The Role of Fusobacterium nucleatum in Oral and Colorectal Carcinogenesis
Abstract
:1. Introduction
2. Fusobacterium nucleatum
3. Interrelationships between P. gingivalis and F. nucleatum
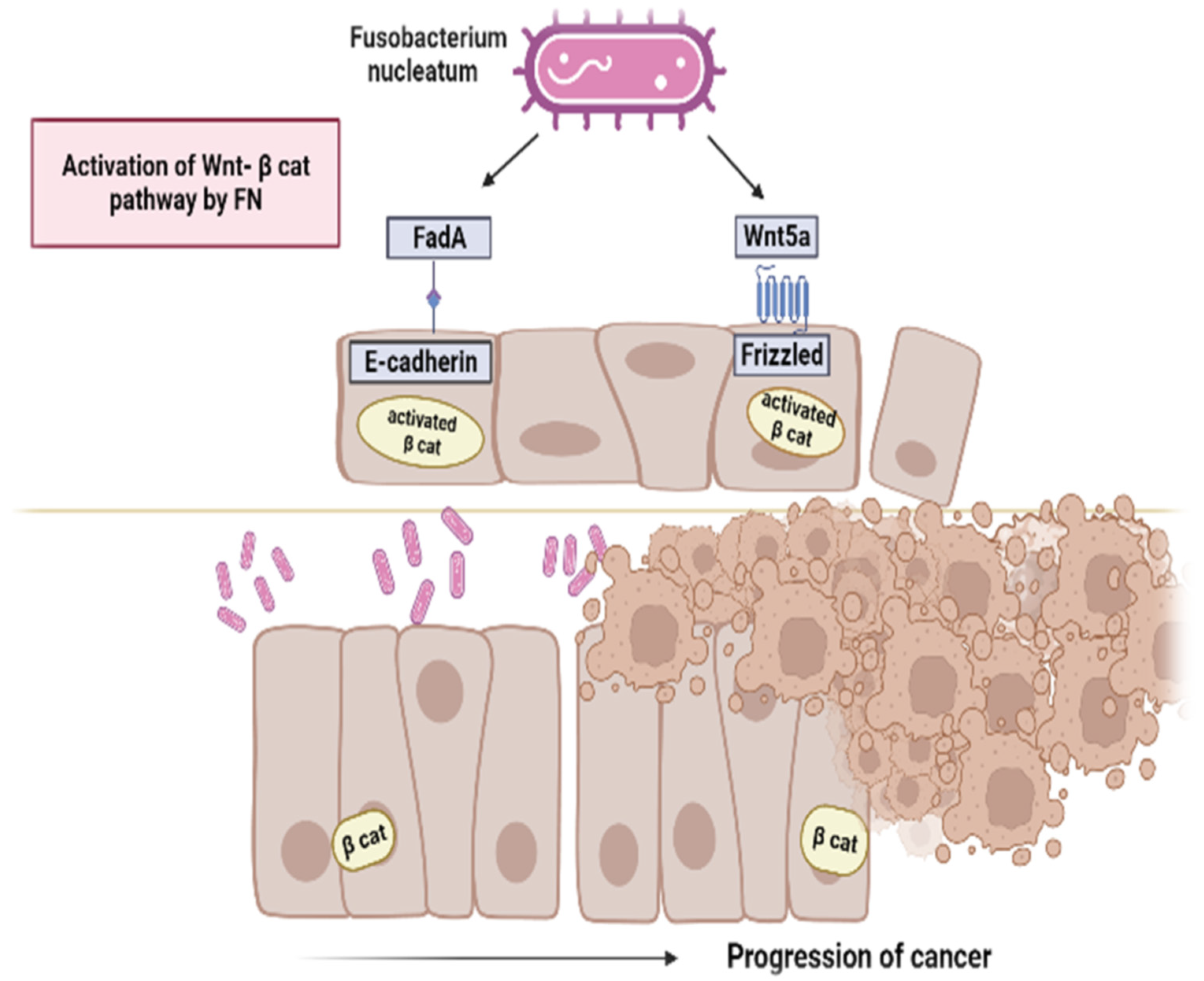
4. Mechanisms of Carcinogenesis of F. nucleatum
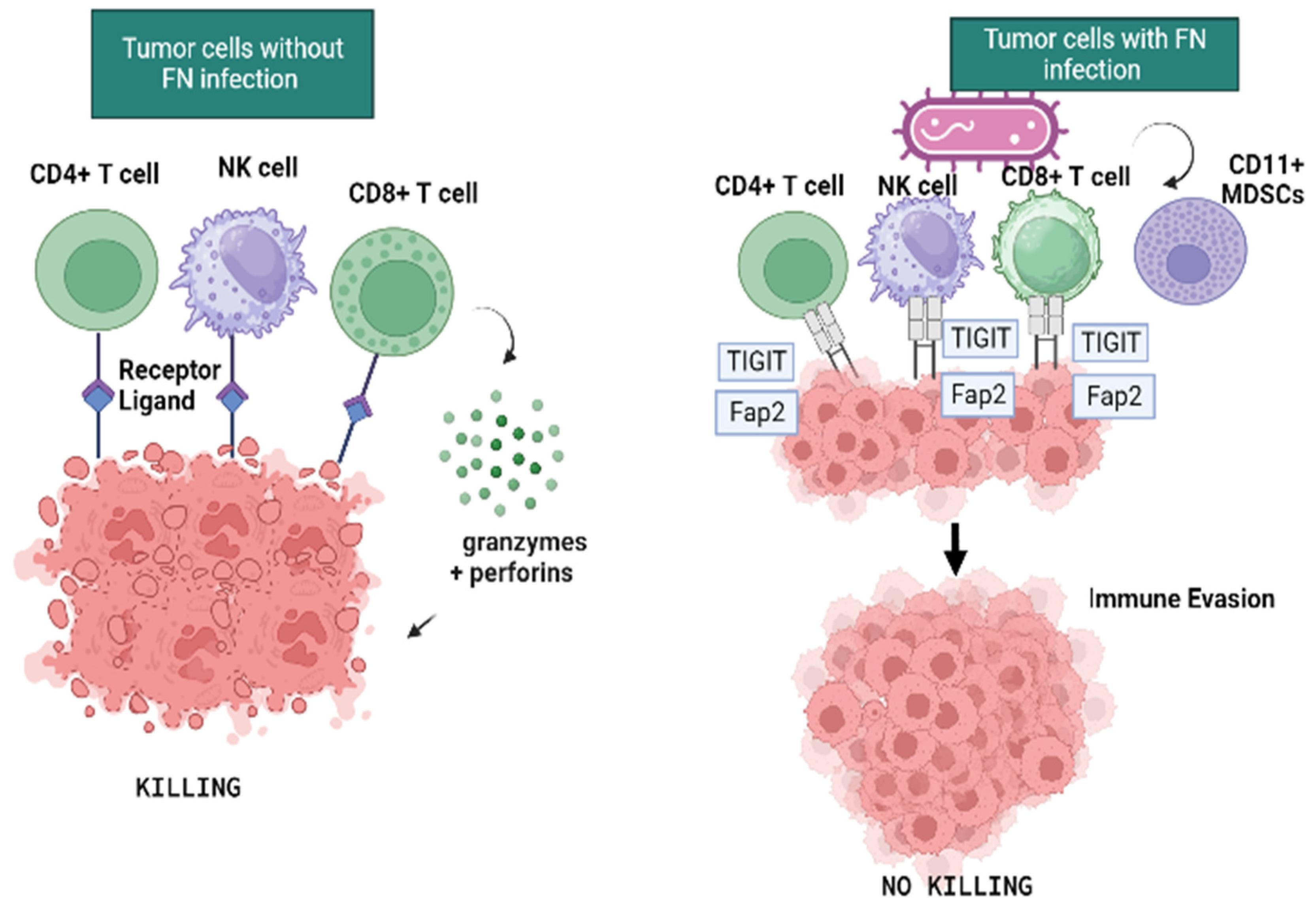
5. F. nucleatum and Immune Response
6. F. nucleatum in Oral and Colorectal Cancer
| Reference | Topic | Type of Study | Conclusion |
|---|---|---|---|
| Rubinstein, M.R., 2013 [59] | Molecular mechanisms | In vivo/Animal | FadA of Fn binds to E-cadherin on normal and CRC cells, promoting their attachment and invasion. FadA levels in adenomas and adenocarcinomas are >10–100-fold higher than in normal subjects. Its increased expression correlates with CRC cell growth and oncogenic and inflammatory responses. |
| Castellarin, M., 2012 [37] | Molecular mechanisms | In vitro | Fusobacterium was higher relative abundance in tumors with >50% circumferential involvement. High Fusobacterium tumors were significantly more likely to have regional lymph node metastases. |
| Kostic, A.D., 2013 [47] | Molecular mechanisms | In vivo/Animal | Fn were enriched in human colonic adenomas and in stool samples from CRC patients compared to healthy subjects. It generated a pro-inflammatory microenvironment via the recruitment of tumor-infiltrating myeloid cells and the expression of pro-inflammatory genes. |
| Abed, J., 2020 [52] | Molecular mechanisms | In vivo/Animal | Fn can colonize the CRC by migrating from the oral microbial reservoir through the hematogenous route. |
| Gur, C., 2015 [76] | Immunity | In vitro | The Fap2 protein of Fn inhibited immune cell activity, tumor-infiltrating lymphocytes expressed TIGIT and T-cell activities. |
| Zhao, H., 2017 [86] | OSCC carcinogenesis | In vivo/Human | A group of periodontitis-correlated taxa, including Fusobacterium, was found to be significantly enriched in OSCC samples. The operational taxonomic units (OTUs) associated with Fusobacterium were highly involved in OSCC and demonstrated good diagnostic power. |
| Yost, S.,2018 [87] | OSCC carcinogenesis | In vivo/Human | Fn was the most active bacterium expressing putative virulence factors in the tumor sites. At tumor sites proteolysis, DNA mismatch repair, carbohydrate metabolism, cell redox homeostasis and citrate transport were all over represented. |
| Yang, C.-Y., 2018 [88] | OSCC progression | In vivo/Human | The abundance of Fusobacteria increased significantly with the evolution of oral cancer from the healthy controls to OSCC stage 1 through stage 4. |
| Desai, S., 2022 [89] | OSCC prognosis | In vivo/Human | Fusobacterium presence is associated with an inflamed, innate immune cell-enriched and pro-tumorigenic microenvironment, as opposed to the HPV-positive HNSC tumors. Fn is also associated with poor prognosis, nodal metastases and high extracapsular spread in tongue tumors. |
| Neuzillet, C., 2021 [90] | OSCC prognosis | In vivo/Human | Fn-associated OSCC had a specific immune microenvironment, was more frequent in older, non-drinking patients, with a favorable prognosis. Patients with OSCC had significantly longer overall survival, relapse-free and metastasis-free survival. |
| McCoy, A.N., 2013 [61] | CRC carcinogenesis | In vivo/Human | There was a higher abundance of Fusobacterium species in the sigmoid than right side CRC location. Fn was more abundant in CRA than controls. CRA but not controls had a significant positive correlation between local cytokine gene expression and Fusobacterium quantity. |
| Rezasoltani, S., 2018 [91] | CRC carcinogenesis | In vivo/Human | Higher numbers of Fn were detected in adenomatous polyps in contrast to samples from the normal cases, hyperplastic polyps and sessile serrated polyps. |
| Komiya, Y., 2019 [92] | CRC carcinogenesis | In vivo/Human | Identical Fn strains were detected in both CRC and saliva from 42.9% of the patients. Fn was detected from stages 0 to IV and there were no significant differences in the detection rate among each lesion site. |
| Ito, M., 2015 [93] | CRC carcinogenesis | In vivo/Human | Fn was significantly higher in CRCs than in premalignant lesions of any histological type, in the latter increased gradually from sigmoid colon to cecum, frequently associated with CIMP-high lesions. Fn positivity increased according to histological grade. |
| Genua, F., 2023 [94] | Serum Antibody in CRA and CRC | In vivo/Human | IgG sero-positivity to Fn was associated with an increased CRC risk. Fn abundance in the normal mucosa positively correlated with the IgA response to the Fn antigen. |
| Lee, J.B., 2021 [95] | CC prognosis | In vivo/Human | Fn enriched right-sided metastatic, and recurrent colon cancer was significantly associated with worse progression-free survival, indicating that Fn enriched right-sided colon responded less to palliative cytotoxic chemotherapy. |
| Mima, K., 2016 [50] | CRC prognosis | In vivo/Human | The quantity of Fn DNA in colorectal cancer tissue was positively associated with pT stage and with colorectal cancer-specific mortality, independent of clinical, pathological, and major tumor molecular features. |
| Pignatelli, P., 2021 [81] | CC prognosis | In vivo/ Human | Fn oral concentration influenced colon tissue concentrations. Fn was statistically significantly higher in pathological tissue compared to the matched adjacent non-neoplastic mucosa. The Fn quantity in the colon cancer tissue predicted the staging. |
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mager, D. Bacteria and Cancer: Cause, Coincidence or Cure? A Review. J. Transl. Med. 2006, 4, 14. [Google Scholar] [CrossRef]
- Nagy, K.N.; Sonkodi, I.; Szöke, I.; Nagy, E.; Newman, H.N. The Microflora Associated with Human Oral Carcinomas. Oral Oncol. 1998, 34, 304–308. [Google Scholar] [CrossRef]
- Dejea, C.M.; Fathi, P.; Craig, J.M.; Boleij, A.; Taddese, R.; Geis, A.L.; Wu, X.; DeStefano Shields, C.E.; Hechenbleikner, E.M.; Huso, D.L.; et al. Patients with Familial Adenomatous Polyposis Harbor Colonic Biofilms Containing Tumorigenic Bacteria. Science 2018, 359, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Keku, T.O.; McCoy, A.N.; Azcarate-Peril, A.M. Fusobacterium spp. and Colorectal Cancer: Cause or Consequence? Trends Microbiol. 2013, 21, 506–508. [Google Scholar] [CrossRef] [PubMed]
- Sears, C.L.; Garrett, W.S. Microbes, Microbiota, and Colon Cancer. Cell Host Microbe 2014, 15, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.; Greten, F.R. The Inflammatory Pathogenesis of Colorectal Cancer. Nat. Rev. Immunol. 2021, 21, 653–667. [Google Scholar] [CrossRef]
- Chen, Y.; Peng, Y.; Yu, J.; Chen, T.; Wu, Y.; Shi, L.; Li, Q.; Wu, J.; Fu, X. Invasive Fusobacterium Nucleatum Activates Beta-Catenin Signaling in Colorectal Cancer via a TLR4/P-PAK1 Cascade. Oncotarget 2017, 8, 31802–31814. [Google Scholar] [CrossRef]
- Matly, A.; Quinn, J.A.; McMillan, D.C.; Park, J.H.; Edwards, J. The Relationship between β-Catenin and Patient Survival in Colorectal Cancer Systematic Review and Meta-Analysis. Crit. Rev. Oncol. Hematol. 2021, 163, 103337. [Google Scholar] [CrossRef]
- Wang, S.; Liu, Y.; Li, J.; Zhao, L.; Yan, W.; Lin, B.; Guo, X.; Wei, Y. Fusobacterium Nucleatum Acts as a Pro-Carcinogenic Bacterium in Colorectal Cancer: From Association to Causality. Front. Cell Dev. Biol. 2021, 9, 710165. [Google Scholar] [CrossRef]
- Morgan, E.; Arnold, M.; Gini, A.; Lorenzoni, V.; Cabasag, C.J.; Laversanne, M.; Vignat, J.; Ferlay, J.; Murphy, N.; Bray, F. Global Burden of Colorectal Cancer in 2020 and 2040: Incidence and Mortality Estimates from GLOBOCAN. Gut 2023, 72, 338–344. [Google Scholar] [CrossRef]
- Soerjomataram, I.; Bray, F. Planning for Tomorrow: Global Cancer Incidence and the Role of Prevention 2020–2070. Nat. Rev. Clin. Oncol. 2021, 18, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T.; et al. Analysis of Fusobacterium Persistence and Antibiotic Response in Colorectal Cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef]
- Kostic, A.D.; Xavier, R.J.; Gevers, D. The Microbiome in Inflammatory Bowel Disease: Current Status and the Future Ahead. Gastroenterology 2014, 146, 1489–1499. [Google Scholar] [CrossRef] [PubMed]
- Gholizadeh, P.; Eslami, H.; Kafil, H.S. Carcinogenesis Mechanisms of Fusobacterium Nucleatum. Biomed. Pharmacother. 2017, 89, 918–925. [Google Scholar] [CrossRef]
- Hagelskjaer Kristensen, L.; Prag, J. Lemierre’s Syndrome and Other Disseminated Fusobacterium Necrophorum Infections in Denmark: A Prospective Epidemiological and Clinical Survey. Eur. J. Clin. Microbiol. Infect. Dis. 2008, 27, 779–789. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Buhimschi, C.S.; Temoin, S.; Bhandari, V.; Han, Y.W.; Buhimschi, I.A. Comparative Microbial Analysis of Paired Amniotic Fluid and Cord Blood from Pregnancies Complicated by Preterm Birth and Early-Onset Neonatal Sepsis. PLoS ONE 2013, 8, e56131. [Google Scholar] [CrossRef]
- Han, X.Y.; Weinberg, J.S.; Prabhu, S.S.; Hassenbusch, S.J.; Fuller, G.N.; Tarrand, J.J.; Kontoyiannis, D.P. Fusobacterial Brain Abscess: A Review of Five Cases and an Analysis of Possible Pathogenesis. J. Neurosurg. 2003, 99, 693–700. [Google Scholar] [CrossRef]
- Rizzato, C.; Torres, J.; Kasamatsu, E.; Camorlinga-Ponce, M.; Bravo, M.M.; Canzian, F.; Kato, I. Potential Role of Biofilm Formation in the Development of Digestive Tract Cancer With Special Reference to Helicobacter Pylori Infection. Front. Microbiol. 2019, 10, 846. [Google Scholar] [CrossRef]
- Han, Y.W. Fusobacterium Nucleatum: A Commensal-Turned Pathogen. Curr. Opin. Microbiol. 2015, 23, 141–147. [Google Scholar] [CrossRef]
- Kaplan, C.W.; Ma, X.; Paranjpe, A.; Jewett, A.; Lux, R.; Kinder-Haake, S.; Shi, W. Fusobacterium Nucleatum Outer Membrane Proteins Fap2 and RadD Induce Cell Death in Human Lymphocytes. Infect. Immun. 2010, 78, 4773–4778. [Google Scholar] [CrossRef]
- Kaplan, C.W.; Lux, R.; Haake, S.K.; Shi, W. The Fusobacterium Nucleatum Outer Membrane Protein RadD Is an Arginine-Inhibitable Adhesin Required for Inter-Species Adherence and the Structured Architecture of Multispecies Biofilm. Mol. Microbiol. 2009, 71, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.A.; Garrett, W.S. Fusobacterium Nucleatum—Symbiont, Opportunist and Oncobacterium. Nat. Rev. Microbiol. 2019, 17, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Shokeen, B.; He, X.; Shi, W.; Lux, R. Streptococcus Mutans SpaP Binds to RadD of Fusobacterium Nucleatum Ssp. Polymorphum. Mol. Oral Microbiol. 2017, 32, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Kolenbrander, P.E.; Palmer, R.J.; Periasamy, S.; Jakubovics, N.S. Oral Multispecies Biofilm Development and the Key Role of Cell-Cell Distance. Nat. Rev. Microbiol. 2010, 8, 471–480. [Google Scholar] [CrossRef]
- Kaplan, A.; Kaplan, C.W.; He, X.; McHardy, I.; Shi, W.; Lux, R. Characterization of Aid1, a Novel Gene Involved in Fusobacterium Nucleatum Interspecies Interactions. Microb. Ecol. 2014, 68, 379–387. [Google Scholar] [CrossRef]
- Ferreira, M.C.; Dias-Pereira, A.C.; Branco-de-Almeida, L.S.; Martins, C.C.; Paiva, S.M. Impact of Periodontal Disease on Quality of Life: A Systematic Review. J. Periodontal Res. 2017, 52, 651–665. [Google Scholar] [CrossRef]
- Laine, M.L.; Moustakis, V.; Koumakis, L.; Potamias, G.; Loos, B.G. Modeling Susceptibility to Periodontitis. J. Dent. Res. 2013, 92, 45–50. [Google Scholar] [CrossRef]
- Papapanou, P.N.; Sanz, M.; Buduneli, N.; Dietrich, T.; Feres, M.; Fine, D.H.; Flemmig, T.F.; Garcia, R.; Giannobile, W.V.; Graziani, F.; et al. Periodontitis: Consensus Report of Workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Periodontol. 2018, 89 (Suppl. S1), S173–S182. [Google Scholar] [CrossRef]
- Berglundh, T.; Liljenberg, B.; Tarkowski, A.; Lindhe, J. The Presence of Local and Circulating Autoreactive B Cells in Patients with Advanced Periodontitis. J. Clin. Periodontol. 2002, 29, 281–286. [Google Scholar] [CrossRef]
- Polak, D.; Shapira, L.; Weiss, E.I.; Houri-Haddad, Y. The Role of Coaggregation between Porphyromonas Gingivalis and Fusobacterium Nucleatum on the Host Response to Mixed Infection. J. Clin. Periodontol. 2012, 39, 617–625. [Google Scholar] [CrossRef]
- Zijnge, V.; van Leeuwen, M.B.M.; Degener, J.E.; Abbas, F.; Thurnheer, T.; Gmür, R.; Harmsen, H.J.M. Oral Biofilm Architecture on Natural Teeth. PLoS ONE 2010, 5, e9321. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.-H.; Chun, S.; Park, C.; Lee, J.-H.; Lee, S.-W.; Lee, T.-H. Transcriptome Profiling Analysis of Senescent Gingival Fibroblasts in Response to Fusobacterium Nucleatum Infection. PLoS ONE 2017, 12, e0188755. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-R.; Kim, D.-J.; Han, S.-H.; Kang, M.-J.; Lee, J.-Y.; Jeong, Y.-J.; Lee, S.-J.; Kim, T.-H.; Ahn, S.-G.; Yoon, J.-H.; et al. Diverse Toll-like Receptors Mediate Cytokine Production by Fusobacterium Nucleatum and Aggregatibacter Actinomycetemcomitans in Macrophages. Infect. Immun. 2014, 82, 1914–1920. [Google Scholar] [CrossRef]
- Engevik, M.A.; Danhof, H.A.; Ruan, W.; Engevik, A.C.; Chang-Graham, A.L.; Engevik, K.A.; Shi, Z.; Zhao, Y.; Brand, C.K.; Krystofiak, E.S.; et al. Fusobacterium Nucleatum Secretes Outer Membrane Vesicles and Promotes Intestinal Inflammation. mBio 2021, 12, e02706-20. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, M.R.; Baik, J.E.; Lagana, S.M.; Han, R.P.; Raab, W.J.; Sahoo, D.; Dalerba, P.; Wang, T.C.; Han, Y.W. Fusobacterium Nucleatum Promotes Colorectal Cancer by Inducing Wnt/β-Catenin Modulator Annexin A1. EMBO Rep. 2019, 20, e47638. [Google Scholar] [CrossRef]
- Wu, S.; Rhee, K.-J.; Albesiano, E.; Rabizadeh, S.; Wu, X.; Yen, H.-R.; Huso, D.L.; Brancati, F.L.; Wick, E.; McAllister, F.; et al. A Human Colonic Commensal Promotes Colon Tumorigenesis via Activation of T Helper Type 17 T Cell Responses. Nat. Med. 2009, 15, 1016–1022. [Google Scholar] [CrossRef]
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A.; et al. Fusobacterium Nucleatum Infection Is Prevalent in Human Colorectal Carcinoma. Genome Res. 2012, 22, 299–306. [Google Scholar] [CrossRef]
- Fan, Z.; Tang, P.; Li, C.; Yang, Q.; Xu, Y.; Su, C.; Li, L. Fusobacterium Nucleatum and Its Associated Systemic Diseases: Epidemiologic Studies and Possible Mechanisms. J. Oral Microbiol. 2023, 15, 2145729. [Google Scholar] [CrossRef]
- Saito, Y.; Fujii, R.; Nakagawa, K.-I.; Kuramitsu, H.K.; Okuda, K.; Ishihara, K. Stimulation of Fusobacterium Nucleatum Biofilm Formation by Porphyromonas Gingivalis. Oral Microbiol. Immunol. 2008, 23, 1–6. [Google Scholar] [CrossRef]
- Diaz, P.I.; Zilm, P.S.; Rogers, A.H. Fusobacterium Nucleatum Supports the Growth of Porphyromonas Gingivalis in Oxygenated and Carbon-Dioxide-Depleted Environments. Microbiology 2002, 148, 467–472. [Google Scholar] [CrossRef]
- Weiss, E.I.; Shaniztki, B.; Dotan, M.; Ganeshkumar, N.; Kolenbrander, P.E.; Metzger, Z. Attachment of Fusobacterium Nucleatum PK1594 to Mammalian Cells and Its Coaggregation with Periodontopathogenic Bacteria Are Mediated by the Same Galactose-Binding Adhesin. Oral Microbiol. Immunol. 2000, 15, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Darenfed, H.; Grenier, D.; Mayrand, D. Acquisition of Plasmin Activity by Fusobacterium Nucleatum Subsp. Nucleatum and Potential Contribution to Tissue Destruction during Periodontitis. Infect. Immun. 1999, 67, 6439–6444. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Darveau, R.P.; Samaranayake, L.P.; Wang, C.-Y.; Jin, L. Differential Modulation of Human {beta}-Defensins Expression in Human Gingival Epithelia by Porphyromonas Gingivalis Lipopolysaccharide with Tetra- and Penta-Acylated Lipid A Structures. Innate Immun. 2009, 15, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, H.; Wang, X.; Lu, Y.; Yang, C.; Yang, P. Coinfection with Fusobacterium Nucleatum Can Enhance the Attachment and Invasion of Porphyromonas Gingivalis or Aggregatibacter Actinomycetemcomitans to Human Gingival Epithelial Cells. Arch. Oral Biol. 2015, 60, 1387–1393. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D. Periodontal Microbial Ecology. Periodontol. 2000 2005, 38, 135–187. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D. Dental Biofilms: Difficult Therapeutic Targets. Periodontol. 2000 2002, 28, 12–55. [Google Scholar] [CrossRef]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium Nucleatum Potentiates Intestinal Tumorigenesis and Modulates the Tumor-Immune Microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef]
- Kostic, A.D.; Gevers, D.; Pedamallu, C.S.; Michaud, M.; Duke, F.; Earl, A.M.; Ojesina, A.I.; Jung, J.; Bass, A.J.; Tabernero, J.; et al. Genomic Analysis Identifies Association of Fusobacterium with Colorectal Carcinoma. Genome Res. 2012, 22, 292–298. [Google Scholar] [CrossRef]
- Tahara, T.; Yamamoto, E.; Suzuki, H.; Maruyama, R.; Chung, W.; Garriga, J.; Jelinek, J.; Yamano, H.; Sugai, T.; An, B.; et al. Fusobacterium in Colonic Flora and Molecular Features of Colorectal Carcinoma. Cancer Res. 2014, 74, 1311–1318. [Google Scholar] [CrossRef]
- Mima, K.; Sukawa, Y.; Nishihara, R.; Qian, Z.R.; Yamauchi, M.; Inamura, K.; Kim, S.A.; Masuda, A.; Nowak, J.A.; Nosho, K.; et al. Fusobacterium Nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015, 1, 653–661. [Google Scholar] [CrossRef]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium Nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017, 170, 548–563.e16. [Google Scholar] [CrossRef]
- Abed, J.; Maalouf, N.; Manson, A.L.; Earl, A.M.; Parhi, L.; Emgård, J.E.M.; Klutstein, M.; Tayeb, S.; Almogy, G.; Atlan, K.A.; et al. Colon Cancer-Associated Fusobacterium Nucleatum May Originate From the Oral Cavity and Reach Colon Tumors via the Circulatory System. Front. Cell Infect. Microbiol. 2020, 10, 400. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Liu, F.; Ling, Z.; Tong, X.; Xiang, C. Human Intestinal Lumen and Mucosa-Associated Microbiota in Patients with Colorectal Cancer. PLoS ONE 2012, 7, e39743. [Google Scholar] [CrossRef]
- Shang, F.-M.; Liu, H.-L. Fusobacterium Nucleatum and Colorectal Cancer: A Review. World J. Gastrointest. Oncol. 2018, 10, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Fardini, Y.; Wang, X.; Témoin, S.; Nithianantham, S.; Lee, D.; Shoham, M.; Han, Y.W. Fusobacterium Nucleatum Adhesin FadA Binds Vascular Endothelial Cadherin and Alters Endothelial Integrity. Mol. Microbiol. 2011, 82, 1468–1480. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.W.; Ikegami, A.; Rajanna, C.; Kawsar, H.I.; Zhou, Y.; Li, M.; Sojar, H.T.; Genco, R.J.; Kuramitsu, H.K.; Deng, C.X. Identification and Characterization of a Novel Adhesin Unique to Oral Fusobacteria. J. Bacteriol. 2005, 187, 5330–5340. [Google Scholar] [CrossRef]
- Xu, M.; Yamada, M.; Li, M.; Liu, H.; Chen, S.G.; Han, Y.W. FadA from Fusobacterium Nucleatum Utilizes Both Secreted and Nonsecreted Forms for Functional Oligomerization for Attachment and Invasion of Host Cells. J. Biol. Chem. 2007, 282, 25000–25009. [Google Scholar] [CrossRef]
- Gumbiner, B.M. Regulation of Cadherin-Mediated Adhesion in Morphogenesis. Nat. Rev. Mol. Cell Biol. 2005, 6, 622–634. [Google Scholar] [CrossRef]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium Nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/β-Catenin Signaling via Its FadA Adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef]
- Bryant, D.M.; Stow, J.L. The Ins and Outs of E-Cadherin Trafficking. Trends Cell Biol. 2004, 14, 427–434. [Google Scholar] [CrossRef]
- McCoy, A.N.; Araújo-Pérez, F.; Azcárate-Peril, A.; Yeh, J.J.; Sandler, R.S.; Keku, T.O. Fusobacterium Is Associated with Colorectal Adenomas. PLoS ONE 2013, 8, e53653. [Google Scholar] [CrossRef] [PubMed]
- Quah, S.Y.; Bergenholtz, G.; Tan, K.S. Fusobacterium Nucleatum Induces Cytokine Production through Toll-like-Receptor-Independent Mechanism. Int. Endod. J. 2014, 47, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Dharmani, P.; Strauss, J.; Ambrose, C.; Allen-Vercoe, E.; Chadee, K. Fusobacterium Nucleatum Infection of Colonic Cells Stimulates MUC2 Mucin and Tumor Necrosis Factor Alpha. Infect. Immun. 2011, 79, 2597–2607. [Google Scholar] [CrossRef]
- von Kleist, L.; Stahlschmidt, W.; Bulut, H.; Gromova, K.; Puchkov, D.; Robertson, M.J.; MacGregor, K.A.; Tomilin, N.; Pechstein, A.; Chau, N.; et al. Role of the Clathrin Terminal Domain in Regulating Coated Pit Dynamics Revealed by Small Molecule Inhibition. Cell 2011, 146, 471–484. [Google Scholar] [CrossRef]
- Gerke, V.; Moss, S.E. Annexins: From Structure to Function. Physiol. Rev. 2002, 82, 331–371. [Google Scholar] [CrossRef] [PubMed]
- Su, N.; Xu, X.-Y.; Chen, H.; Gao, W.-C.; Ruan, C.-P.; Wang, Q.; Sun, Y.-P. Increased Expression of Annexin A1 Is Correlated with K-Ras Mutation in Colorectal Cancer. Tohoku J. Exp. Med. 2010, 222, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Duncan, R.; Carpenter, B.; Main, L.C.; Telfer, C.; Murray, G.I. Characterisation and Protein Expression Profiling of Annexins in Colorectal Cancer. Br. J. Cancer 2008, 98, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Fearon, E.R.; Vogelstein, B. A Genetic Model for Colorectal Tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Li, R.; Xiao, L.; Gong, T.; Liu, J.; Li, Y.; Zhou, X.; Li, Y.; Zheng, X. Role of Oral Microbiome in Oral Oncogenesis, Tumor Progression, and Metastasis. Mol. Oral Microbiol. 2023, 38, 9–22. [Google Scholar] [CrossRef]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The Human Tumor Microbiome Is Composed of Tumor Type-Specific Intracellular Bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef]
- D’Antonio, D.L.; Marchetti, S.; Pignatelli, P.; Piattelli, A.; Curia, M.C. The Oncobiome in Gastroenteric and Genitourinary Cancers. Int. J. Mol. Sci. 2022, 23, 9664. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, N.; Kurzrock, R. Targeting the Wnt/Beta-Catenin Pathway in Cancer: Update on Effectors and Inhibitors. Cancer Treat. Rev. 2018, 62, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, L.; Schmid, J.; Ebert, M.; Soucek, P.; Kunicka, T.; Liska, V.; Bruha, J.; Neary, P.; Dezeeuw, N.; Tommasino, M.; et al. Fusobacterium Nucleatum Associates with Stages of Colorectal Neoplasia Development, Colorectal Cancer and Disease Outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Mima, K.; Nishihara, R.; Qian, Z.R.; Cao, Y.; Sukawa, Y.; Nowak, J.A.; Yang, J.; Dou, R.; Masugi, Y.; Song, M.; et al. Fusobacterium Nucleatum in Colorectal Carcinoma Tissue and Patient Prognosis. Gut 2016, 65, 1973–1980. [Google Scholar] [CrossRef]
- Onozawa, H.; Saito, M.; Saito, K.; Kanke, Y.; Watanabe, Y.; Hayase, S.; Sakamoto, W.; Ishigame, T.; Momma, T.; Ohki, S.; et al. Annexin A1 Is Involved in Resistance to 5-FU in Colon Cancer Cells. Oncol. Rep. 2017, 37, 235–240. [Google Scholar] [CrossRef]
- Gur, C.; Ibrahim, Y.; Isaacson, B.; Yamin, R.; Abed, J.; Gamliel, M.; Enk, J.; Bar-On, Y.; Stanietsky-Kaynan, N.; Coppenhagen-Glazer, S.; et al. Binding of the Fap2 Protein of Fusobacterium Nucleatum to Human Inhibitory Receptor TIGIT Protects Tumors from Immune Cell Attack. Immunity 2015, 42, 344–355. [Google Scholar] [CrossRef]
- Zitvogel, L.; Ayyoub, M.; Routy, B.; Kroemer, G. Microbiome and Anticancer Immunosurveillance. Cell 2016, 165, 276–287. [Google Scholar] [CrossRef]
- Chauvin, J.-M.; Pagliano, O.; Fourcade, J.; Sun, Z.; Wang, H.; Sander, C.; Kirkwood, J.M.; Chen, T.T.; Maurer, M.; Korman, A.J.; et al. TIGIT and PD-1 Impair Tumor Antigen-Specific CD8+ T Cells in Melanoma Patients. J. Clin. Investig. 2015, 125, 2046–2058. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Bronte, V. Coordinated Regulation of Myeloid Cells by Tumours. Nat. Rev. Immunol. 2012, 12, 253–268. [Google Scholar] [CrossRef]
- Hieken, T.J.; Chen, J.; Hoskin, T.L.; Walther-Antonio, M.; Johnson, S.; Ramaker, S.; Xiao, J.; Radisky, D.C.; Knutson, K.L.; Kalari, K.R.; et al. The Microbiome of Aseptically Collected Human Breast Tissue in Benign and Malignant Disease. Sci. Rep. 2016, 6, 30751. [Google Scholar] [CrossRef]
- Pignatelli, P.; Iezzi, L.; Pennese, M.; Raimondi, P.; Cichella, A.; Bondi, D.; Grande, R.; Cotellese, R.; Di Bartolomeo, N.; Innocenti, P.; et al. The Potential of Colonic Tumor Tissue Fusobacterium Nucleatum to Predict Staging and Its Interplay with Oral Abundance in Colon Cancer Patients. Cancers 2021, 13, 1032. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, Y.; Zheng, H.J.; Zhang, C.P. The Oral Microbiota May Have Influence on Oral Cancer. Front. Cell Infect. Microbiol. 2019, 9, 476. [Google Scholar] [CrossRef] [PubMed]
- Hooper, S.J.; Crean, S.J.; Lewis, M.A.O.; Spratt, D.A.; Wade, W.G.; Wilson, M.J. Viable Bacteria Present within Oral Squamous Cell Carcinoma Tissue. J. Clin. Microbiol. 2006, 44, 1719–1725. [Google Scholar] [CrossRef] [PubMed]
- Al-Hebshi, N.N.; Nasher, A.T.; Idris, A.M.; Chen, T. Robust Species Taxonomy Assignment Algorithm for 16S RRNA NGS Reads: Application to Oral Carcinoma Samples. J. Oral. Microbiol. 2015, 7, 28934. [Google Scholar] [CrossRef] [PubMed]
- Pushalkar, S.; Ji, X.; Li, Y.; Estilo, C.; Yegnanarayana, R.; Singh, B.; Li, X.; Saxena, D. Comparison of Oral Microbiota in Tumor and Non-Tumor Tissues of Patients with Oral Squamous Cell Carcinoma. BMC Microbiol. 2012, 12, 144. [Google Scholar] [CrossRef]
- Zhao, H.; Chu, M.; Huang, Z.; Yang, X.; Ran, S.; Hu, B.; Zhang, C.; Liang, J. Variations in Oral Microbiota Associated with Oral Cancer. Sci. Rep. 2017, 7, 11773. [Google Scholar] [CrossRef]
- Yost, S.; Stashenko, P.; Choi, Y.; Kukuruzinska, M.; Genco, C.A.; Salama, A.; Weinberg, E.O.; Kramer, C.D.; Frias-Lopez, J. Increased Virulence of the Oral Microbiome in Oral Squamous Cell Carcinoma Revealed by Metatranscriptome Analyses. Int. J. Oral Sci. 2018, 10, 32. [Google Scholar] [CrossRef]
- Yang, C.-Y.; Yeh, Y.-M.; Yu, H.-Y.; Chin, C.-Y.; Hsu, C.-W.; Liu, H.; Huang, P.-J.; Hu, S.-N.; Liao, C.-T.; Chang, K.-P.; et al. Oral Microbiota Community Dynamics Associated With Oral Squamous Cell Carcinoma Staging. Front. Microbiol. 2018, 9, 862. [Google Scholar] [CrossRef]
- Desai, S.; Dharavath, B.; Manavalan, S.; Rane, A.; Redhu, A.K.; Sunder, R.; Butle, A.; Mishra, R.; Joshi, A.; Togar, T.; et al. Fusobacterium Nucleatum Is Associated with Inflammation and Poor Survival in Early-Stage HPV-Negative Tongue Cancer. NAR Cancer 2022, 4, zcac006. [Google Scholar] [CrossRef]
- Neuzillet, C.; Marchais, M.; Vacher, S.; Hilmi, M.; Schnitzler, A.; Meseure, D.; Leclere, R.; Lecerf, C.; Dubot, C.; Jeannot, E.; et al. Prognostic Value of Intratumoral Fusobacterium Nucleatum and Association with Immune-Related Gene Expression in Oral Squamous Cell Carcinoma Patients. Sci. Rep. 2021, 11, 7870. [Google Scholar] [CrossRef]
- Rezasoltani, S.; Asadzadeh Aghdaei, H.; Dabiri, H.; Akhavan Sepahi, A.; Modarressi, M.H.; Nazemalhosseini Mojarad, E. The Association between Fecal Microbiota and Different Types of Colorectal Polyp as Precursors of Colorectal Cancer. Microb. Pathog. 2018, 124, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Komiya, Y.; Shimomura, Y.; Higurashi, T.; Sugi, Y.; Arimoto, J.; Umezawa, S.; Uchiyama, S.; Matsumoto, M.; Nakajima, A. Patients with Colorectal Cancer Have Identical Strains of Fusobacterium Nucleatum in Their Colorectal Cancer and Oral Cavity. Gut 2019, 68, 1335–1337. [Google Scholar] [CrossRef]
- Ito, M.; Kanno, S.; Nosho, K.; Sukawa, Y.; Mitsuhashi, K.; Kurihara, H.; Igarashi, H.; Takahashi, T.; Tachibana, M.; Takahashi, H.; et al. Association of Fusobacterium Nucleatum with Clinical and Molecular Features in Colorectal Serrated Pathway. Int. J. Cancer 2015, 137, 1258–1268. [Google Scholar] [CrossRef] [PubMed]
- Genua, F.; Butt, J.; Waterboer, T.; Hughes, D.J. Association of Antibody Responses to Fusobacterium Nucleatum and Streptococcus Gallolyticus Proteins with Colorectal Adenoma and Colorectal Cancer. Dig. Dis. Sci. 2023, 68, 3300–3311. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.B.; Kim, K.-A.; Cho, H.Y.; Kim, D.; Kim, W.K.; Yong, D.; Lee, H.; Yoon, S.S.; Han, D.H.; Han, Y.D.; et al. Association between Fusobacterium Nucleatum and Patient Prognosis in Metastatic Colon Cancer. Sci. Rep. 2021, 11, 20263. [Google Scholar] [CrossRef]
- Schmidt, B.L.; Kuczynski, J.; Bhattacharya, A.; Huey, B.; Corby, P.M.; Queiroz, E.L.S.; Nightingale, K.; Kerr, A.R.; DeLacure, M.D.; Veeramachaneni, R.; et al. Changes in Abundance of Oral Microbiota Associated with Oral Cancer. PLoS ONE 2014, 9, e106297. [Google Scholar] [CrossRef]
- Amer, A.; Galvin, S.; Healy, C.M.; Moran, G.P. The Microbiome of Potentially Malignant Oral Leukoplakia Exhibits Enrichment for Fusobacterium, Leptotrichia, Campylobacter, and Rothia Species. Front. Microbiol. 2017, 8, 2391. [Google Scholar] [CrossRef]
- Binder Gallimidi, A.; Fischman, S.; Revach, B.; Bulvik, R.; Maliutina, A.; Rubinstein, A.M.; Nussbaum, G.; Elkin, M. Periodontal Pathogens Porphyromonas Gingivalis and Fusobacterium Nucleatum Promote Tumor Progression in an Oral-Specific Chemical Carcinogenesis Model. Oncotarget 2015, 6, 22613–22623. [Google Scholar] [CrossRef]
- Chen, Z.; Wong, P.Y.; Ng, C.W.K.; Lan, L.; Fung, S.; Li, J.W.; Cai, L.; Lei, P.; Mou, Q.; Wong, S.H.; et al. The Intersection between Oral Microbiota, Host Gene Methylation and Patient Outcomes in Head and Neck Squamous Cell Carcinoma. Cancers 2020, 12, 3425. [Google Scholar] [CrossRef]
- Park, D.-G.; Woo, B.H.; Lee, B.-J.; Yoon, S.; Cho, Y.; Kim, Y.-D.; Park, H.R.; Song, J.M. Serum Levels of Interleukin-6 and Titers of Antibodies Against Porphyromonas Gingivalis Could Be Potential Biomarkers for the Diagnosis of Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2019, 20, 2749. [Google Scholar] [CrossRef]
- Chattopadhyay, I.; Verma, M.; Panda, M. Role of Oral Microbiome Signatures in Diagnosis and Prognosis of Oral Cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033819867354. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Koido, S.; Odamaki, T.; Kajihara, M.; Kato, K.; Horiuchi, S.; Adachi, S.; Arakawa, H.; Yoshida, S.; Akasu, T.; et al. Metagenomic Analyses of the Gut Microbiota Associated with Colorectal Adenoma. PLoS ONE 2019, 14, e0212406. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Chen, Y.; Fu, X.; Zhou, X.; Peng, Y.; Shi, L.; Chen, T.; Wu, Y. Invasive Fusobacterium Nucleatum May Play a Role in the Carcinogenesis of Proximal Colon Cancer through the Serrated Neoplasia Pathway. Int. J. Cancer 2016, 139, 1318–1326. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-F.; Li, L.-F.; Guo, S.-H.; Zeng, Q.-Y.; Ning, F.; Liu, W.-L.; Zhang, G. Evaluation of Antibody Level against Fusobacterium Nucleatum in the Serological Diagnosis of Colorectal Cancer. Sci. Rep. 2016, 6, 33440. [Google Scholar] [CrossRef]
- Xiao, L.; Zhang, Q.; Peng, Y.; Wang, D.; Liu, Y. The Effect of Periodontal Bacteria Infection on Incidence and Prognosis of Cancer: A Systematic Review and Meta-Analysis. Medicine 2020, 99, e19698. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pignatelli, P.; Nuccio, F.; Piattelli, A.; Curia, M.C. The Role of Fusobacterium nucleatum in Oral and Colorectal Carcinogenesis. Microorganisms 2023, 11, 2358. https://doi.org/10.3390/microorganisms11092358
Pignatelli P, Nuccio F, Piattelli A, Curia MC. The Role of Fusobacterium nucleatum in Oral and Colorectal Carcinogenesis. Microorganisms. 2023; 11(9):2358. https://doi.org/10.3390/microorganisms11092358
Chicago/Turabian StylePignatelli, Pamela, Federica Nuccio, Adriano Piattelli, and Maria Cristina Curia. 2023. "The Role of Fusobacterium nucleatum in Oral and Colorectal Carcinogenesis" Microorganisms 11, no. 9: 2358. https://doi.org/10.3390/microorganisms11092358
APA StylePignatelli, P., Nuccio, F., Piattelli, A., & Curia, M. C. (2023). The Role of Fusobacterium nucleatum in Oral and Colorectal Carcinogenesis. Microorganisms, 11(9), 2358. https://doi.org/10.3390/microorganisms11092358








