Comparative Genome Analyses Provide Insight into the Antimicrobial Activity of Endophytic Burkholderia
Abstract
1. Introduction
2. Materials and Methods
2.1. Burkholderia Strains Used for Genome Comparison
2.2. Calculation of Whole Genome Nucleotide Identity
2.3. Pan-Genome Analysis
2.4. Identification of the Genes Associated with the Production of Antimicrobial Compounds and Siderophores
2.5. Identification of the Genes Associated with Toxin Production, Virulence-Related Features, and Antibiotic Resistance
2.6. Identification of Secretion Systems
2.7. Identification of CRISPR-Cas System
3. Results
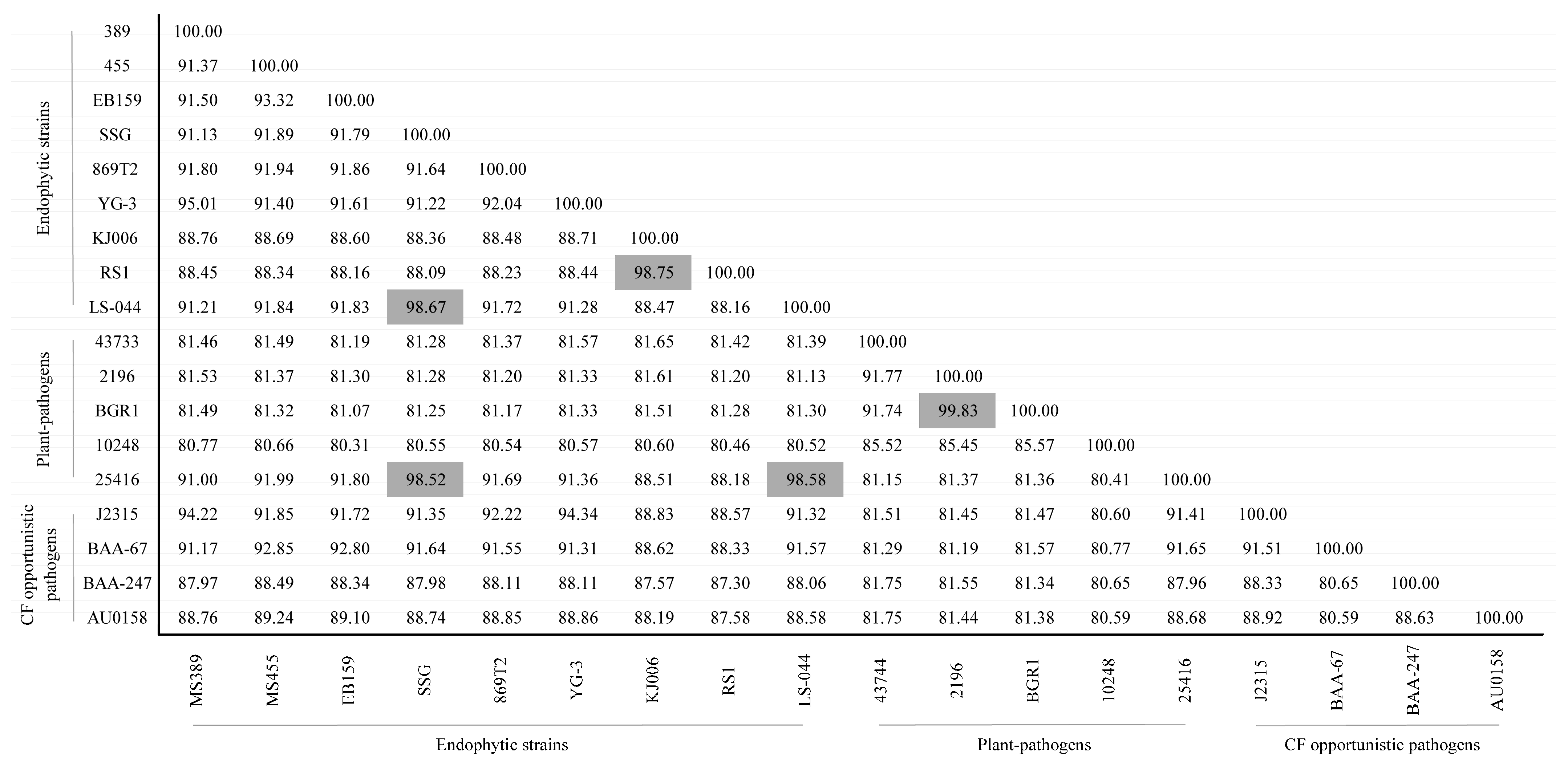
3.1. Whole-Genome Nucleotide Identity
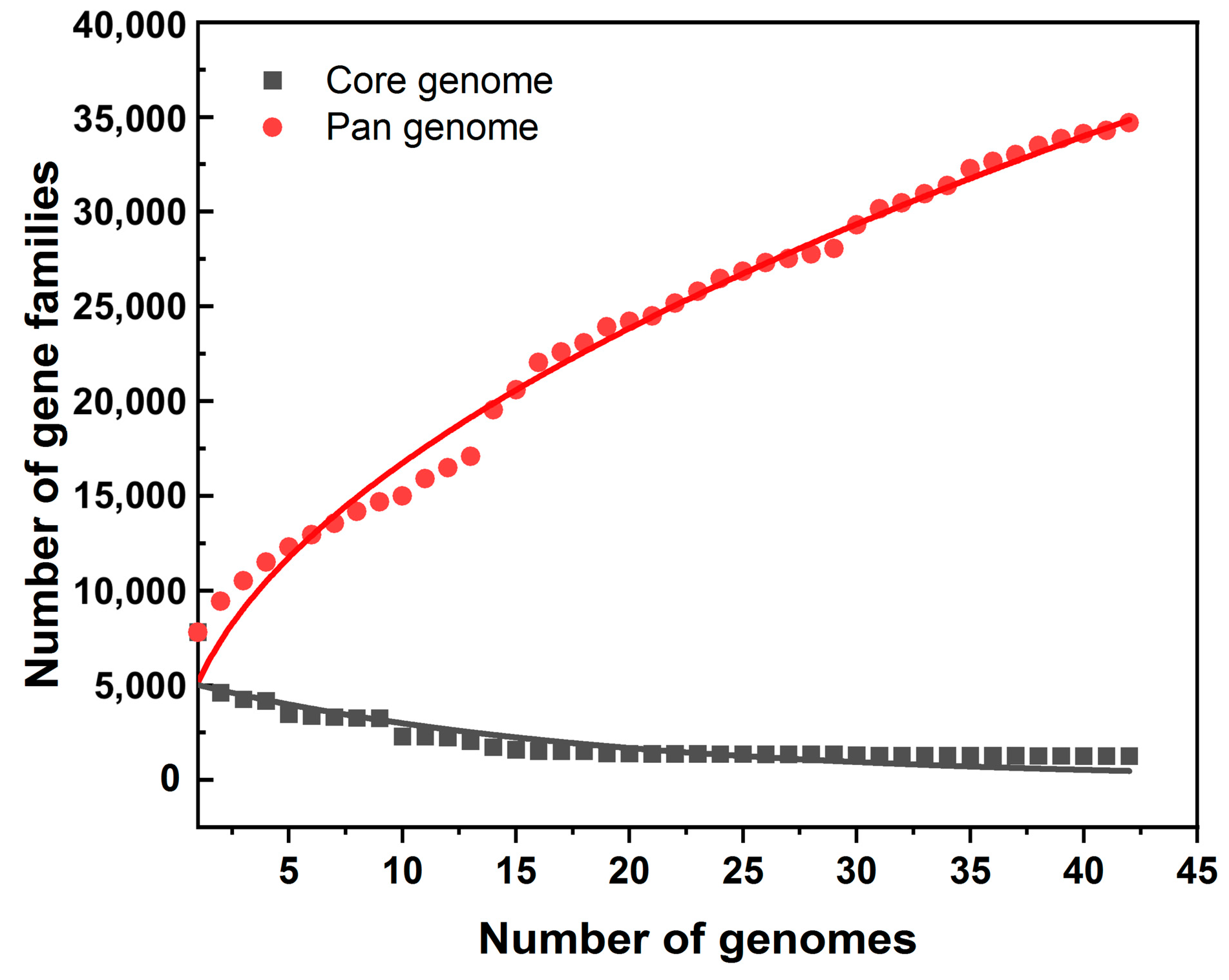
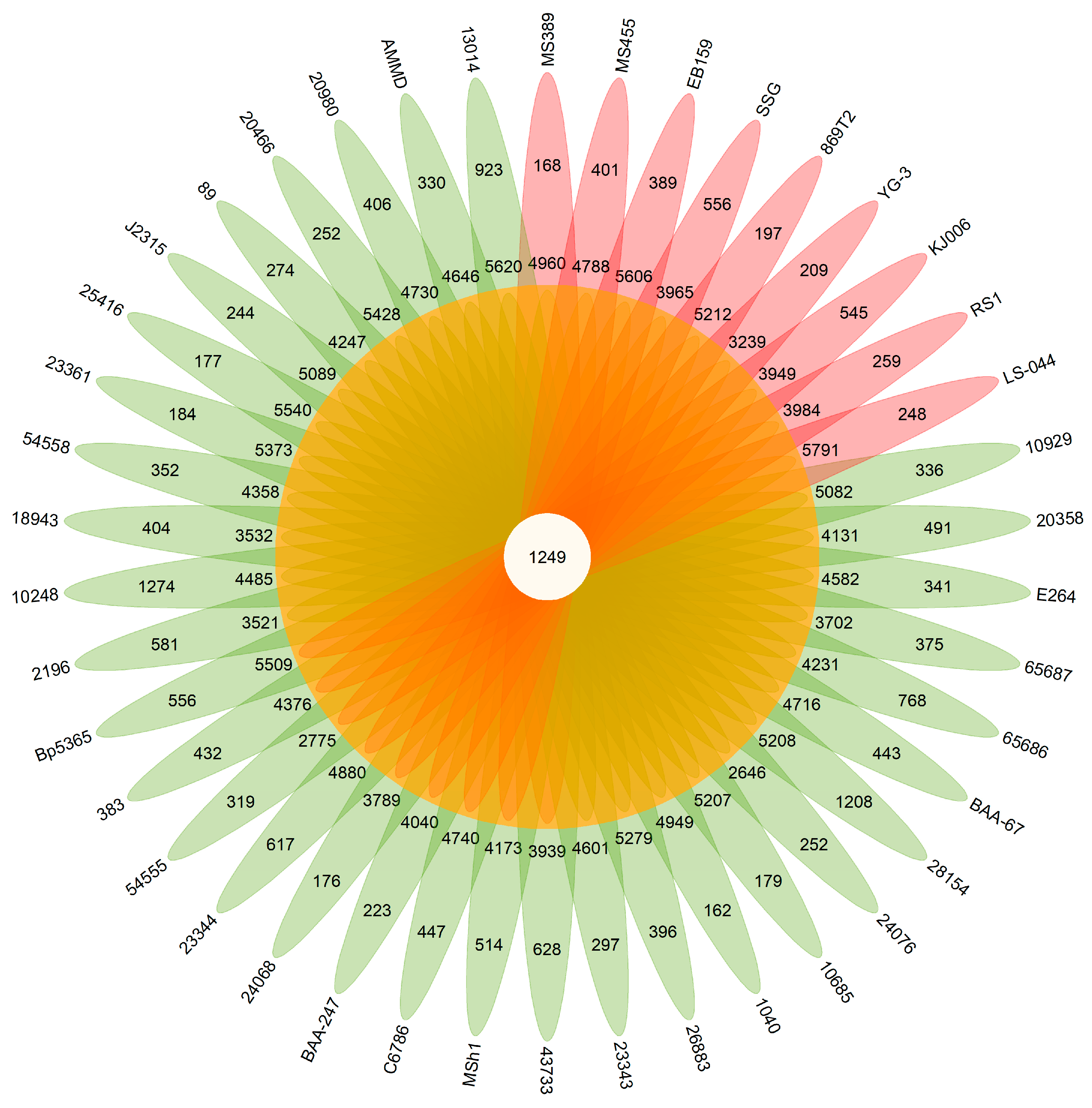
3.2. Pan-Genomic Analysis
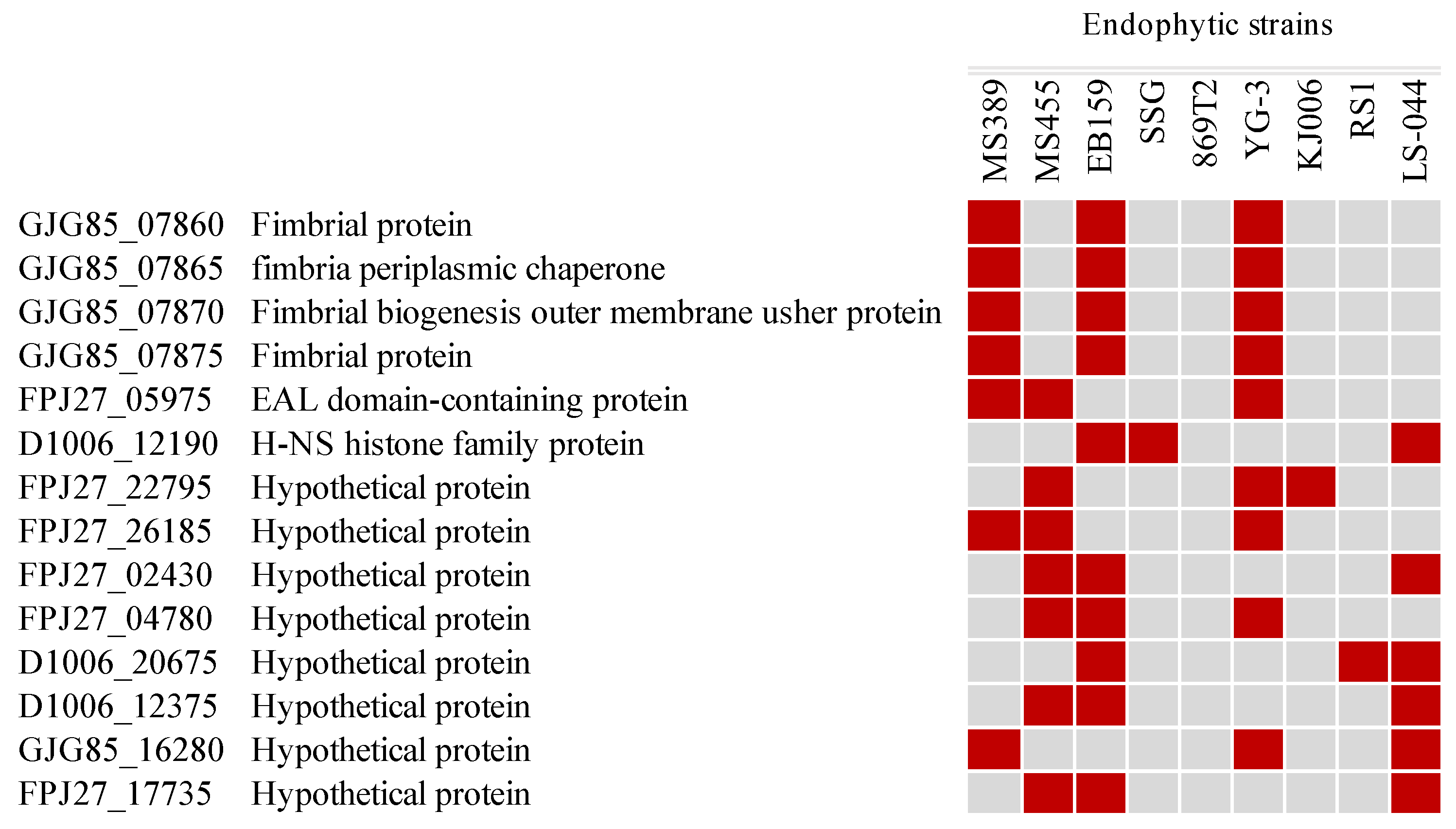
3.3. Production of Antimicrobial Compounds and Siderophores
3.4. Toxin Production and Virulence-Related Features
3.5. Antibiotic Resistance
3.6. Secretion Systems
3.7. Prediction of CRISPR/Cas System
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Olanrewaju, O.S.; Glick, B.R.; Babalola, O.O. Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 2017, 33, 197. [Google Scholar] [CrossRef]
- Compant, S.; Reiter, B.; Sessitsch, A.; Nowak, J.; Clément, C.; Barka, E.A. Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Appl. Environ. Microbiol. 2005, 71, 1685–1693. [Google Scholar] [CrossRef]
- Pereira, S.; Monteiro, C.; Vega, A.; Castro, P.M. Endophytic culturable bacteria colonizing Lavandula dentata L. plants: Isolation, characterization and evaluation of their plant growth-promoting activities. Ecol. Eng. 2016, 87, 91–97. [Google Scholar] [CrossRef]
- Marques, A.P.; Pires, C.; Moreira, H.; Rangel, A.O.; Castro, P.M. Assessment of the plant growth promotion abilities of six bacterial isolates using Zea mays as indicator plant. Soil Biol. Biochem. 2010, 42, 1229–1235. [Google Scholar] [CrossRef]
- Costacurta, A.; Vanderleyden, J. Synthesis of phytohormones by plant-associated bacteria. Crit. Rev. Microbiol. 1995, 21, 1–18. [Google Scholar] [CrossRef]
- Zinniel, D.K.; Lambrecht, P.; Harris, N.B.; Feng, Z.; Kuczmarski, D.; Higley, P.; Ishimaru, C.A.; Arunakumari, A.; Barletta, R.G.; Vidaver, A.K. Isolation and characterization of endophytic colonizing bacteria from agronomic crops and prairie plants. Appl. Environ. Microbiol. 2002, 68, 2198–2208. [Google Scholar] [CrossRef]
- Lodewyckx, C.; Vangronsveld, J.; Porteous, F.; Moore, E.R.; Taghavi, S.; Mezgeay, M.; der Lelie, D.V. Endophytic bacteria and their potential applications. Crit. Rev. Plant. Sci. 2002, 21, 583–606. [Google Scholar] [CrossRef]
- Hardoim, P.R.; van Overbeek, L.S.; van Elsas, J.D. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008, 16, 463–471. [Google Scholar] [CrossRef]
- Ma, Y.; Prasad, M.; Rajkumar, M.; Freitas, H. Plant growth promoting rhizobacteria and endophytes accelerate phytoremediation of metalliferous soils. Biotechnol. Adv. 2011, 29, 248–258. [Google Scholar] [CrossRef]
- Sessitsch, A.; Coenye, T.; Sturz, A.; Vandamme, P.; Barka, E.A.; Salles, J.; Van Elsas, J.; Faure, D.; Reiter, B.; Glick, B. Burkholderia phytofirmans sp. nov., a novel plant-associated bacterium with plant-beneficial properties. Int. J. Syst. Evol. 2005, 55, 1187–1192. [Google Scholar] [CrossRef]
- Liu, X.; Dou, G.; Ma, Y. Potential of endophytes from medicinal plants for biocontrol and plant growth promotion. J. Gen. Plant Pathol. 2016, 82, 165–173. [Google Scholar] [CrossRef]
- Vendan, R.T.; Yu, Y.J.; Lee, S.H.; Rhee, Y.H. Diversity of endophytic bacteria in ginseng and their potential for plant growth promotion. J Microbiol. 2010, 48, 559–565. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, L.A.; Mahenthiralingam, E. Biotechnological potential within the genus Burkholderia. Lett. Appl. Microbiol. 2005, 41, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Winsor, G.L.; Khaira, B.; Van Rossum, T.; Lo, R.; Whiteside, M.D.; Brinkman, F.S. The Burkholderia Genome Database: Facilitating flexible queries and comparative analyses. Bioinformatics 2008, 24, 2803–2804. [Google Scholar] [CrossRef] [PubMed]
- Lessie, T.G.; Hendrickson, W.; Manning, B.D.; Devereux, R. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol. Lett. 1996, 144, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Nowak, J.; Coenye, T.; Clément, C.; Ait Barka, E. Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol. Rev. 2008, 32, 607–626. [Google Scholar] [CrossRef]
- Dobritsa, A.P.; Samadpour, M. Transfer of eleven species of the genus Burkholderia to the genus Paraburkholderia and proposal of Caballeronia gen. nov. to accommodate twelve species of the genera Burkholderia and Paraburkholderia. Int. J. Syst. Evol. Microbiol. 2016, 66, 2836–2846. [Google Scholar] [CrossRef] [PubMed]
- Estrada-de Los Santos, P.; Palmer, M.; Chávez-Ramírez, B.; Beukes, C.; Steenkamp, E.T.; Briscoe, L.; Khan, N.; Maluk, M.; Lafos, M.; Humm, E. Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): Implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes 2018, 9, 389. [Google Scholar] [CrossRef]
- Lopes-Santos, L.; Castro, D.B.A.; Ferreira-Tonin, M.; Corrêa, D.B.A.; Weir, B.S.; Park, D.; Ottoboni, L.M.M.; Neto, J.R.; Destéfano, S.A.L. Reassessment of the taxonomic position of Burkholderia andropogonis and description of Robbsia andropogonis gen. nov., comb. nov. Antonie Van Leeuwenhoek 2017, 110, 727–736. [Google Scholar] [CrossRef]
- Lin, Q.-H.; Lv, Y.-Y.; Gao, Z.-H.; Qiu, L.-H. Pararobbsia silviterrae gen. nov., sp. nov., isolated from forest soil and reclassification of Burkholderia alpina as Pararobbsia alpina comb. nov. Int. J. Syst. Evol. 2020, 70, 1412–1420. [Google Scholar] [CrossRef]
- Beukes, C.W.; Palmer, M.; Manyaka, P.; Chan, W.Y.; Avontuur, J.R.; van Zyl, E.; Huntemann, M.; Clum, A.; Pillay, M.; Palaniappan, K. Genome data provides high support for generic boundaries in Burkholderia sensu lato. Front. Microbiol. 2017, 8, 1154. [Google Scholar] [CrossRef]
- Mahenthiralingam, E.; Urban, T.A.; Goldberg, J.B. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 2005, 3, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Mahenthiralingam, E.; Baldwin, A.; Vandamme, P. Burkholderia cepacia complex infection in patients with cystic fibrosis. J. Med. Microbiol. 2002, 51, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Eberl, L.; Vandamme, P. Members of the genus Burkholderia: Good and bad guys. F1000 Res. 2016, 5, F1000 Faculty Rev-1007. [Google Scholar] [CrossRef] [PubMed]
- Chiarini, L.; Bevivino, A.; Dalmastri, C.; Tabacchioni, S.; Visca, P. Burkholderia cepacia complex species: Health hazards and biotechnological potential. Trends Microbiol. 2006, 14, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Parke, J.L.; Gurian-Sherman, D. Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Annu. Rev. Phytopathol 2001, 39, 225–258. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Ford, E.; Baird, S.M.; Lu, S.-E. Complete Genome Sequence Resource for the Endophytic Burkholderia sp. Strain MS389 Isolated from a Healthy Soybean Growing Adjacent to Charcoal Rot Disease Patch. Plant Dis. 2021, 105, 2704–2707. [Google Scholar] [CrossRef]
- Jia, J.; Ford, E.; Hobbs, S.M.; Baird, S.M.; Lu, S.-E. Occidiofungin is the key metabolite for antifungal activity of the endophytic bacterium Burkholderia sp. MS455 against Aspergillus flavus. Phytopathology 2022, 112, 481–491. [Google Scholar] [CrossRef]
- Kim, H.; Mohanta, T.K.; Park, Y.-H.; Park, S.-C.; Shanmugam, G.; Park, J.-S.; Jeon, J.; Bae, H. Complete genome sequence of the mountain-cultivated ginseng endophyte Burkholderia stabilis and its antimicrobial compounds against ginseng root rot disease. Biol. Control 2020, 140, 104126. [Google Scholar] [CrossRef]
- Kong, P.; Hong, C. Complete genome sequence of a boxwood endophyte Burkholderia sp. SSG with broad biotechnological application potential. Biotechnol. Rep. 2020, 26, e00455. [Google Scholar] [CrossRef]
- Ho, Y.-N.; Huang, C.-C. Draft genome sequence of Burkholderia cenocepacia strain 869T2, a plant-beneficial endophytic bacterium. Genome Announc. 2015, 3. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Liu, X.; Huang, Z.; Niu, S.; Xu, T.; Zeng, J.; Li, H.; Wang, T.; Gao, Y. Physiological, biochemical and proteomic insight into integrated strategies of an endophytic bacterium Burkholderia cenocepacia strain YG-3 response to cadmium stress. Metallomics 2019, 11, 1252–1264. [Google Scholar] [CrossRef]
- Kwak, M.-J.; Song, J.Y.; Kim, S.-Y.; Jeong, H.; Kang, S.G.; Kim, B.K.; Kwon, S.-K.; Lee, C.H.; Yu, D.S.; Park, S.-H. Complete genome sequence of the endophytic bacterium Burkholderia sp. strain KJ006. J. Bacteriol. 2012, 194. [Google Scholar] [CrossRef]
- Shinjo, R.; Uesaka, K.; Ihara, K.; Sakazaki, S.; Yano, K.; Kondo, M.; Tanaka, A. Draft genome sequence of Burkholderia vietnamiensis strain rs1, a nitrogen-fixing endophyte isolated from sweet potato. Microbiol. Resour. Announc. 2018, 7, 10–1128. [Google Scholar] [CrossRef]
- Hameed, A.; Shahina, M.; Lai, W.-A.; Stothard, P.; Young, L.-S.; Lin, S.-Y.; Young, C.-C. Draft genome sequence reveals co-occurrence of multiple antimicrobial resistance and plant probiotic traits in rice root endophytic strain Burkholderia sp. LS-044 affiliated to Burkholderia cepacia complex. J. Glob. Antimicrob. Resist. 2020, 20, 28–30. [Google Scholar] [CrossRef]
- Seo, Y.-S.; Lim, J.Y.; Park, J.; Kim, S.; Lee, H.-H.; Cheong, H.; Kim, S.-M.; Moon, J.S.; Hwang, I. Comparative genome analysis of rice-pathogenic Burkholderia provides insight into capacity to adapt to different environments and hosts. BMC Genom. 2015, 16, 349. [Google Scholar] [CrossRef]
- Johnson, S.L.; Bishop-Lilly, K.A.; Ladner, J.T.; Daligault, H.E.; Davenport, K.W.; Jaissle, J.; Frey, K.G.; Koroleva, G.I.; Bruce, D.C.; Coyne, S.R. Complete genome sequences for 59 Burkholderia isolates, both pathogenic and near neighbor. Genome Announc. 2015, 3. [Google Scholar] [CrossRef]
- Lim, J.; Lee, T.-H.; Nahm, B.H.; Do Choi, Y.; Kim, M.; Hwang, I. Complete genome sequence of Burkholderia glumae BGR1. J. Bacteriol. 2009, 191, 3758–3759. [Google Scholar] [CrossRef]
- Daligault, H.; Davenport, K.; Minogue, T.; Bishop-Lilly, K.; Broomall, S.; Bruce, D.; Chain, P.; Coyne, S.; Frey, K.; Gibbons, H. Whole-genome assemblies of 56 burkholderia species. Genome Announc. 2014, 2, 10-1128. [Google Scholar] [CrossRef]
- Holden, M.T.; Seth-Smith, H.M.; Crossman, L.C.; Sebaihia, M.; Bentley, S.D.; Cerdeño-Tárraga, A.M.; Thomson, N.R.; Bason, N.; Quail, M.A.; Sharp, S. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J. Bacteriol. 2009, 191, 261–277. [Google Scholar] [CrossRef]
- Vandamme, P.; Mahenthiralingam, E.; Holmes, B.; Coenye, T.; Hoste, B.; De Vos, P.; Henry, D.; Speert, D. Identification and Population Structure of Burkholderia stabilis sp. nov.(formerly Burkholderia cepacia Genomovar IV). J. Clin. Microbiol. 2000, 38, 1042–1047. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Kim, Y.O.; Park, S.-C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Oh, H.-S.; Park, S.-C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. 2014, 64, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA-an ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed]
- Rost, B. Twilight zone of protein sequence alignments. Protein Eng. 1999, 12, 85–94. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Cury, J.; Jové, T.; Touchon, M.; Néron, B.; Rocha, E.P. Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic Acids Res. 2016, 44, 4539–4550. [Google Scholar] [CrossRef]
- Abby, S.S.; Rocha, E.P. Identification of protein secretion systems in bacterial genomes using MacSyFinder. In Bacterial Protein Secretion Systems; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–21. [Google Scholar]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef] [PubMed]
- Biswas, A.; Gagnon, J.N.; Brouns, S.J.; Fineran, P.C.; Brown, C.M. CRISPRTarget: Bioinformatic prediction and analysis of crRNA targets. RNA Biol. 2013, 10, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.-E.; Novak, J.; Austin, F.W.; Gu, G.; Ellis, D.; Kirk, M.; Wilson-Stanford, S.; Tonelli, M.; Smith, L. Occidiofungin, a unique antifungal glycopeptide produced by a strain of Burkholderia contaminans. Biochemistry 2009, 48, 8312–8321. [Google Scholar] [CrossRef] [PubMed]
- Hammer, P.E.; Hill, D.S.; Lam, S.T.; Van Pée, K.-H.; Ligon, J.M. Four genes from Pseudomonas fluorescens that encode the biosynthesis of pyrrolnitrin. Appl. Environ. Microbiol. 1997, 63, 2147–2154. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.; Blom, J.F.; Pernthaler, J.; Berg, G.; Baldwin, A.; Mahenthiralingam, E.; Eberl, L. Production of the antifungal compound pyrrolnitrin is quorum sensing-regulated in members of the Burkholderia cepacia complex. Environ. Microbiol. 2009, 11, 1422–1437. [Google Scholar] [CrossRef] [PubMed]
- Nowak-Thompson, B.; Chaney, N.; Wing, J.S.; Gould, S.J.; Loper, J.E. Characterization of the pyoluteorin biosynthetic gene cluster of Pseudomonas fluorescens Pf-5. J. Bacteriol. 1999, 181, 2166–2174. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Carlson, R.; Tharpe, W.; Schell, M.A. Characterization of genes involved in biosynthesis of a novel antibiotic from Burkholderia cepacia BC11 and their role in biological control of Rhizoctonia solani. Appl. Environ. Microbiol. 1998, 64, 3939–3947. [Google Scholar] [CrossRef]
- Mullins, A.J.; Murray, J.A.; Bull, M.J.; Jenner, M.; Jones, C.; Webster, G.; Green, A.E.; Neill, D.R.; Connor, T.R.; Parkhill, J. Genome mining identifies cepacin as a plant-protective metabolite of the biopesticidal bacterium Burkholderia ambifaria. Nat. Microbiol. 2019, 4, 996–1005. [Google Scholar] [CrossRef]
- Serino, L.; Reimmann, C.; Visca, P.; Beyeler, M.; Chiesa, V.; Haas, D. Biosynthesis of pyochelin and dihydroaeruginoic acid requires the iron-regulated pchDCBA operon in Pseudomonas aeruginosa. J. Bacteriol. 1997, 179, 248–257. [Google Scholar] [CrossRef]
- Agnoli, K.; Lowe, C.A.; Farmer, K.L.; Husnain, S.I.; Thomas, M.S. The ornibactin biosynthesis and transport genes of Burkholderia cenocepacia are regulated by an extracytoplasmic function σ factor which is a part of the Fur regulon. J. Bacteriol. 2006, 188, 3631–3644. [Google Scholar] [CrossRef]
- Jenul, C.; Sieber, S.; Daeppen, C.; Mathew, A.; Lardi, M.; Pessi, G.; Hoepfner, D.; Neuburger, M.; Linden, A.; Gademann, K. Biosynthesis of fragin is controlled by a novel quorum sensing signal. Nat. Commun. 2018, 9, 1297. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Oliver, R.A.; Townsend, C.A. Identification and characterization of the sulfazecin monobactam biosynthetic gene cluster. Cell Chem. Biol. 2017, 24, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Carr, G.; Seyedsayamdost, M.R.; Chandler, J.R.; Greenberg, E.P.; Clardy, J. Sources of diversity in bactobolin biosynthesis by Burkholderia thailandensis E264. Org. Lett. 2011, 13, 3048–3051. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhou, H.; Chen, H.; Jing, X.; Zheng, W.; Li, R.; Sun, T.; Liu, J.; Fu, J.; Huo, L. Discovery of recombinases enables genome mining of cryptic biosynthetic gene clusters in Burkholderiales species. Proc. Natl. Acad. Sci. USA 2018, 115, E4255–E4263. [Google Scholar] [CrossRef]
- Dose, B.; Niehs, S.P.; Scherlach, K.; Flórez, L.V.; Kaltenpoth, M.; Hertweck, C. Unexpected bacterial origin of the antibiotic icosalide: Two-tailed depsipeptide assembly in multifarious Burkholderia symbionts. ACS Chem. Biol. 2018, 13, 2414–2420. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, F.; Sawada, H.; Azegami, K.; Tsuchiya, K. Molecular characterization of the tox operon involved in toxoflavin biosynthesis of Burkholderia glumae. J. Gen. Plant Pathol. 2004, 70, 97–107. [Google Scholar] [CrossRef]
- Kim, J.; Kim, J.G.; Kang, Y.; Jang, J.Y.; Jog, G.J.; Lim, J.Y.; Kim, S.; Suga, H.; Nagamatsu, T.; Hwang, I. Quorum sensing and the LysR-type transcriptional activator ToxR regulate toxoflavin biosynthesis and transport in Burkholderia glumae. Mol. Microbiol. 2004, 54, 921–934. [Google Scholar] [CrossRef]
- Miwa, S.; Kihira, E.; Yoshioka, A.; Nakasone, K.; Okamoto, S.; Hatano, M.; Igarashi, M.; Eguchi, Y.; Kato, A.; Ichikawa, N. Identification of the three genes involved in controlling production of a phytotoxin tropolone in Burkholderia plantarii. J. Bacteriol. 2016, 198, 1604–1609. [Google Scholar] [CrossRef]
- Okazaki, S.; Sugawara, M.; Minamisawa, K. Bradyrhizobium elkanii rtxC gene is required for expression of symbiotic phenotypes in the final step of rhizobitoxine biosynthesis. Appl. Environ. Microbiol. 2004, 70, 535–541. [Google Scholar] [CrossRef][Green Version]
- Biggins, J.B.; Ternei, M.A.; Brady, S.F. Malleilactone, a polyketide synthase-derived virulence factor encoded by the cryptic secondary metabolome of Burkholderia pseudomallei group pathogens. J. Am. Chem. Soc. 2012, 134, 13192–13195. [Google Scholar] [CrossRef]
- Partida-Martinez, L.P.; Hertweck, C. A gene cluster encoding rhizoxin biosynthesis in “Burkholderia rhizoxina”, the bacterial endosymbiont of the fungus Rhizopus microsporus. ChemBioChem 2007, 8, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Moebius, N.; Ross, C.; Scherlach, K.; Rohm, B.; Roth, M.; Hertweck, C. Biosynthesis of the respiratory toxin bongkrekic acid in the pathogenic bacterium Burkholderia gladioli. Chem. Biol. 2012, 19, 1164–1174. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, C.F.; Pettit, E.A.; Valadez, V.A.; Provin, E.M. Mobilization, cloning, and sequence determination of a plasmid-encoded polygalacturonase from a phytopathogenic Burkholderia (Pseudomonas) cepacia. Mol. Plant Microbe Interact. 1997, 10, 840–851. [Google Scholar] [CrossRef] [PubMed]
- Degrassi, G.; Devescovi, G.; Kim, J.; Hwang, I.; Venturi, V. Identification, characterization and regulation of two secreted polygalacturonases of the emerging rice pathogen Burkholderia glumae. FEMS Microbiol. Ecol. 2008, 65, 251–262. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ortega, X.; Hunt, T.A.; Loutet, S.; Vinion-Dubiel, A.D.; Datta, A.; Choudhury, B.; Goldberg, J.B.; Carlson, R.; Valvano, M.A. Reconstitution of O-specific lipopolysaccharide expression in Burkholderia cenocepacia strain J2315, which is associated with transmissible infections in patients with cystic fibrosis. J. Bacteriol. 2005, 187, 1324–1333. [Google Scholar] [CrossRef]
- Rhodes, K.A.; Schweizer, H.P. Antibiotic resistance in Burkholderia species. Drug Resist. Updat. 2016, 28, 82–90. [Google Scholar] [CrossRef]
- Yuan, C.; Wei, Y.; Zhang, S.; Cheng, J.; Cheng, X.; Qian, C.; Wang, Y.; Zhang, Y.; Yin, Z.; Chen, H. Comparative Genomic Analysis Reveals Genetic Mechanisms of the Variety of Pathogenicity, Antibiotic Resistance, and Environmental Adaptation of Providencia Genus. Front. Microbiol. 2020, 11, 2512. [Google Scholar] [CrossRef]
- Green, E.R.; Mecsas, J. Bacterial secretion systems: An overview. Virulence Mech. Bact. Pathog. 2016, 213–239. [Google Scholar] [CrossRef]
- Angus, A.A.; Agapakis, C.M.; Fong, S.; Yerrapragada, S.; Estrada-de Los Santos, P.; Yang, P.; Song, N.; Kano, S.; Caballero-Mellado, J.; De Faria, S.M. Plant-associated symbiotic Burkholderia species lack hallmark strategies required in mammalian pathogenesis. PLoS ONE 2014, 9, e83779. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Hurek, T. Living inside plants: Bacterial endophytes. Curr. Opin. Plant. Biol. 2011, 14, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Louwen, R.; Staals, R.H.; Endtz, H.P.; van Baarlen, P.; van der Oost, J. The role of CRISPR-Cas systems in virulence of pathogenic bacteria. Microbiol. Mol. Biol. Rev. 2014, 78, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Haft, D.H. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Zhou, J.; Zhou, J.; Hu, M.; Zhang, Q.; Kong, N.; Ren, H.; Liang, L.; Yue, J. Genome-based classification of Burkholderia cepacia complex provides new insight into its taxonomic status. Biol. Direct 2020, 15, 6. [Google Scholar] [CrossRef] [PubMed]
- Mitter, B.; Petric, A.; Shin, M.W.; Chain, P.S.; Hauberg-Lotte, L.; Reinhold-Hurek, B.; Nowak, J.; Sessitsch, A. Comparative genome analysis of Burkholderia phytofirmans PsJN reveals a wide spectrum of endophytic lifestyles based on interaction strategies with host plants. Front. Plant Sci. 2013, 4, 120. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Duan, J.; Charles, T.C.; Glick, B.R. A bioinformatics approach to the determination of genes involved in endophytic behavior in Burkholderia spp. J. Theor. Biol. 2014, 343, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Weening, E.H.; Barker, J.D.; Laarakker, M.C.; Humphries, A.D.; Tsolis, R.M.; Bäumler, A.J. The Salmonella enterica serotype Typhimurium lpf, bcf, stb, stc, std, and sth fimbrial operons are required for intestinal persistence in mice. Infect. Immun. 2005, 73, 3358–3366. [Google Scholar] [CrossRef]
- Hinton, D.M.; Bacon, C.W. Enterobacter cloacae is an endophytic symbiont of corn. Mycopathologia 1995, 129, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Römling, U.; Gomelsky, M.; Galperin, M.Y. C-di-GMP: The dawning of a novel bacterial signalling system. Mol. Microbiol. 2005, 57, 629–639. [Google Scholar] [CrossRef]
- Mark, G.L.; Dow, J.M.; Kiely, P.D.; Higgins, H.; Haynes, J.; Baysse, C.; Abbas, A.; Foley, T.; Franks, A.; Morrissey, J. Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe-plant interactions. Proc. Natl. Acad. Sci. USA 2005, 102, 17454–17459. [Google Scholar] [CrossRef]
- Rimsky, S. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr. Opin. Microbiol. 2004, 7, 109–114. [Google Scholar] [CrossRef]
- English, M.; Coulson, T.; Horsman, S.; Patten, C. Overexpression of hns in the plant growth-promoting bacterium Enterobacter cloacae UW5 increases root colonization. J. Appl. Microbiol. 2010, 108, 2180–2190. [Google Scholar] [PubMed]
- Depoorter, E.; Bull, M.J.; Peeters, C.; Coenye, T.; Vandamme, P.; Mahenthiralingam, E. Burkholderia: An update on taxonomy and biotechnological potential as antibiotic producers. Appl. Microbiol. Biotechnol. 2016, 100, 5215–5229. [Google Scholar] [CrossRef]
- Hider, R.C.; Kong, X. Chemistry and biology of siderophores. Nat. Prod. Rep. 2010, 27, 637–657. [Google Scholar] [CrossRef] [PubMed]
- Visser, M.; Majumdar, S.; Hani, E.; Sokol, P. Importance of the ornibactin and pyochelin siderophore transport systems in Burkholderia cenocepacia lung infections. Infect. Immun. 2004, 72, 2850–2857. [Google Scholar] [CrossRef] [PubMed]
- Mathew, A.; Eberl, L.; Carlier, A.L. A novel siderophore-independent strategy of iron uptake in the genus B urkholderia. Mol. Microbiol. 2014, 91, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Azegami, K.; Nishiyama, K.; Kato, H. Effect of iron limitation on “Pseudomonas plantarii” growth and tropolone and protein production. Appl. Environ. Microbiol. 1988, 54, 844–847. [Google Scholar] [CrossRef] [PubMed]
- Schaefers, M.M. Regulation of virulence by two-component systems in pathogenic Burkholderia. Infect. Immun. 2020, 88, e00927-19. [Google Scholar] [CrossRef]
- Gonzalez, C.F.; Vidaver, A.K. Bacteriocin, plasmid and pectolytic diversity in Pseudomonas cepacia of clinical and plant origin. Microbiology 1979, 110, 161–170. [Google Scholar] [CrossRef]
- Kong, P.; Hong, C. A potent Burkholderia endophyte against boxwood blight caused by Calonectria pseudonaviculata. Microorganisms 2020, 8, 310. [Google Scholar] [CrossRef]
- Ramírez, M.S.; Vargas, L.J.; Cagnoni, V.; Tokumoto, M.; Centrón, D. Class 2 integron with a novel cassette array in a Burkholderia cenocepacia isolate. Antimicrob. Agents Chemother. 2005, 49, 4418. [Google Scholar] [CrossRef]
- Tseng, S.-P.; Tsai, W.-C.; Liang, C.-Y.; Lin, Y.-S.; Huang, J.-W.; Chang, C.-Y.; Tyan, Y.-C.; Lu, P.-L. The contribution of antibiotic resistance mechanisms in clinical Burkholderia cepacia complex isolates: An emphasis on efflux pump activity. PLoS ONE 2014, 9, e104986. [Google Scholar] [CrossRef]
- Kim, N.; Kim, J.J.; Kim, I.; Mannaa, M.; Park, J.; Kim, J.; Lee, H.H.; Lee, S.B.; Park, D.S.; Sul, W.J. Type VI secretion systems of plant-pathogenic Burkholderia glumae BGR1 play a functionally distinct role in interspecies interactions and virulence. Mol. Plant Pathol. 2020, 21, 1055–1069. [Google Scholar] [CrossRef] [PubMed]
- Dolores, J.S.; Agarwal, S.; Egerer, M.; Satchell, K.J. Vibrio cholerae MARTX toxin heterologous translocation of beta-lactamase and roles of individual effector domains on cytoskeleton dynamics. Mol. Microbiol. 2015, 95, 590–604. [Google Scholar] [CrossRef]
- Letoffe, S.; Delepelaire, P.; Wandersman, C. Protein secretion in gram-negative bacteria: Assembly of the three components of ABC protein-mediated exporters is ordered and promoted by substrate binding. EMBO J. 1996, 15, 5804–5811. [Google Scholar] [CrossRef]
- Xia, Y.; DeBolt, S.; Dreyer, J.; Scott, D.; Williams, M.A. Characterization of culturable bacterial endophytes and their capacity to promote plant growth from plants grown using organic or conventional practices. Front. Plant Sci. 2015, 6, 490. [Google Scholar] [CrossRef] [PubMed]
- Piromyou, P.; Songwattana, P.; Greetatorn, T.; Okubo, T.; Kakizaki, K.C.; Prakamhang, J.; Tittabutr, P.; Boonkerd, N.; Teaumroong, N.; Minamisawa, K. The type III secretion system (T3SS) is a determinant for rice-endophyte colonization by non-photosynthetic Bradyrhizobium. Microbes Environ. 2015, 30, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Pratama, A.A.; Chaib De Mares, M.; Van Elsas, J.D. Evolutionary history of bacteriophages in the genus Paraburkholderia. Front. microbiol. 2018, 9, 835. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-H.; Park, J.; Jung, H.; Seo, Y.-S. Pan-Genome Analysis Reveals Host-Specific Functional Divergences in Burkholderia gladioli. Microorganisms 2021, 9, 1123. [Google Scholar] [CrossRef]
- Burstein, D.; Sun, C.L.; Brown, C.T.; Sharon, I.; Anantharaman, K.; Probst, A.J.; Thomas, B.C.; Banfield, J.F. Major bacterial lineages are essentially devoid of CRISPR-Cas viral defence systems. Nat. Commun. 2016, 7, 10613. [Google Scholar] [CrossRef]
- Wheatley, R.M.; MacLean, R.C. CRISPR-Cas systems restrict horizontal gene transfer in Pseudomonas aeruginosa. ISME J. 2021, 15, 1420–1433. [Google Scholar] [CrossRef]
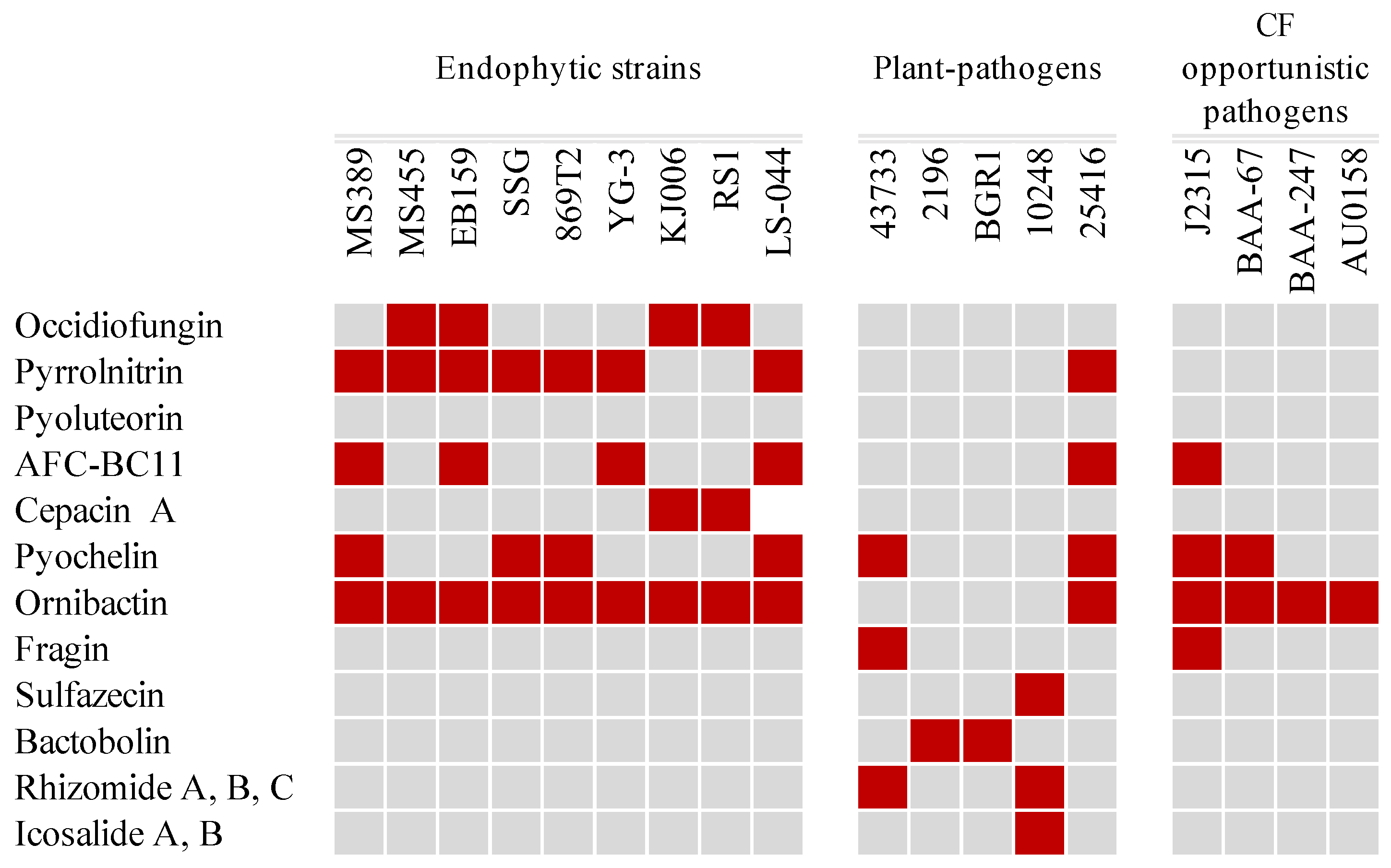

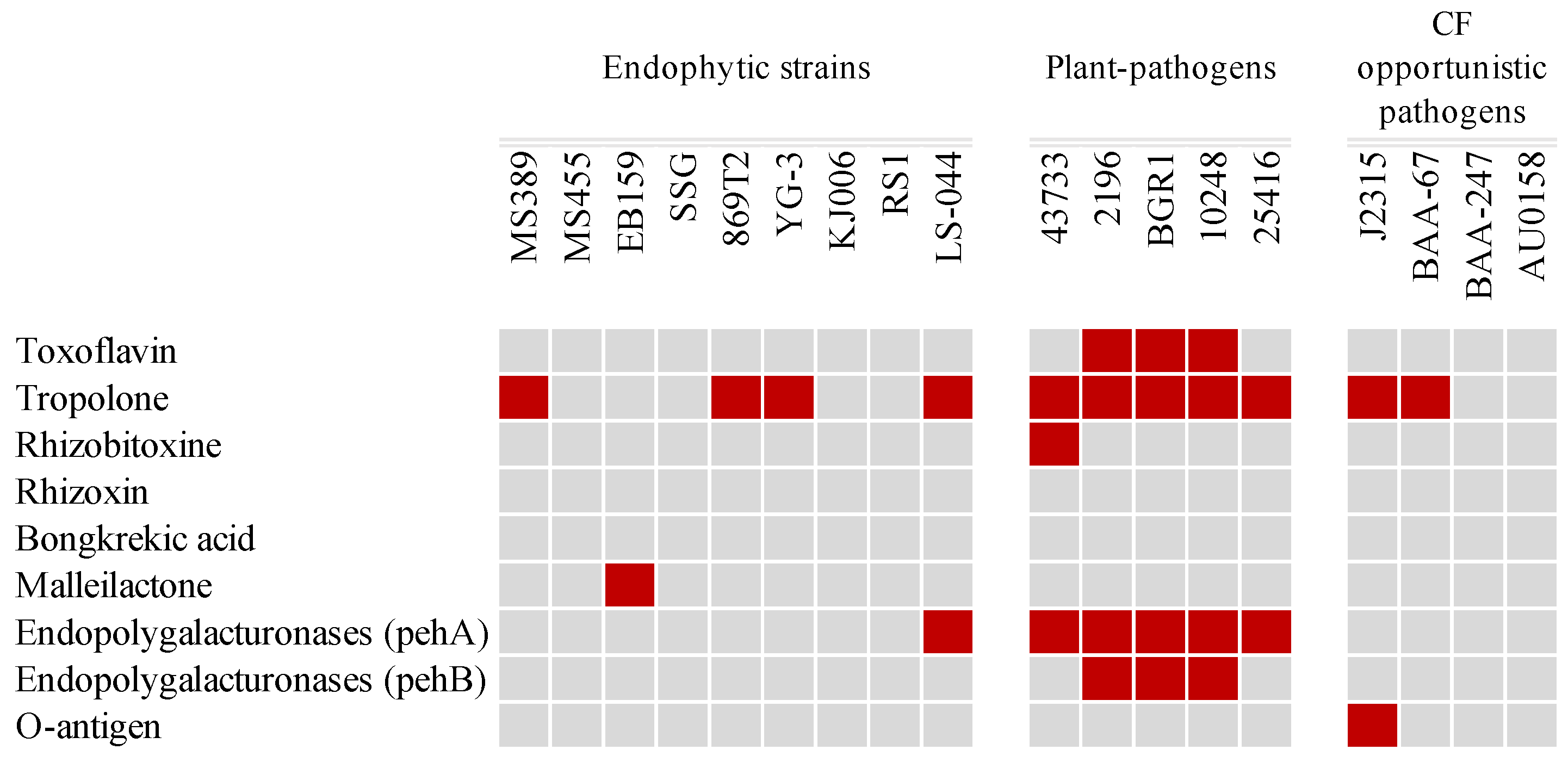
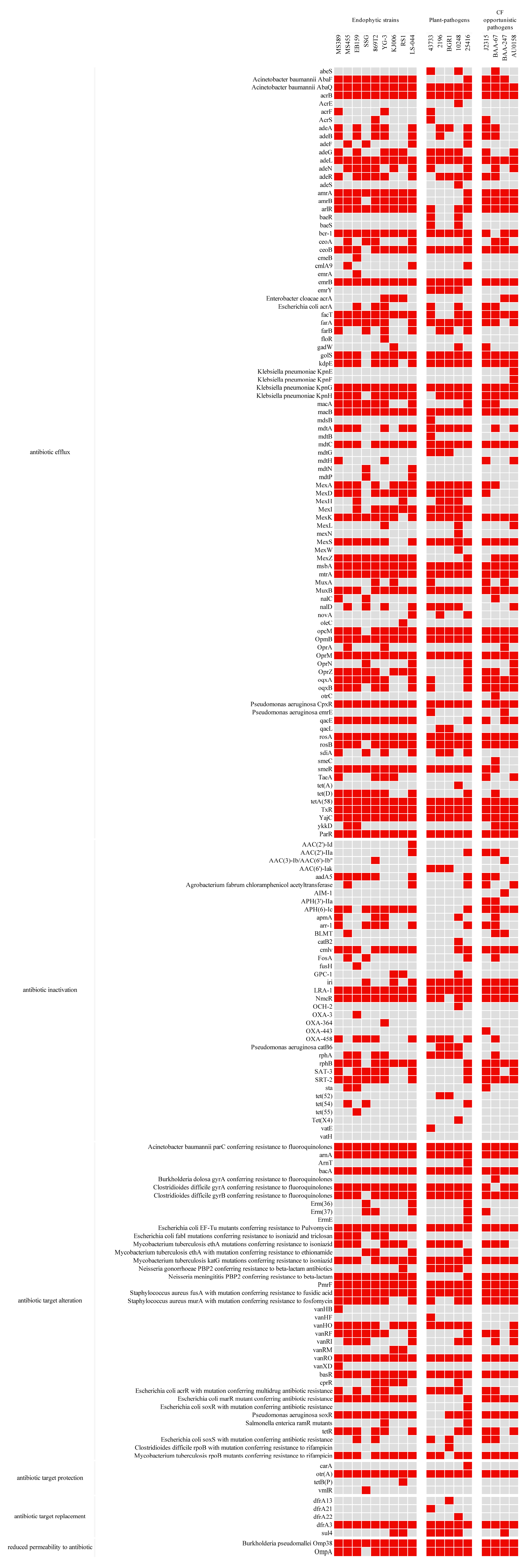
| Strain | # Chr | # Plasmid | Size (bp) | GC% | Isolation Source | Accession Number | |
|---|---|---|---|---|---|---|---|
| Endophytic strains | Burkholderia sp. MS389 | 3 | - | 7,746,250 | 66.73 | Soybean plant | GCA_016899425 |
| Burkholderia sp. MS455 | 3 | 1 | 8,055,789 | 66.06 | Soybean plant | GCA_016899445 | |
| B. stabilis EB159 | - | - | 9,093,480 | 66.00 | Mountain cultivated ginseng | GCA_004125865 | |
| B. cepacia SSG | - | - | 8,571,737 | 66.94 | Boxwood leaves | GCA_012510315 | |
| B. cenocepacia 869T2 | - | - | 7,979,184 | 67.07 | Vetiver grass roots | GCA_000705535 | |
| B. cenocepacia YG-3 | 3 | - | 8,036,463 | 66.81 | Poplar tree | GCA_003966315 | |
| B. vietnamiensis KJ006 | 3 | 1 | 6,629,912 | 67.18 | Rice root | GCA_000262695 | |
| B. vietnamiensis RS1 | - | - | 6,542,727 | 67.61 | Tuberous roots of sweet potato | GCA_003203575 | |
| Burkholderia sp. LS-044 | - | - | 8,782,274 | 66.52 | Rice root | GCA_004803625 | |
| Plant pathogens | B. plantarii ATCC 43733 | 2 | 1 | 8,081,051 | 68.55 | Diseased rice sheath | GCA_001411805 |
| B. glumae LMG 2196 | 2 | 2 | 6,820,727 | 68.18 | Diseased rice grain | GCA_000960995 | |
| B. glumae BGR1 | 2 | 4 | 7,284,636 | 67.93 | Diseased rice grain | GCA_000022645 | |
| B. gladioli ATCC 10248 | 2 | 3 | 8,899,459 | 67.63 | Diseased gladiolus corms | GCA_000959725 | |
| B. cepacia ATCC 25416 | 3 | 1 | 8,605,945 | 66.61 | Rotten onion | GCA_001411495 | |
| CF opportunistic pathogens | B. cenocepacia J2315 | 3 | 1 | 8,055,782 | 66.90 | Sputum of cystic fibrosis patients | GCA_000009485 |
| B. stabilis ATCC BAA-67 | 3 | - | 8,527,947 | 66.42 | Sputum of cystic fibrosis patients | GCA_001742165 | |
| B. multivorans ATCC BAA-247 | 3 | - | 6,322,746 | 67.24 | Sputum of cystic fibrosis patients | GCA_000959525 | |
| B. dolosa AU0158 | 3 | - | 6,409,095 | 67.01 | Sputum of cystic fibrosis patients | GCA_000959505 |
| Strain | # Chr | # Plasmid | Size (bp) | GC% | Isolation Source | Accession Number |
|---|---|---|---|---|---|---|
| B. aenigmatica LMG 13014 | - | - | 8,897,342 | 65.95 | Contaminant of hand cream | GCA_902499175 |
| B. ambifaria AMMD | 3 | - | 7,528,567 | 66.77 | Rhizosphere of pea | GCA_000203915 |
| B. anthina LMG 20980 | - | - | 7,608,177 | 66.74 | Rhizosphere of a house-plant | GCA_902498995 |
| B. arboris LMG 24066 | - | - | 8,271,305 | 66.84 | Morris Arboretum soil | GCA_902499125 |
| B. catarinensis 89 | - | - | 8,137,374 | 66.46 | Native grassland soil | GCA_001883705 |
| B. cenocepacia J2315 | 3 | 1 | 8,055,782 | 66.9 | Sputum of cystic fibrosis patients | GCA_000009485 |
| B. cepacia ATCC 25416 | 3 | 1 | 8,605,945 | 66.61 | Rotten onion | GCA_001411495 |
| B. contaminans LMG 23361 | 4 | - | 10,352,616 | 65.68 | Milk of dairy sheep with mastitis | GCA_001758385 |
| B. diffusa CCUG 54558 | - | - | 7,111,827 | 66.25 | Sputum of cystic fibrosis patients | GCA_008802145 |
| B. dolosa LMG 18943 | - | - | 6,079,078 | 66.97 | Sputum of cystic fibrosis patients | GCA_902499135 |
| B. gladioli ATCC 10248 | 2 | 3 | 8,899,459 | 67.63 | Diseased gladiolus corms | GCA_000959725 |
| B. glumae LMG 2196 | 2 | 2 | 6,820,727 | 68.18 | Diseased rice grain | GCA_000960995 |
| B. humptydooensis MSMB43 | 2 | 1 | 7,287,809 | 67.14 | Bore water sample | GCA_001513745 |
| B. lata 383 | 3 | - | 8,676,277 | 66.27 | Forest soil | GCA_000012945 |
| B. latens CCUG 54555 | - | - | 7,103,502 | 66.82 | Sputum of cystic fibrosis patients | GCA_008802135 |
| B. mallei ATCC 23344 | - | - | 5,835,527 | 68.49 | Etiological agent of glanders | GCA_000011705 |
| B. metallica LMG 24068 | - | - | 7,532,498 | 67.04 | Sputum of cystic fibrosis patients | GCA_902499065 |
| B. multivorans ATCC BAA-247 | 3 | - | 6,322,746 | 67.24 | Sputum of cystic fibrosis patients | GCA_000959525 |
| B. oklahomensis C6786 | 2 | - | 7,135,022 | 67.07 | Deep leg wound heavily contaminated with soil | GCA_000959365 |
| B. paludis MSh1 | - | - | 8,633,651 | 67.08 | Southeast Pahang tropical peat swamp forest soil | GCA_000732615 |
| B. plantarii ATCC 43733 | 2 | 1 | 8,081,051 | 68.55 | Diseased rice sheath | GCA_001411805 |
| B. pseudomallei ATCC 23343 | - | - | 7,037,422 | 68.27 | Etiological agent of melioidosis | GCA_001182285 |
| B. pseudomultivorans LMG 26883 | - | - | 7,396,511 | 66.99 | Human respiratory specimens | GCA_902499075 |
| B. puraquae CAMPA 1040 | - | - | 8,098,134 | 66.59 | Hemodialysis water | GCA_002099195 |
| B. pyrrocinia DSM 10685 | 3 | 1 | 7,961,346 | 66.46 | Environmental soil | GCA_001028665 |
| B. seminalis LMG 24067 | - | - | 7,928,753 | 67.07 | Sputum of cystic fibrosis patients | GCA_902499165 |
| B. singularis LMG 28154 | - | - | 5,538,112 | 64.34 | Sputum of cystic fibrosis patients | GCA_900176645 |
| B. stabilis ATCC BAA-67 | 3 | - | 8,527,947 | 66.42 | Sputum of cystic fibrosis patients | GCA_001742165 |
| B. stagnalis CCUG 65686 | - | - | 8,125,942 | 66.94 | Environmental soil | GCA_008802125 |
| B. territorii CCUG 65687 | - | - | 6,834,211 | 66.55 | Environmental groundwater | GCA_008802115 |
| B. thailandensis E264 | 2 | - | 6,723,972 | 67.63 | Rice field soil | GCA_000012365 |
| B. ubonensis LMG 20358 | - | - | 7,692,957 | 67.24 | Surface soil of roadside | GCA_902499185 |
| B. vietnamiensis LMG 10929 | 3 | 1 | 6,930,496 | 66.83 | Rice rhizosphere soil | GCA_000959445 |
| Name | Antibiotic Efflux | Antibiotic Inactivation | Antibiotic Target Alteration | Antibiotic Target Protection | Antibiotic Target Replacement | Reduced Permeability to Antibiotic | Total | |
|---|---|---|---|---|---|---|---|---|
| Endophytic strains | MS389 | 101 | 13 | 29 | 1 | 1 | 13 | 158 |
| MS455 | 88 | 14 | 25 | 1 | 1 | 14 | 143 | |
| EB159 | 98 | 15 | 28 | 1 | 1 | 19 | 162 | |
| SSG | 63 | 12 | 21 | 2 | 1 | 11 | 110 | |
| 869T2 | 109 | 14 | 30 | 1 | 1 | 13 | 168 | |
| YG-3 | 105 | 12 | 31 | 1 | 1 | 12 | 162 | |
| KJ006 | 67 | 10 | 23 | 1 | 2 | 10 | 113 | |
| RS1 | 62 | 10 | 25 | 2 | 2 | 9 | 110 | |
| LS-044 | 113 | 17 | 27 | 1 | 1 | 17 | 176 | |
| Plant pathogens | 43733 | 90 | 10 | 23 | 2 | 3 | 14 | 142 |
| 2196 | 68 | 8 | 19 | 2 | 2 | 9 | 108 | |
| BGR1 | 65 | 7 | 21 | 2 | 3 | 9 | 107 | |
| 10248 | 105 | 13 | 28 | 1 | 3 | 20 | 170 | |
| 25416 | 108 | 18 | 32 | 2 | 1 | 17 | 178 | |
| CF opportunistic pathogens | J2315 | 92 | 18 | 27 | 1 | 1 | 10 | 149 |
| BAA-67 | 96 | 20 | 27 | 1 | 1 | 18 | 163 | |
| BAA-247 | 68 | 13 | 20 | 1 | 2 | 12 | 116 | |
| AU0158 | 74 | 10 | 25 | 1 | 1 | 10 | 121 |
| Name | T1SS | T2SS | T3SS | T4SS | T5SS | T6SS | Sec | Tat | Flagellar Apparatus | |
|---|---|---|---|---|---|---|---|---|---|---|
| Endophytic strains | MS389 | - | 1 | 1 | - | 15 | 3 | 1 | 1 | 1 |
| MS455 | - | 1 | 1 | 1 | 9 | 3 | 1 | 1 | 1 | |
| EB159 | 4 | 1 | 1 | 3 | 19 | 2 | 1 | 1 | 1 | |
| SSG | - | 1 | - | - | 3 | 2 | 1 | 1 | 1 | |
| 869T2 | 4 | 1 | 1 | 1 | 10 | 1 | 1 | 1 | 1 | |
| YG-3 | 2 | 1 | 1 | - | 13 | 3 | 1 | 1 | 1 | |
| KJ006 | - | 1 | 1 | 1 | 12 | 2 | 1 | 1 | 1 | |
| RS1 | - | 1 | 1 | - | 12 | 2 | 1 | 1 | 1 | |
| LS-044 | 1 | 1 | - | 1 | 9 | 3 | 1 | 1 | 1 | |
| Plant pathogens | 43733 | 2 | 1 | 1 | 2 | 6 | 3 | 1 | 1 | 1 |
| 2196 | - | 1 | 1 | 1 | 7 | 3 | 1 | 1 | 1 | |
| BGR1 | 1 | 1 | 1 | 2 | 9 | 4 | 1 | 1 | 1 | |
| 10248 | 1 | 1 | 1 | 1 | 10 | 2 | 1 | 1 | 1 | |
| 25416 | - | 1 | - | 1 | 9 | 2 | 1 | 1 | 1 | |
| CF opportunistic pathogens | J2315 | 2 | 1 | 1 | 2 | 8 | 1 | 1 | 1 | 1 |
| BAA-67 | 1 | 1 | 1 | 1 | 9 | 2 | 1 | 1 | 1 | |
| BAA-247 | 1 | 1 | 1 | 2 | 7 | 2 | 1 | 1 | 1 | |
| AU0158 | - | 1 | 1 | - | 14 | 2 | 1 | 1 | 1 |
| Name | CRISPR | Cas | Spacer | |
|---|---|---|---|---|
| Endophytic strains | MS389 | - | - | - |
| MS455 | 3 | - | 2 | |
| EB159 | 1 | - | 1 | |
| SSG | 2 | - | 4 | |
| 869T2 | 3 | - | 3 | |
| YG-3 | 2 | - | 2 | |
| KJ006 | 3 | - | 2 | |
| RS1 | 2 | - | 1 | |
| LS-044 | 6 | - | 8 | |
| Plant pathogens | 43733 | 5 | 1 | 22 |
| 2196 | 2 | - | 3 | |
| BGR1 | 1 | - | 2 | |
| 10248 | 4 | - | 1 | |
| 25416 | 2 | - | 2 | |
| CF opportunistic pathogens | J2315 | 1 | - | 1 |
| BAA-67 | 2 | - | 1 | |
| BAA-247 | 2 | - | 3 | |
| AU0158 | 4 | - | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, J.; Lu, S.-E. Comparative Genome Analyses Provide Insight into the Antimicrobial Activity of Endophytic Burkholderia. Microorganisms 2024, 12, 100. https://doi.org/10.3390/microorganisms12010100
Jia J, Lu S-E. Comparative Genome Analyses Provide Insight into the Antimicrobial Activity of Endophytic Burkholderia. Microorganisms. 2024; 12(1):100. https://doi.org/10.3390/microorganisms12010100
Chicago/Turabian StyleJia, Jiayuan, and Shi-En Lu. 2024. "Comparative Genome Analyses Provide Insight into the Antimicrobial Activity of Endophytic Burkholderia" Microorganisms 12, no. 1: 100. https://doi.org/10.3390/microorganisms12010100
APA StyleJia, J., & Lu, S.-E. (2024). Comparative Genome Analyses Provide Insight into the Antimicrobial Activity of Endophytic Burkholderia. Microorganisms, 12(1), 100. https://doi.org/10.3390/microorganisms12010100




