Increased Expression of lncRNA AC000120.7 and SENP3-EIF4A1 in Patients with Acute Respiratory Distress Syndrome Induced by SARS-CoV-2 Infection: A Pilot Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Subjects
2.3. SARS-CoV-2 Detection in the NSD and PSD Groups
2.4. Isolation of Leukocytes in the NSD and PSD Groups
2.5. Total RNA Isolation and cDNA Synthesis in the NSD and PSD Groups
2.6. cDNA Synthesis and qRT-PCR Analysis in the NSD and PSD Groups
2.7. Total RNA Isolation in the ARDS and ARDS-CoV-2 Groups
2.8. RT-qPCR Analysis in the ARDS and ARDS-CoV-2 Groups
2.9. Statistical Analysis
2.10. Pathway Network Interaction Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ganesh, B.; Rajakumar, T.; Malathi, M.; Manikandan, N.; Nagaraj, J.; Santhakumar, A.; Elangovan, A.; Malik, Y.S. Epidemiology and Pathobiology of SARS-CoV-2 (COVID-19) in Comparison with SARS, MERS: An Updated Overview of Current Knowledge and Future Perspectives. Clin. Epidemiol. Glob. Health 2021, 10, 100694. [Google Scholar] [CrossRef]
- Wiersinga, W.J.; Rhodes, A.; Cheng, A.C.; Peacock, S.J.; Prescott, H.C. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA 2020, 324, 782–793. [Google Scholar] [CrossRef]
- Alkhathami, M.G.; Advani, S.; Al Shehri, M.K.; Abalkhail, A.; Alkhathami, F.; Al Salamah, J.A.; Albeashy, E. Prevalence and Mortality of Lung Comorbidities Among Patients with COVID-19: A Systematic Review and Meta-Analysis. Lung India 2021, 38, S31–S40. [Google Scholar] [CrossRef]
- Mohamadian, M.; Chiti, H.; Shoghli, A.; Biglari, S.; Parsamanesh, N.; Esmaeilzadeh, A. COVID-19: Virology, Biology and Novel Laboratory Diagnosis. J. Gene Med. 2021, 23, e3303. [Google Scholar] [CrossRef]
- Krynytska, I.; Marushchak, M.; Birchenko, I.; Dovgalyuk, A.; Tokarskyy, O. COVID-19-Associated Acute Respiratory Distress Syndrome versus Classical Acute Respiratory Distress Syndrome (a Narrative Review). Iran. J. Microbiol. 2021, 13, 737–747. [Google Scholar] [CrossRef]
- Li, X.; Xu, S.; Yu, M.; Wang, K.; Tao, Y.; Zhou, Y.; Shi, J.; Zhou, M.; Wu, B.; Yang, Z.; et al. Risk Factors for Severity and Mortality in Adult COVID-19 Inpatients in Wuhan. J. Allergy Clin. Immunol. 2020, 146, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Chillón, I.; Marcia, M. The Molecular Structure of Long Non-Coding RNAs: Emerging Patterns and Functional Implications. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 662–690. [Google Scholar] [CrossRef] [PubMed]
- Montoya, G.L.; Ramírez, J.G.; Basilio, J.S.; Higuera, I.S.; Espinoza, M.I.; González, R.G.; Higuera, N.S. Long Non-Coding RNAs: Regulators of the Activity of Myeloid-Derived Suppressor Cells. Front. Immunol. 2019, 10, 1734. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long Non-Coding RNAs: Insights into Functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Guzmán-Martín, C.A.; Juárez-Vicuña, Y.; Domínguez-López, A.; González-Ramírez, J.; Amezcua-Guerra, L.M.; Martínez-Martínez, L.A.; Sánchez-Muñoz, F. LncRNAs Dysregulation in Monocytes from Primary Antiphospholipid Syndrome Patients: A Bioinformatic and an Experimental Proof-of-Concept Approach. Mol. Biol. Rep. 2022, 50, 937–941. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Sun, Q.; Zhang, B.; Zhao, W.; Shen, C. The Regulation of LncRNAs and MiRNAs in SARS-CoV-2 Infection. Front. Cell Dev. Biol. 2023, 11, 1229393. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Ashley, C.L.; Steain, M.; Ataide, S.F. Assessing the Suitability of Long Non-Coding RNAs as Therapeutic Targets and Biomarkers in SARS-CoV-2 Infection. Front. Mol. Biosci. 2022, 9, 975322. [Google Scholar] [CrossRef] [PubMed]
- Moazzam-Jazi, M.; Lanjanian, H.; Maleknia, S.; Hedayati, M.; Daneshpour, M.S. Interplay between SARS-CoV-2 and Human Long Non-Coding RNAs. J. Cell. Mol. Med. 2021, 25, 5823–5827. [Google Scholar] [CrossRef]
- Huang, K.; Wang, C.; Vagts, C.; Raguveer, V.; Finn, P.W.; Perkins, D.L. Long Non-Coding RNAs (LncRNAs) NEAT1 and MALAT1 Are Differentially Expressed in Severe COVID-19 Patients: An Integrated Single-Cell Analysis. PLoS ONE 2022, 17, e0261242. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Zhou, X.; Feng, W.; Jia, M.; Zhang, X.; An, T.; Luan, M.; Pan, Y.; Zhang, S.; Zhou, Z.; et al. Risk Stratification by Long Non-Coding RNAs Profiling in COVID-19 Patients. J. Cell. Mol. Med. 2021, 25, 4753–4764. [Google Scholar] [CrossRef]
- Zampetaki, A.; Albrecht, A.; Steinhofel, K. Long Non-Coding RNA Structure and Function: Is There a Link? Front. Physiol. 2018, 9, 1201. [Google Scholar] [CrossRef]
- Ouyang, J.; Hu, J.; Chen, J.L. LncRNAs Regulate the Innate Immune Response to Viral Infection. Wiley Interdiscip. Rev. RNA 2016, 7, 129–143. [Google Scholar] [CrossRef]
- Aghamohamadi, N.; Shahba, F.; Zarezadeh Mehrabadi, A.; Khorramdelazad, H.; Karimi, M.; Falak, R.; Emameh, R.Z. Age-Dependent Immune Responses in COVID-19-Mediated Liver Injury: Focus on Cytokines. Front. Endocrinol. 2023, 14, 1139692. [Google Scholar] [CrossRef]
- Zaira, B.; Yulianti, T.; Levita, J. Correlation between Hepatocyte Growth Factor (HGF) with D-Dimer and Interleukin-6 as Prognostic Markers of Coagulation and Inflammation in Long COVID-19 Survivors. Curr. Issues Mol. Biol. 2023, 45, 5725–5740. [Google Scholar] [CrossRef]
- Springall, R.; González-Flores, J.; García-Ávila, C.; Juárez-Vicuña, Y.; Hernández-Diazcouder, A.; Márquez-Velasco, R.; Cásares-Alvarado, S.; Sánchez-Muñoz, F.; Basilio-Gálvez, E.; Castillo-Salazar, M.; et al. Elevated Levels of Soluble CD147 Are Associated with Hyperinflammation and Disease Severity in COVID-19: A Proof-of-Concept Clinical Study. Arch. Immunol. Ther. Exp. 2022, 70, 18. [Google Scholar] [CrossRef]
- Zhou, M.; Wang, X.; Shi, H.; Cheng, L.; Wang, Z.; Zhao, H.; Yang, L.; Sun, J. Characterization of Long Non-Coding RNA-Associated CeRNA Network to Reveal Potential Prognostic LncRNA Biomarkers in Human Ovarian Cancer. Oncotarget 2016, 7, 12598–12611. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Muñoz, F.; Martínez-Coronilla, G.; Leija-Montoya, A.G.; Rieke-Campoy, U.; Angelina Lopez-Carrasco, R.; de Lourdes Montaño-Pérez, M.; Beltrán-Partida, E.; Bojórquez-Anaya, Y.; Serafin-Higuera, N.; González-Ramírez, J. Periodontitis May Modulate Long-Non Coding RNA Expression. Arch. Oral Biol. 2018, 95, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.X.; Gong, L.; et al. CD147-Spike Protein Is a Novel Route for SARS-CoV-2 Infection to Host Cells. Signal Transduct. Target. Ther. 2020, 5, 283. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Miao, X.; Si, C.; Qin, A.; Zhang, Y.; Chu, C.; Li, Z.; Wang, T.; Liu, X. Long Non-Coding RNA SENP3-EIF4A1 Functions as a Sponge of MiR-195-5p to Drive Triple-Negative Breast Cancer Progress by Overexpressing CCNE1. Front. Cell Dev. Biol. 2021, 9, 647527. [Google Scholar] [CrossRef]
- Xu, Y.; Jiang, W.; Zhong, L.; Li, H.; Bai, L.; Chen, X.; Lin, Y.; Zheng, D. miR-195-5p alleviates acute kidney injury through repression of inflammation and oxidative stress by targeting vascular endothelial growth factor A. Aging 2020, 12, 10235–10245. [Google Scholar] [CrossRef]
- Lee, Y.C. The Involvement of VEGF in Endothelial Permeability: A Target for Anti-Inflammatory Therapy. Curr. Opin. Investig. Drugs 2005, 6, 1124–1130. [Google Scholar]
- Yin, X.X.; Zheng, X.R.; Peng, W.; Wu, M.L.; Mao, X.Y.; Mao, X.Y.; Mao, X.Y.; Mao, X.Y. Vascular Endothelial Growth Factor (VEGF) as a Vital Target for Brain Inflammation during the COVID-19 Outbreak. ACS Chem. Neurosci. 2020, 11, 1704–1705. [Google Scholar] [CrossRef]
- Bras, J.P.; Silva, A.M.; Calin, G.A.; Barbosa, M.A.; Santos, S.G.; Almeida, M.I. MiR-195 Inhibits Macrophages pro-Inflammatory Profile and Impacts the Crosstalk with Smooth Muscle Cells. PLoS ONE 2017, 12, e0188530. [Google Scholar] [CrossRef]
- Nishiga, M.; Wang, D.W.; Han, Y.; Lewis, D.B.; Wu, J.C. COVID-19 and Cardiovascular Disease: From Basic Mechanisms to Clinical Perspectives. Nat. Rev. Cardiol. 2020, 17, 543–558. [Google Scholar] [CrossRef]
- Li, S.; Jiang, L.; Yang, Y.; Cao, J.; Zhang, Q.; Zhang, J.; Wang, R.; Deng, X.; Li, Y. MiR-195-5p Inhibits the Development of Chronic Obstructive Pulmonary Disease via Targeting Siglec1. Hum. Exp. Toxicol. 2020, 39, 1333–1344. [Google Scholar] [CrossRef]
- Ding, J.; Chen, J.; Yin, X.; Zhou, J. Current Understanding on Long Non-Coding RNAs in Immune Response to COVID-19. Virus Res. 2023, 323, 198956. [Google Scholar] [CrossRef] [PubMed]
- Enguita, F.J.; Leitão, A.L.; McDonald, J.T.; Zaksas, V.; Das, S.; Galeano, D.; Taylor, D.; Wurtele, E.S.; Saravia-Butler, A.; Baylin, S.B.; et al. The Interplay between LncRNAs, RNA-Binding Proteins and Viral Genome during SARS-CoV-2 Infection Reveals Strong Connections with Regulatory Events Involved in RNA Metabolism and Immune Response. Theranostics 2022, 12, 3946–3962. [Google Scholar] [CrossRef] [PubMed]
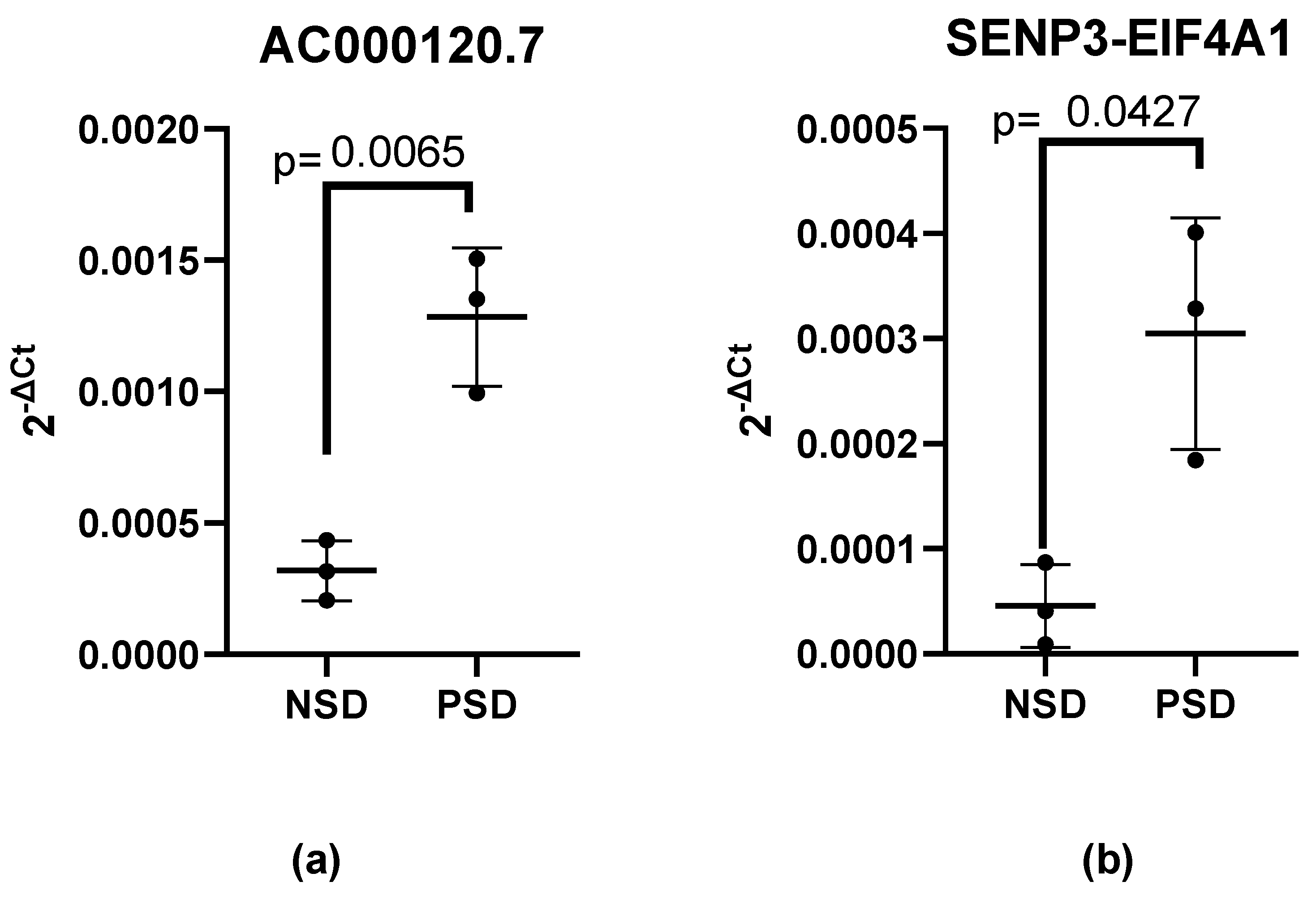
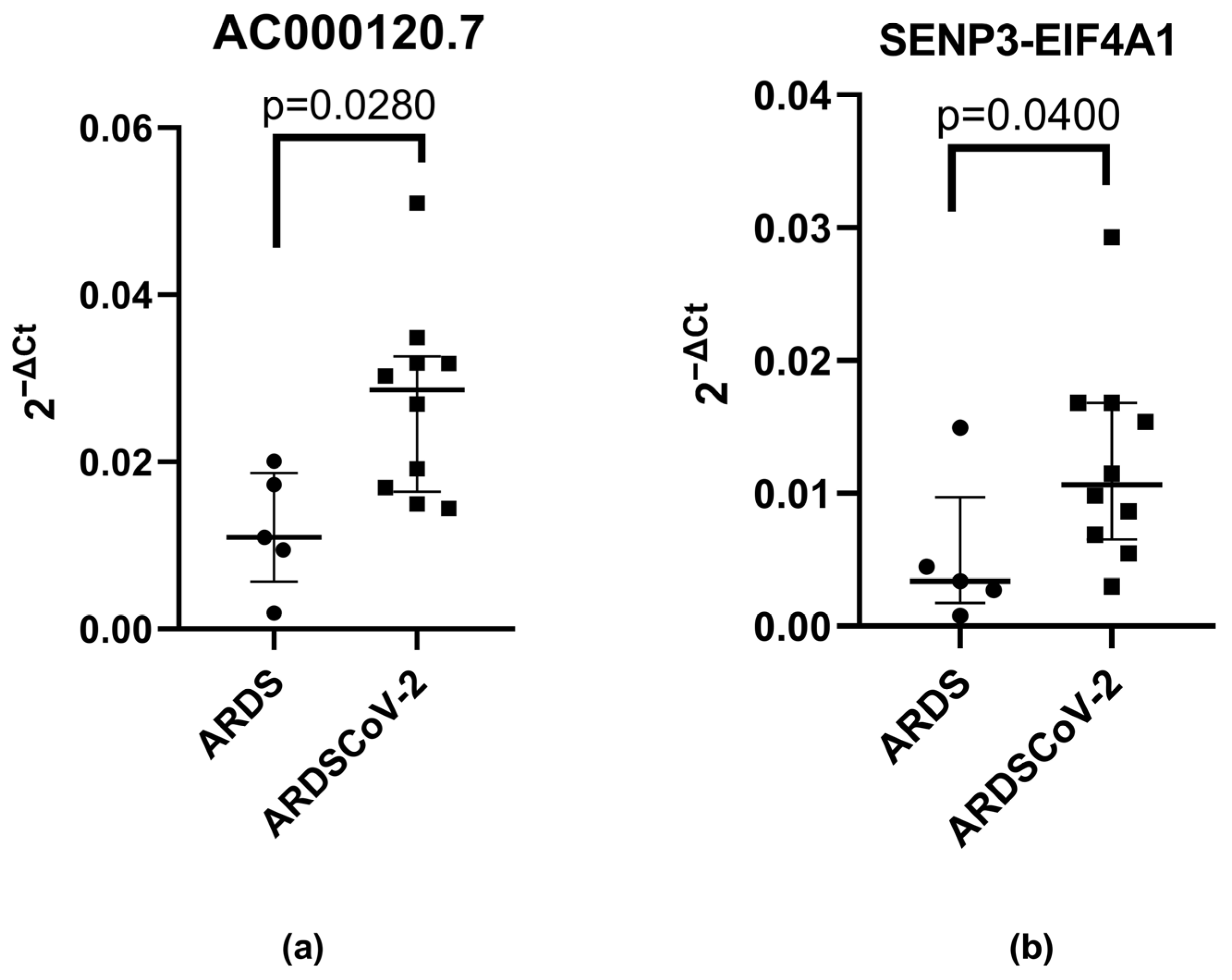

| RT-PCR Positive or Negative Cases | NSD | PSD | |||||
|---|---|---|---|---|---|---|---|
| General | Female | x | x | x | x | x | |
| Male | x | ||||||
| Age | 54 | 25 | 29 | 45 | 54 | 23 | |
| Symptoms | Sudden onset of symptoms | ||||||
| Fever | x | x | |||||
| Cough | x | x | x | x | x | ||
| Headache | x | x | x | x | x | ||
| Dyspnea | x | x | |||||
| Irritability | x | x | x | x | |||
| Diarrhea | x | x | |||||
| Chest pain | x | x | |||||
| Chills | x | x | |||||
| Odynophagia | x | x | |||||
| Myalgia | x | x | x | x | |||
| Arthralgia | x | x | x | x | |||
| Malaise | x | x | x | x | |||
| Rhinorrhea | x | x | x | ||||
| Abdominal pain | x | x | |||||
| Anosmia | x | ||||||
| Comorbidities | Hypertension | x | |||||
| Smoking | x | ||||||
| Treatment | Antipyretics | x | x | x | x | ||
| Epidemiological history | Contact with influenza or COVID-19 cases in the last 2 weeks | x | x | ||||
| Negative for COVID-19 (n = 5) | Positive for COVID-19 (n = 10) | |
|---|---|---|
| Age, years | 64 (53.5–67) | 64 (52.5–72.7) |
| Female, n (%) | 2 (40) | 4 (40) |
| Male, n (%) | 3 (60) | 6 (60) |
| Invasive mechanical ventilation | 3 (60) | 3 (30) |
| Death (%) | 2 (40) | 2 (20) |
| Diabetes (%) | 1 (20) | 3 (30) |
| Systemic hypertension | 3 (60) | 5 (50) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Ramírez, J.; Leija-Montoya, A.G.; Serafín-Higuera, N.; Guzmán-Martín, C.A.; Amezcua-Guerra, L.M.; Olvera-Sandoval, C.; Machado-Contreras, J.R.; Ruiz-Hernández, A.; Hernández-Díazcouder, A.; Estrada-Guzmán, J.D.; et al. Increased Expression of lncRNA AC000120.7 and SENP3-EIF4A1 in Patients with Acute Respiratory Distress Syndrome Induced by SARS-CoV-2 Infection: A Pilot Study. Microorganisms 2023, 11, 2342. https://doi.org/10.3390/microorganisms11092342
González-Ramírez J, Leija-Montoya AG, Serafín-Higuera N, Guzmán-Martín CA, Amezcua-Guerra LM, Olvera-Sandoval C, Machado-Contreras JR, Ruiz-Hernández A, Hernández-Díazcouder A, Estrada-Guzmán JD, et al. Increased Expression of lncRNA AC000120.7 and SENP3-EIF4A1 in Patients with Acute Respiratory Distress Syndrome Induced by SARS-CoV-2 Infection: A Pilot Study. Microorganisms. 2023; 11(9):2342. https://doi.org/10.3390/microorganisms11092342
Chicago/Turabian StyleGonzález-Ramírez, Javier, Ana Gabriela Leija-Montoya, Nicolás Serafín-Higuera, Carlos A. Guzmán-Martín, Luis M. Amezcua-Guerra, Carlos Olvera-Sandoval, Jesús René Machado-Contreras, Armando Ruiz-Hernández, Adrián Hernández-Díazcouder, Julia Dolores Estrada-Guzmán, and et al. 2023. "Increased Expression of lncRNA AC000120.7 and SENP3-EIF4A1 in Patients with Acute Respiratory Distress Syndrome Induced by SARS-CoV-2 Infection: A Pilot Study" Microorganisms 11, no. 9: 2342. https://doi.org/10.3390/microorganisms11092342
APA StyleGonzález-Ramírez, J., Leija-Montoya, A. G., Serafín-Higuera, N., Guzmán-Martín, C. A., Amezcua-Guerra, L. M., Olvera-Sandoval, C., Machado-Contreras, J. R., Ruiz-Hernández, A., Hernández-Díazcouder, A., Estrada-Guzmán, J. D., & Sánchez-Muñoz, F. (2023). Increased Expression of lncRNA AC000120.7 and SENP3-EIF4A1 in Patients with Acute Respiratory Distress Syndrome Induced by SARS-CoV-2 Infection: A Pilot Study. Microorganisms, 11(9), 2342. https://doi.org/10.3390/microorganisms11092342







