Efficacy of Life Protection Probably from Newly Isolated Bacteria against Cisplatin-Induced Lethal Toxicity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Materials
2.3. Real-Time PCR
2.4. Western Blotting
2.5. Statistical Analysis
3. Results
3.1. Survival Curve of High-Dose Cisplatin-Treated Mice with or without Newly Isolated Bacteria
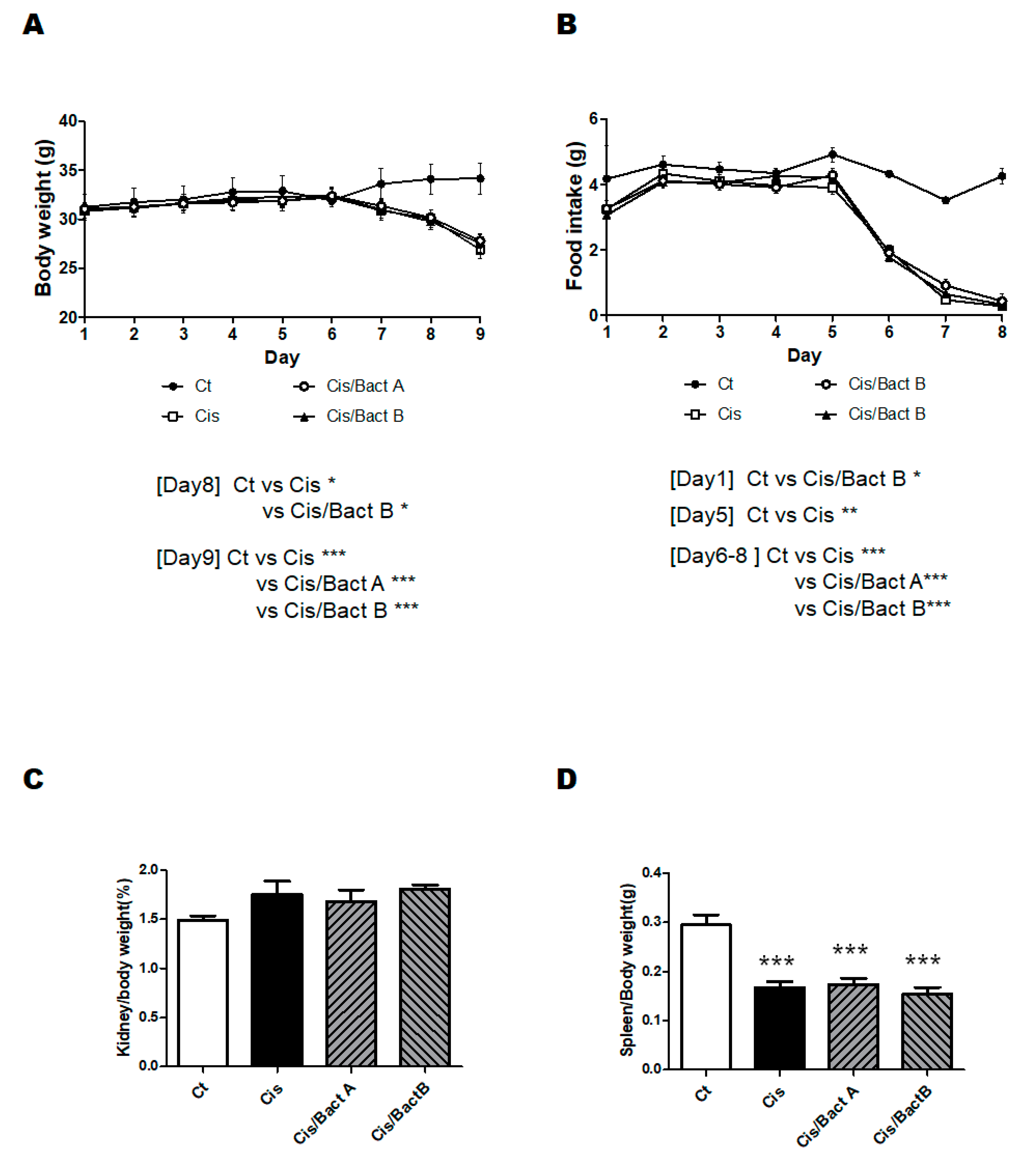
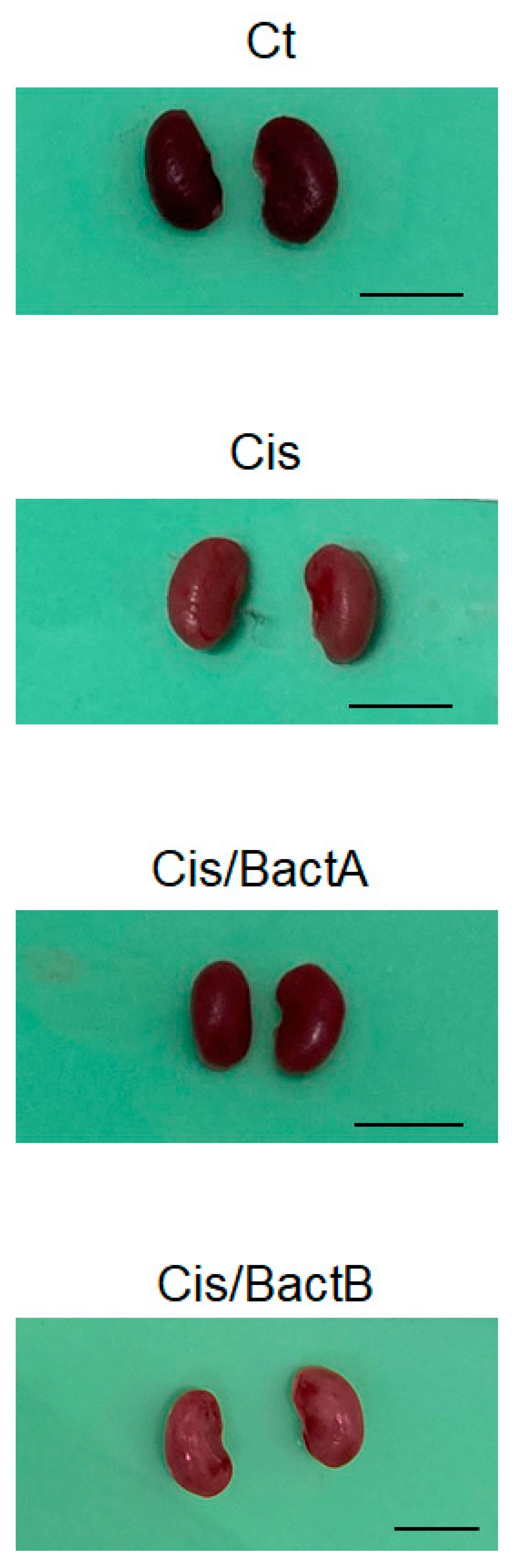
3.2. Characteristics of the High-Dose Cisplatin-Treated Mice
3.3. Analyses for Genes and Protein Expression in the High-Dose Cisplatin-Treated Mice
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AKT | Protein kinase B |
| AKI | acute kidney injury |
| CKD | chronic kidney disease |
| DSS | dextran sulfate sodium |
| Ho-1 | heme oxygenase 1 |
| KIM-1 | Kidney injury molecule-1 |
| Nrf2 | nuclear factor erythroid 2-related factor 2 |
| PI3K | phosphatidylinositol-3 kinase |
| QOL | quality of life |
| ROS | reactive oxygen species |
| Sod1 | superoxide dismutase 1 |
| Sod2 | superoxide dismutase 2 |
| Sod3 | superoxide dismutase 3 |
References
- Miller, R.P.; Tadagavadi, R.K.; Ramesh, G.; Reeves, W.B. Mechanisms of Cisplatin nephrotoxicity. Toxins 2010, 2, 2490–2518. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Guo, F.; Huang, Z.; Xia, Z.; Liu, J.; Tao, S.; Li, L.; Feng, Y.; Du, X.; Ma, L.; et al. Pharmacological and genetic inhibition of fatty acid-binding protein 4 alleviated cisplatin-induced acute kidney injury. J. Cell. Mol. Med. 2019, 23, 6260–6270. [Google Scholar] [CrossRef] [PubMed]
- Vanmassenhove, J.; Kielstein, J.; Jörres, A.; Biesen, W.V. Management of patients at risk of acute kidney injury. Lancet 2017, 389, 2139–2151. [Google Scholar] [CrossRef]
- Chawla, L.S.; Bellomo, R.; Bihorac, A.; Goldstein, S.L.; Siew, E.D.; Bagshaw, S.M.; Bittleman, D.; Cruz, D.; Endre, Z.; Fitzgerald, R.L.; et al. Acute kidney disease and renal recovery: Consensus report of the Acute Disease Quality Initiative (ADQI) 16 Workgroup. Nat. Rev. Nephrol. 2017, 13, 241–257. [Google Scholar] [CrossRef]
- Pabla, N.; Dong, Z. Cisplatin nephrotoxicity: Mechanisms and renoprotective strategies. Kidney Int. 2008, 73, 994–1007. [Google Scholar] [CrossRef]
- Santos, N.A.; Bezerra, C.S.; Martins, N.M.; Curti, C.; Bianchi, M.L.; Santos, A.C. Hydroxyl radical scavenger ameliorates cisplatin-induced nephrotoxicity by preventing oxidative stress, redox state unbalance, impairment of energetic metabolism and apoptosis in rat kidney mitochondria. Cancer Chemother. Pharmacol. 2008, 61, 145–155. [Google Scholar] [CrossRef]
- Zhu, H.; Jiang, W.; Zhao, H.; He, C.; Tang, X.; Xu, S.; Xu, C.; Feng, R.; Li, J.; Ma, T.; et al. PSTPIP2 inhibits cisplatin-induced acute kidney injury by suppressing apoptosis of renal tubular epithelial cells. Cell. Death Dis. 2020, 11, 1057. [Google Scholar] [CrossRef]
- Chen, D.; Jin, D.; Huang, S.; Wu, J.; Xu, M.; Liu, T.; Dong, W.; Liu, X.; Wang, S.; Zhong, W.; et al. Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett. 2020, 469, 456–467. [Google Scholar] [CrossRef]
- Ansari, M.A. Sinapic acid modulates Nrf2/HO-1 signaling pathway in cisplatin-induced nephrotoxicity in rats. Biomed. Pharmacother. 2017, 93, 646–653. [Google Scholar] [CrossRef]
- Hsiao, Y.P.; Chen, H.L.; Tsai, J.N.; Lin, M.Y.; Liao, J.W.; Wei, M.S.; Ko, J.L.; Ou, C.C. Administration of Lactobacillus reuteri Combined with Clostridium butyricum Attenuates Cisplatin-Induced Renal Damage by Gut Microbiota Reconstitution, Increasing Butyric Acid Production, and Suppressing Renal Inflammation. Nutrients 2021, 13, 2792. [Google Scholar] [CrossRef]
- Chen, X.; Xu, J.; Su, Y.; Zhu, W. Effects of Intravenous Infusion with Sodium Butyrate on Colonic Microbiota, Intestinal Development- and Mucosal Immune-Related Gene Expression in Normal Growing Pigs. Front. Microbiol. 2018, 9, 1652. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.; Xia, Y.; Li, B.; Yu, W.; Rao, T.; Ye, Z.; Yan, X.; Song, B.; Li, L.; Lin, F.; et al. Gut microbiota in patients with kidney stones: A systematic review and meta-analysis. BMC Microbiol. 2023, 23, 143. [Google Scholar] [CrossRef] [PubMed]
- Przybyciński, J.; Drożdżal, S.; Wilk, A.; Dziedziejko, V.; Szumilas, K.; Pawlik, A. The Effect of the Gut Microbiota on Transplanted Kidney Function. Int. J. Mol. Sci. 2023, 24, 1260. [Google Scholar] [CrossRef]
- Christova, T.Y.; Gorneva, G.A.; Taxirov, S.I.; Duridanova, D.B.; Setchenska, M.S. Effect of cisplatin and cobalt chloride on antioxidant enzymes in the livers of Lewis lung carcinoma-bearing mice: Protective role of heme oxygenase. Toxicol. Lett. 2003, 138, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Hecquet, B.; Vennin, P.; Fournier, C.; Poissonnier, B. Evaluation of the pharmacological benefit and determination of the influencing factors of intraarterial cis-diamminedichloroplatinum administration in patients with uterine cervical cancer. Cancer Res. 1987, 47, 6134–6137. [Google Scholar]
- van Timmeren, M.M.; van den Heuvel, M.C.; Bailly, V.; Bakker, S.J.; van Goor, H.; Stegeman, C.A. Tubular kidney injury molecule-1 (KIM-1) in human renal disease. J. Pathol. 2007, 212, 209–217. [Google Scholar] [CrossRef]
- Chen, L.S.; Zheng, D.S. Safflor Yellow A Protects Beas-2B Cells Against LPS-Induced Injury via Activating Nrf2. Rev. Bras. Farmacogn. 2023, 1–10. [Google Scholar] [CrossRef] [PubMed]
- van Moorsel, C.J.; Pinedo, H.M.; Veerman, G.; Vermorken, J.B.; Postmus, P.E.; Peters, G.J. Scheduling of gemcitabine and cisplatin in Lewis lung tumour bearing mice. Eur. J. Cancer 1999, 35, 808–814. [Google Scholar] [CrossRef]
- Gales, A.; Monteiro-Pai, S.; Hyndman, K.A. Endothelin system expression in the kidney following cisplatin-induced acute kidney injury in male and female mice. Can. J. Physiol. Pharmacol. 2022, 100, 868–879. [Google Scholar] [CrossRef]
- Wardill, H.R.; Gibson, R.J.; Van Sebille, Y.Z.; Secombe, K.R.; Coller, J.K.; White, I.A.; Manavis, J.; Hutchinson, M.R.; Staikopoulos, V.; Logan, R.M.; et al. Irinotecan-Induced Gastrointestinal Dysfunction and Pain Are Mediated by Common TLR4-Dependent Mechanisms. Mol. Cancer Ther. 2016, 15, 1376–1386. [Google Scholar] [CrossRef]
- Wang, T.; Zheng, J.; Dong, S.; Ismael, M.; Shan, Y.; Wang, X.; Lü, X. Lacticaseibacillus rhamnosus LS8 Ameliorates Azoxymethane/Dextran Sulfate Sodium-Induced Colitis-Associated Tumorigenesis in Mice via Regulating Gut Microbiota and Inhibiting Inflammation. Probiotics Antimicrob. Proteins 2022, 14, 947–959. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Du, H.; Zhang, M.; Xu, H.; Pu, X.; Chen, Q.; Luo, R.; Hu, Y.; Wang, Y.; Tu, H.; et al. Anti-inflammatory effect of Rhein on ulcerative colitis via inhibiting PI3K/Akt/mTOR signaling pathway and regulating gut microbiota. Phytother. Res. 2022, 36, 2081–2094. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Lv, L.; Liu, B.; Wang, S.; Zhang, S.; Wu, Z.; Yang, L.; Bian, X.; Wang, Q.; Wang, K.; et al. Akkermansia muciniphila Ameliorates Acetaminophen-Induced Liver Injury by Regulating Gut Microbial Composition and Metabolism. Microbiol. Spectr. 2022, 10, e0159621. [Google Scholar] [CrossRef]
- Jiang, P.; Yang, W.; Jin, Y.; Huang, H.; Shi, C.; Jiang, Y.; Wang, J.; Kang, Y.; Wang, C.; Yang, G. Lactobacillus reuteri protects mice against Salmonella typhimurium challenge by activating macrophages to produce nitric oxide. Microb. Pathog. 2019, 137, 103754. [Google Scholar] [CrossRef]
- Li, Z.; Fang, X.; Hu, X.; Li, C.; Wan, Y.; Yu, D. Amelioration of alcohol-induced acute liver injury in C57BL/6 mice by a mixture of TCM phytochemicals and probiotics with antioxidative and anti-inflammatory effects. Front. Nutr. 2023, 10, 1144589. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Dhaneshwar, S. Role of prebiotics, probiotics, and synbiotics in management of inflammatory bowel disease: Current perspectives. World J. Gastroenterol. 2023, 29, 2078–2100. [Google Scholar] [CrossRef]
- Schroeder, B.O.; Birchenough, G.M.H.; Ståhlman, M.; Arike, L.; Johansson, M.E.V.; Hansson, G.C.; Bäckhed, F. Bifidobacteria or Fiber Protects against Diet-Induced Microbiota-Mediated Colonic Mucus Deterioration. Cell. Host Microbe. 2018, 23, 27–40.e7. [Google Scholar] [CrossRef]
- Tsai, Y.S.; Chen, Y.P.; Lin, S.W.; Chen, Y.L.; Chen, C.C.; Huang, G.J. Lactobacillus rhamnosus GKLC1 ameliorates cisplatin-induced chronic nephrotoxicity by inhibiting cell inflammation and apoptosis. Biomed. Pharmacother. 2022, 147, 112701. [Google Scholar] [CrossRef]
- Mapuskar, K.A.; Vasquez-Martinez, G.; Mayoral-Andrade, G.; Tomanek-Chalkley, A.; Zepeda-Orozco, D.; Allen, B.G. Mitochondrial Oxidative Metabolism: An Emerging Therapeutic Target to Improve CKD Outcomes. Biomedicines. 2023, 11, 1573. [Google Scholar] [CrossRef]
- Yang, T.; Li, W.; Peng, A.; Liu, J.; Wang, Q. Exosomes Derived from Bone Marrow-Mesenchymal Stem Cells Attenuates Cisplatin-Induced Ototoxicity in a Mouse Model. J. Clin. Med. 2022, 11, 4743. [Google Scholar] [CrossRef]




| Gene | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
|---|---|---|
| β-actin | TTCTACAATGAGCTGCGTGTG | CTTTTCACGGTTGGCCTTAG |
| Kim-1 | ACATATCGTGGAATCACAACGAC | ACAAGCAGAAGATGGGCATTG |
| Sod1 | AAGAGAGGCATGTTGGAGACC | CGGCCAATGATGGAATGCTC |
| Sod2 | TGGAGAACCCAAAGGAGAGTTG | CAGGCAGCAATCTGTAAGCG |
| Sod3 | CTGACAGGTGCAGAGAACCTC | GCGTGTCGCCTATCTTCTCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ikeda, Y.; Suga, N.; Matsuda, S. Efficacy of Life Protection Probably from Newly Isolated Bacteria against Cisplatin-Induced Lethal Toxicity. Microorganisms 2023, 11, 2246. https://doi.org/10.3390/microorganisms11092246
Ikeda Y, Suga N, Matsuda S. Efficacy of Life Protection Probably from Newly Isolated Bacteria against Cisplatin-Induced Lethal Toxicity. Microorganisms. 2023; 11(9):2246. https://doi.org/10.3390/microorganisms11092246
Chicago/Turabian StyleIkeda, Yuka, Naoko Suga, and Satoru Matsuda. 2023. "Efficacy of Life Protection Probably from Newly Isolated Bacteria against Cisplatin-Induced Lethal Toxicity" Microorganisms 11, no. 9: 2246. https://doi.org/10.3390/microorganisms11092246
APA StyleIkeda, Y., Suga, N., & Matsuda, S. (2023). Efficacy of Life Protection Probably from Newly Isolated Bacteria against Cisplatin-Induced Lethal Toxicity. Microorganisms, 11(9), 2246. https://doi.org/10.3390/microorganisms11092246






