [Google Scholar] [CrossRef] [PubMed]
- Khanna, S.; Voth, E. Therapeutics for Clostridioides difficile infection: Molecules and microbes. Expert. Rev. Gastroenterol. Hepatol. 2023, 17, 903–911. [Google Scholar] [CrossRef]
- Biliński, J.; Winter, K.; Jasiński, M.; Szczęś, A.; Bilinska, N.; Mullish, B.H.; Małecka-Panas, E.; Basak, G.W. Rapid resolution of COVID-19 after faecal microbiota transplantation. Gut 2022, 71, 230–232. [Google Scholar] [CrossRef]
- Liu, F.; Ye, S.; Zhu, X.; He, X.; Wang, S.; Li, Y.; Lin, J.; Wang, J.; Lin, Y.; Ren, X.; et al. Gastrointestinal disturbance and effect of fecal microbiota transplantation in discharged COVID-19 patients. J. Med. Case Rep. 2021, 15, 60. [Google Scholar] [CrossRef]

| Author, Country | COVID-19 Patients, Type of Study | Timing | Microbiome Changes | Measurement Method |
|---|---|---|---|---|
| Righi, Italy [4] | 82 patients | Samples collected <2 weeks of hospitalization | 16S rRNA gene sequencing. Increased opportunistic pathogens higher in severe COVID-19. Enterococcus: highest number of correlations with therapy (including antibiotics). Dorea, Agathobacter, Roseburia, and Barnesiella negatively correlated with antibiotic therapy | Alpha (Shannon’s index) and beta diversity metrics (Jaccard distance, weighted UniFrac distance) |
| Gaibani, Italy [20] | 57 (32% ICU) and 100 controls | Cross sectional, case—control. Samples collected <week 1 of hospitalization | 16S rRNA gene sequencing. Dysbiosis and over-representation of Enterococcus, especially in ICU; loss of alpha diversity | Alpha diversity metrics, number of observed ASV, and inverse Simpson index |
| Mazzarelli, Italy [21] | 15 hospitalized patients (6 in ICU), 8 controls | Cross sectional, case—control. Swabs collected at hospitalization | 16S rRNA gene sequencing. Patients in the ICU had lower microbial richness. Increase in Proteobacteria and decrease in Fusobacteria and Spirochetes in infected vs. controls | Alpha diversity and richness were assessed using Shannon indices and Chao1 |
| Moreira-Rosário, Portugal [22] | Hospitalized and outpatients (WHO Clinical Progression Scale: 19 mild, 37 moderate, 59 severe) | Cross-sectional. Samples collected after enrolment | 16S rRNA gene sequencing. Moderate and severe disease vs. mild had dysbiosis with lower Firmicutes/Bacteroidetes ratio, higher abundance of Proteobacteria, and lower abundance of Roseburia and Lachnospira | Shannon diversity index |
| Khan, India [23] | 10 asymptomatic, 10 mild symptoms (no oxygen), 10 severe; 10 controls | Cross sectional, case–control. Samples collected at admission | 16S rRNA gene sequencing. Decrease in diversity with increasing COVID-19 severity. Reduction in Prevotella, affecting the gut mucin glycoprotein maintenance, which interacts with the immune system | Alpha diversity and richness (InvSimpson and Fisher indices); Beta Diversity analyzed by LDA Effect Size |
| Reinold, Germany [24] | 26 patients classified as severe, 12 as critical, 79 non-severe (WHO) | 117 patients and 95 controls. Cross sectional. Timing of collection unknown | 16S rRNA gene sequencing. Patients had lower richness with increased Proteobacteria and Bacteroidetes and decrease vs. control. | Shannon diversity index, observed features (ASVs), and Pielou’s evenness index |
| Newsome, United States [25] | 94 patients (50 acute infection, 9 recovered, 34 controls ICU admitted) | Cross sectional, case–control. Samples collected <3 days of ICU admission | 16S rRNA gene sequencing. Patients clustered in the recovered and non-infected groups. Recovered similar to non-infected. Similar Shannon index across groups | Shannon diversity index |
| Righi, Italy * (unpublished) | Healthcare workers (n = 6). Day 14 and >day 30 after negativization collection | 6 to 20 weeks | 16S rRNA gene sequencing. Bacteroides stable; Alistipes increase from day 14 after negativization. Faecalibacterium increased after negativization. Barnesiella increased. Roseburia decreased from day 14 after negativization | Alpha (Shannon’s index) and beta diversity metrics (Jaccard distance, weighted UniFrac distance) |
| Kim, Korea [26] | Two paired samples from 12 patients; negative conversion for SARS-CoV-2 | 10 days median interval | 16S rRNA gene sequencing. Depletion of Bacteroidetes with tendency to rapidly reverse. Firmicutes/Bacteroidetes ratio higher in infected vs. recovered. Gut microbiota recovered after SARS-CoV-2 conversion | Faith’s phylogenetic diversity, Shannon’s index, and Pielou’s evenness |
| Author, Country | Population, Type of Study | Timing | Microbiome Changes | Measurement Method |
|---|---|---|---|---|
| Liu, China [6] | 68 patients with >1 symptom 4 weeks after SARS-CoV-2 clearance | 6 months | 16S rRNA gene sequencing. Bacteria diversity and richness lower vs. controls. Significantly lower C. aerofaciens, F. prausnitzii, B. obeum and higher R. gnavus and B. vulgatus. | Shannon index diversity and Chao1 richness index |
| Su, China [7] | 155 patients (79% > 1 symptom 4 weeks after recovery from COVID-19) | 14 months | Shotgun metagenomic sequencing Bacteria diversity (p = 0.004) and richness (p = 0.0003) lower than controls. Recovered patients had increased E. ramosum and R. gnavus and depletion of B. adolescentis and B. pseudocatenulatum. | Alpha diversity metrics (Shannon diversity, Chao1 richness) |
| Chen, China [32] | 30 patients postconvalescence | 6 months | 16S rRNA gene sequencing. Richness lower in acute phase (median 217, IQR 164–266) vs. controls (median 432, IQR 332–468). Non-significant increase in Chao 1 index from acute to convalescence and postconvalescence phases. Low richness associated with reduced pulmonary function. | Chao 1 index |
| Zhou, China [33] | 15 patients (12 with > 1 symptom) | 3 months | 16S rRNA gene sequencing. F. prausnitzii negatively correlated with chest tightness after activity and I. butyriciproducens with cough. Escherichia positively correlated with fatigue, chest tightness after activity, and myalgia. I. bartlettii positively correlated with anorexia and fatigue. | Spearman’s rank-based correlation test |
| Vestad, Norway [34] | 83 patients (30% with respiratory impairment) | 3 months | 16S rRNA gene sequencing. Reduced alpha diversity. Increased abundance of 5 taxa and reduced of 20 taxa. Reduced Erysipelotrichaceae UCG-003 and increased Veillonella and Flavonifractor. | Alfa-diversity (Faith’s phylogenetic diversity and observed amplicon sequence variants). Beta diversity (Bray–Curtis) |
| Liu, China [35] | 78 (Cluster, 42; Cluster 2, 36) > 1 symptoms at 4 weeks after infection | 6 months | WGS analysis. Cluster 1 (84% long COVID) different vs. Cluster 2 (44% long COVID). Bacteria diversity Cluster 1 lower than Cluster 2. Cluster 1 had increased opportunistic species (E. ramosum, C. bolteae, C. innocuum). | Shannon diversity index, MaAslin analysis |
| Carneiro, US [36] | 34 patients, 15 with continuing or recurring symptoms > 4 weeks after infection | N/A | 16S rRNA gene sequencing. No significant differences between groups. Long COVID had lower ratio of ASV highly related to F. prausnitzii over genus Bacteroides (B. dorei, B. massiliensis, and B. thetaiotaomicron). | SELBAL analysis |
| Zhang, China [37] | 55 patients with >1 symptom | 12 months | 16S rRNA gene sequencing. Decreased richness and diversity; Firmicutes less abundant, Actinobacteriota and Proteobacteria more abundant; decreased Clostridia and Coriobacteriia. SCFAs-producing symbionts significantly depleted. Veillonella enriched. | Sobs and Shannon index, PCoA of Bray–Curtis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Righi, E.; Dalla Vecchia, I.; Auerbach, N.; Morra, M.; Górska, A.; Sciammarella, C.; Lambertenghi, L.; Gentilotti, E.; Mirandola, M.; Tacconelli, E.; et al. Gut Microbiome Disruption Following SARS-CoV-2: A Review. Microorganisms 2024, 12, 131. https://doi.org/10.3390/microorganisms12010131
Righi E, Dalla Vecchia I, Auerbach N, Morra M, Górska A, Sciammarella C, Lambertenghi L, Gentilotti E, Mirandola M, Tacconelli E, et al. Gut Microbiome Disruption Following SARS-CoV-2: A Review. Microorganisms. 2024; 12(1):131. https://doi.org/10.3390/microorganisms12010131
Chicago/Turabian StyleRighi, Elda, Ilaria Dalla Vecchia, Nina Auerbach, Matteo Morra, Anna Górska, Concetta Sciammarella, Lorenza Lambertenghi, Elisa Gentilotti, Massimo Mirandola, Evelina Tacconelli, and et al. 2024. "Gut Microbiome Disruption Following SARS-CoV-2: A Review" Microorganisms 12, no. 1: 131. https://doi.org/10.3390/microorganisms12010131
APA StyleRighi, E., Dalla Vecchia, I., Auerbach, N., Morra, M., Górska, A., Sciammarella, C., Lambertenghi, L., Gentilotti, E., Mirandola, M., Tacconelli, E., & Sartor, A. (2024). Gut Microbiome Disruption Following SARS-CoV-2: A Review. Microorganisms, 12(1), 131. https://doi.org/10.3390/microorganisms12010131








