Antimicrobial Peptides Originating from Expression Libraries of Aurelia aurita and Mnemiopsis leidyi Prevent Biofilm Formation of Opportunistic Pathogens
Abstract
1. Introduction
2. Materials and Methods
2.1. Aurelia aurita Polyp Husbandry
2.2. Sampling of Aurelia aurita Polyps and Mnemiopsis leidyi Medusae
2.3. Direct mRNA Isolation and Construction of the cDNA Expression Libraries
2.4. Preparation of Cell-Free Size-Fractionated Cell Extracts
2.5. Bacterial Biofilm Prevention In Vitro Assay
2.6. Plasmid Preparation, Insert Size Determination, and Sequencing
2.7. Synthesis of Peptides
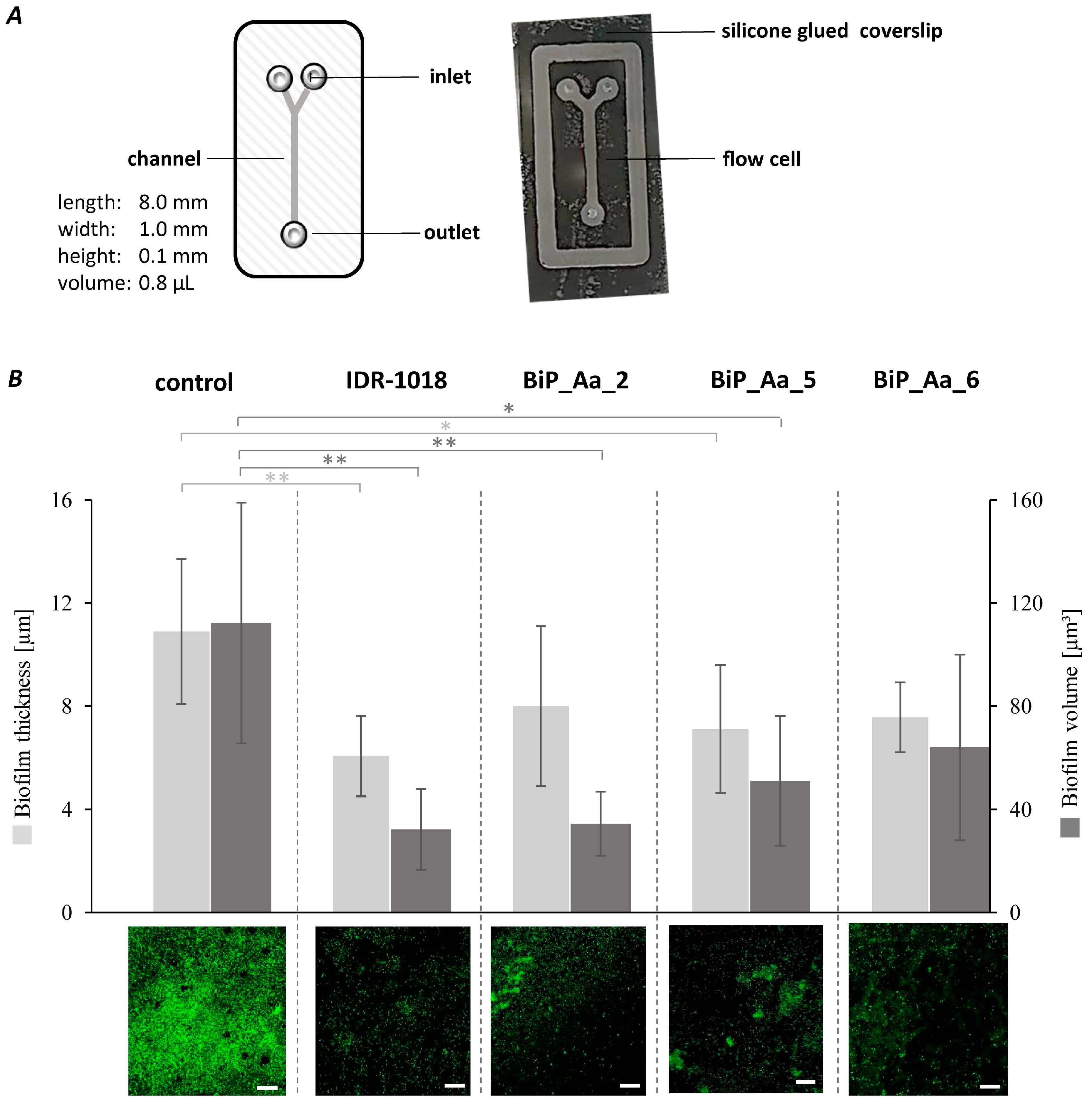
2.8. Effect of Biofilm-Preventing Peptides on Biofilm Formation of K. oxytoca M5aI in a Microfluidic Flow Cell
3. Results
3.1. Construction of cDNA Expression Libraries of Two Basal Metazoans
3.2. Identification of Biofilm-Preventing Clones from the cDNA Expression Libraries
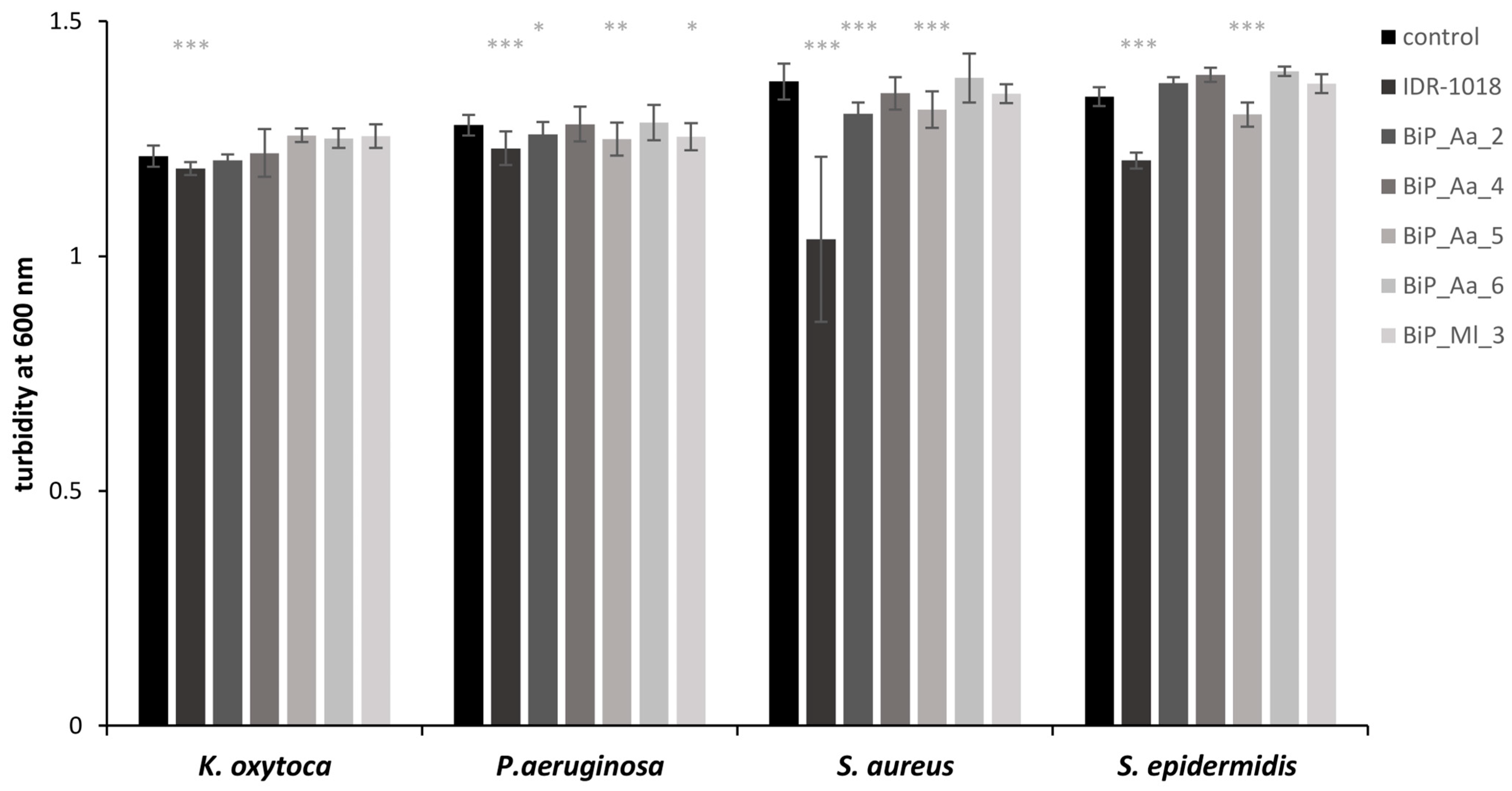
3.3. Synthetic Peptides—Excluding Cytotoxic Effects on Planktonic Bacteria
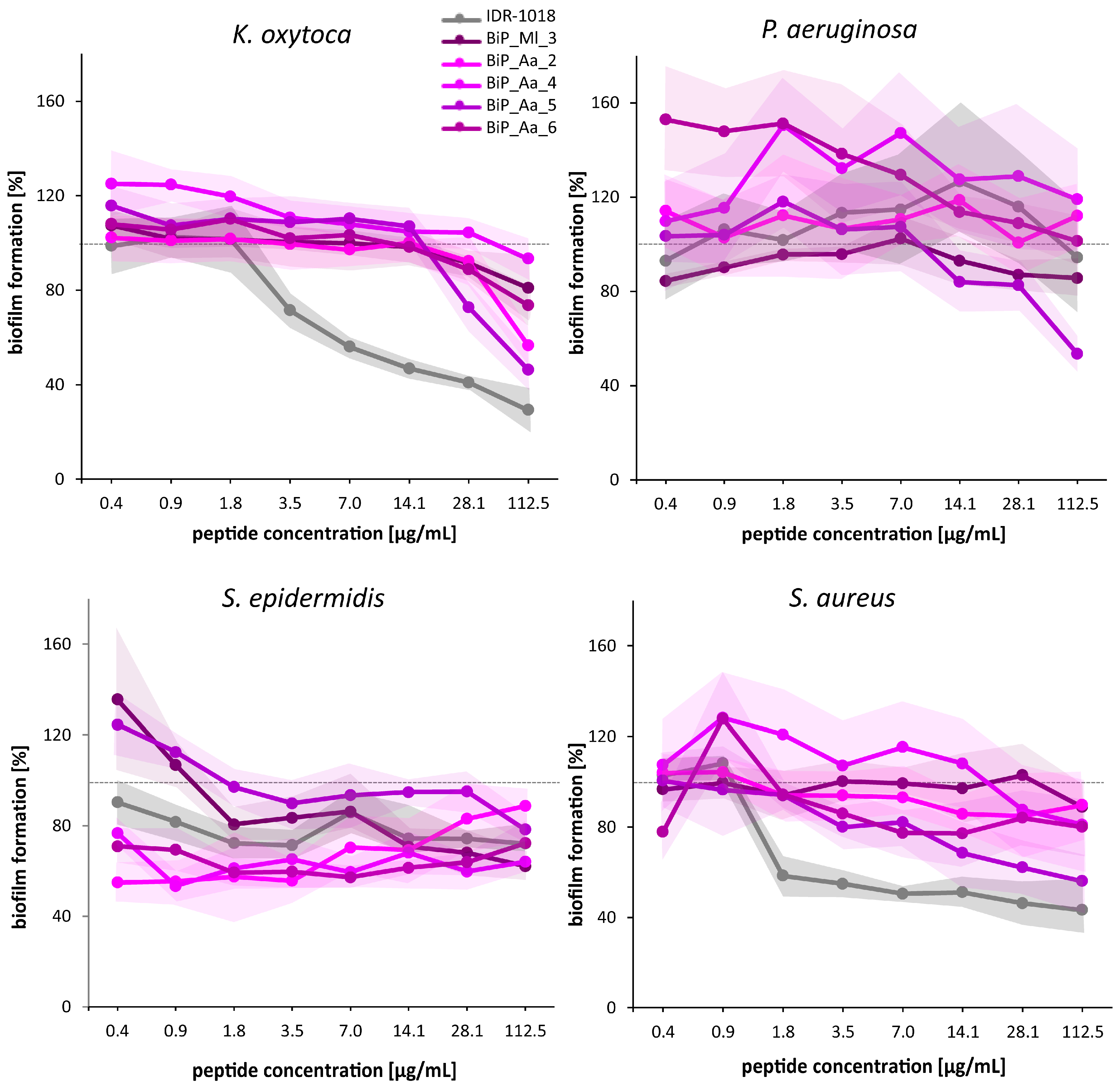
3.4. Synthetic Peptides—Verification of the Biofilm-Preventing Effects on Static Biofilms
3.5. Synthetic Peptides—Biofilm-Preventing Effects on Dynamic K. oxytoca Biofilms
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- McFall-Ngai, M.; Hadfield, M.G.; Bosch, T.C.; Carey, H.V.; Domazet-Lošo, T.; Douglas, A.E.; Dubilier, N.; Eberl, G.; Fukami, T.; Gilbert, S.F. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. USA 2013, 110, 3229–3236. [Google Scholar] [CrossRef]
- Rinkevich, B. Invertebrates versus vertebrates innate immunity: In the light of evolution. Scand. J. Immunol. 1999, 50, 456–460. [Google Scholar] [CrossRef]
- Miller, D.; Hemmrich, G.; Ball, E.; Hayward, D.; Khalturin, K.; Funayama, N.; Agata, K.; Bosch, T. The innate immune repertoire in Cnidaria-ancestral complexity and stochastic gene loss. Genome Biol. 2007, 8, R59. [Google Scholar] [CrossRef]
- Schulenburg, H.; Boehnisch, C.; Michiels, N.K. How do invertebrates generate a highly specific innate immune response? Mol. Immunol. 2007, 44, 3338–3344. [Google Scholar] [CrossRef]
- Auguste, M.; Melillo, D.; Corteggio, A.; Marino, R.; Canesi, L.; Pinsino, A.; Italiani, P.; Boraschi, D. Methodological approaches to assess innate immunity and innate memory in marine invertebrates and humans. Front. Toxicol. 2022, 4, 4. [Google Scholar] [CrossRef]
- Wu, R.; Patocka, J.; Nepovimova, E.; Oleksak, P.; Valis, M.; Wu, W.; Kuca, K. Marine invertebrate peptides: Antimicrobial peptides. Front. Microbiol. 2021, 12, 785085. [Google Scholar] [CrossRef]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019, 11, 3919. [Google Scholar]
- Bahar, A.A.; Ren, D. Antimicrobial peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef]
- Joo, H.-S.; Fu, C.-I.; Otto, M. Bacterial strategies of resistance to antimicrobial peptides. Philos. Trans. R. Soc. B: Biol. Sci. 2016, 371, 20150292. [Google Scholar] [CrossRef]
- Le, C.-F.; Fang, C.-M.; Sekaran, S.D. Intracellular targeting mechanisms by antimicrobial peptides. Antimicrob. Agents Chemother. 2017, 61, 10–1128. [Google Scholar] [CrossRef]
- Juretić, D. Designed multifunctional peptides for intracellular targets. Antibiotics 2022, 11, 1196. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial peptides: Classification, design, application and research progress in multiple fields. Front. Microbiol. 2020, 11, 2559. [Google Scholar] [CrossRef]
- Moretta, A.; Scieuzo, C.; Petrone, A.M.; Salvia, R.; Manniello, M.D.; Franco, A.; Lucchetti, D.; Vassallo, A.; Vogel, H.; Sgambato, A. Antimicrobial peptides: A new hope in biomedical and pharmaceutical fields. Front. Cell. Infect. Microbiol. 2021, 11, 668632. [Google Scholar] [CrossRef]
- Xu, Q.; Hu, X.; Wang, Y. Alternatives to conventional antibiotic therapy: Potential therapeutic strategies of combating antimicrobial-resistance and biofilm-related infections. Mol. Biotechnol. 2021, 63, 1103–1124. [Google Scholar] [CrossRef]
- Shahrour, H.; Ferrer-Espada, R.; Dandache, I.; Bárcena-Varela, S.; Sánchez-Gómez, S.; Chokr, A.; Martinez-de-Tejada, G. AMPs as anti-biofilm agents for human therapy and prophylaxis. In Antimicrobial Peptides: Basics for Clinical Application; Springer: Singapore, 2019; pp. 257–279. [Google Scholar]
- Chung, P.Y.; Khanum, R. Antimicrobial peptides as potential anti-biofilm agents against multidrug-resistant bacteria. J. Microbiol. Immunol. Infect. 2017, 50, 405–410. [Google Scholar] [CrossRef]
- Ghannoum, M.; Parsek, M.; Whiteley, M.; Mukherjee, P.K. Microbial Biofilms; John Wiley & Sons: Hoboken, NJ, USA, 2020. [Google Scholar]
- Singh, S.; Singh, S.K.; Chowdhury, I.; Singh, R. Understanding the Mechanism of Bacterial Biofilms Resistance to Antimicrobial Agents. Open Microbiol. J. 2017, 11, 53–62. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Orhan, I.E.; Banach, M.; Rollinger, J.M.; Barreca, D.; Weckwerth, W.; Bauer, R.; Bayer, E.A.; et al. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Mallapragada, S.; Wadhwa, A.; Agrawal, P. Antimicrobial peptides: The miraculous biological molecules. J. Indian Soc. Periodontol. 2017, 21, 434. [Google Scholar]
- Chen, C.H.; Bepler, T.; Pepper, K.; Fu, D.; Lu, T.K. Synthetic molecular evolution of antimicrobial peptides. Curr. Opin. Biotechnol. 2022, 75, 102718. [Google Scholar] [CrossRef]
- Díaz, G.A. Defensins and cystein rich peptides: Two types of antimicrobial peptides in marine molluscs. Invertebr. Surviv. J. 2010, 7, 157–164. [Google Scholar]
- Charlet, M.; Chernysh, S.; Philippe, H.; Hetru, C.; Hoffmann, J.A.; Bulet, P. Innate immunity. Isolation of several cysteine-rich antimicrobial peptides from the blood of a mollusc, Mytilus edulis. J. Biol. Chem. 1996, 271, 21808–21813. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-H.; Zhao, J.-M.; Song, L.-S. A review of advances in research on marine molluscan antimicrobial peptides and their potential application in aquaculture. Molluscan Res. 2009, 29, 17. [Google Scholar]
- Tasiemski, A. Antimicrobial peptides in annelids. Invertebr. Surviv. J. 2008, 5, 75–82. [Google Scholar]
- Yi, H.-Y.; Chowdhury, M.; Huang, Y.-D.; Yu, X.-Q. Insect antimicrobial peptides and their applications. Appl. Microbiol. Biotechnol. 2014, 98, 5807–5822. [Google Scholar] [CrossRef] [PubMed]
- Rinehart, K.L.; Kishore, V.; Bible, K.C.; Sakai, R.; Sullins, D.W.; Li, K.-M. Didemnins and tunichlorin: Novel natural products from the marine tunicate Trididemnum solidum. J. Nat. Prod. 1988, 51, 1–21. [Google Scholar] [CrossRef]
- Lee, I.H.; Cho, Y.; Lehrer, R.I. Styelins, broad-spectrum antimicrobial peptides from the solitary tunicate, Styela clava. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 1997, 118, 515–521. [Google Scholar] [CrossRef]
- Jo, C.; Khan, F.F.; Khan, M.I.; Iqbal, J. Marine bioactive peptides: Types, structures, and physiological functions. Food Rev. Int. 2017, 33, 44–61. [Google Scholar] [CrossRef]
- Otero-Gonzáiez, A.J.; Magalhaes, B.S.; Garcia-Villarino, M.; López-Abarrategui, C.; Sousa, D.A.; Dias, S.C.; Franco, O.L. Antimicrobial peptides from marine invertebrates as a new frontier for microbial infection control. FASEB J. 2010, 24, 1320–1334. [Google Scholar] [CrossRef]
- Beeton, C.; Pennington, M.W.; Norton, R.S. Analogs of the sea anemone potassium channel blocker ShK for the treatment of autoimmune diseases. Inflamm. Allergy-Drug Targets (Former. Curr. Drug Targets-Inflamm. Allergy) (Discontin.) 2011, 10, 313–321. [Google Scholar] [CrossRef]
- Neshani, A.; Sedighian, H.; Mirhosseini, S.A.; Ghazvini, K.; Zare, H.; Jahangiri, A. Antimicrobial peptides as a promising treatment option against Acinetobacter baumannii infections. Microb. Pathog. 2020, 146, 104238. [Google Scholar] [CrossRef]
- Sierra, J.M.; Fusté, E.; Rabanal, F.; Vinuesa, T.; Viñas, M. An overview of antimicrobial peptides and the latest advances in their development. Expert Opin. Biol. Ther. 2017, 17, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Kang, S.-J.; Park, S.J.; Mishig-Ochir, T.; Lee, B.-J. Antimicrobial peptides: Therapeutic potentials. Expert Rev. Anti-Infect. Ther. 2014, 12, 1477–1486. [Google Scholar] [CrossRef]
- Gordon, Y.J.; Romanowski, E.G.; McDermott, A.M. A review of antimicrobial peptides and their therapeutic potential as anti-infective drugs. Curr. Eye Res. 2005, 30, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial peptides: An emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [PubMed]
- Bellotti, D.; Remelli, M. Lights and shadows on the therapeutic use of antimicrobial peptides. Molecules 2022, 27, 4584. [Google Scholar] [CrossRef]
- Ramazi, S.; Mohammadi, N.; Allahverdi, A.; Khalili, E.; Abdolmaleki, P. A review on antimicrobial peptides databases and the computational tools. Database 2022, 2022, baac011. [Google Scholar] [CrossRef] [PubMed]
- Weiland-Bräuer, N.; Neulinger, S.C.; Pinnow, N.; Kunzel, S.; Baines, J.F.; Schmitz, R.A. Composition of Bacterial Communities Associated with Aurelia aurita Changes with Compartment, Life Stage, and Population. Appl. Environ. Microbiol. 2015, 81, 6038–6052. [Google Scholar] [CrossRef]
- Weiland-Bräuer, N.; Pinnow, N.; Langfeldt, D.; Roik, A.; Güllert, S.; Chibani, C.M.; Reusch, T.B.; Schmitz, R.A. The native microbiome is crucial for offspring generation and fitness of Aurelia aurita. mBio 2020, 11, e02336-20. [Google Scholar] [CrossRef]
- Weiland-Bräuer, N.; Pinnow, N.; Schmitz, R.A. Novel reporter for identification of interference with acyl homoserine lactone and autoinducer-2 quorum sensing. Appl. Environ. Microbiol. 2015, 81, 1477–1489. [Google Scholar] [CrossRef]
- Gerlach, G.F.; Allen, B.L.; Clegg, S. Molecular characterization of the type 3 (MR/K) fimbriae of Klebsiella pneumoniae. J. Bacteriol. 1988, 170, 3547–3553. [Google Scholar] [CrossRef] [PubMed]
- Djordjevic, D.; Wiedmann, M.; McLandsborough, L. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 2002, 68, 2950–2958. [Google Scholar] [CrossRef] [PubMed]
- Mack, D.R.; Blain-Nelson, P.L. Disparate in vitro inhibition of adhesion of enteropathogenic Escherichia coli RDEC-1 by mucins isolated from various regions of the intestinal tract. Pediatr. Res. 1995, 37, 75–80. [Google Scholar] [CrossRef][Green Version]
- Ryan, J.F.; Pang, K.; Schnitzler, C.E.; Nguyen, A.-D.; Moreland, R.T.; Simmons, D.K.; Koch, B.J.; Francis, W.R.; Havlak, P.; Smith, S.A. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 2013, 342, 1242592. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente-Núñez, C.S.; Reffuveille, F.; Haney, E.F.; Straus, S.K.; Hancock, R.E. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog. 2014, 10, e1004152. [Google Scholar] [CrossRef]
- Mansour, S.C.; de la Fuente-Núñez, C.; Hancock, R.E. Peptide IDR-1018: Modulating the immune system and targeting bacterial biofilms to treat antibiotic-resistant bacterial infections. J. Pept. Sci. 2015, 21, 323–329. [Google Scholar] [CrossRef]
- Gold, D.A.; Katsuki, T.; Li, Y.; Yan, X.; Regulski, M.; Ibberson, D.; Holstein, T.; Steele, R.E.; Jacobs, D.K.; Greenspan, R.J. The genome of the jellyfish Aurelia and the evolution of animal complexity. Nat. Ecol. Evol. 2019, 3, 96–104. [Google Scholar] [CrossRef]
- Khalturin, K.; Shinzato, C.; Khalturina, M.; Hamada, M.; Fujie, M.; Koyanagi, R.; Kanda, M.; Goto, H.; Anton-Erxleben, F.; Toyokawa, M. Medusozoan genomes inform the evolution of the jellyfish body plan. Nat. Ecol. Evol. 2019, 3, 811. [Google Scholar] [CrossRef]
- Moreland, R.T.; Nguyen, A.-D.; Ryan, J.F.; Schnitzler, C.E.; Koch, B.J.; Siewert, K.; Wolfsberg, T.G.; Baxevanis, A.D. A customized Web portal for the genome of the ctenophore Mnemiopsis leidyi. BMC Genom. 2014, 15, 316. [Google Scholar] [CrossRef]
- Ovchinnikova, T.V.; Balandin, S.V.; Aleshina, G.M.; Tagaev, A.A.; Leonova, Y.F.; Krasnodembsky, E.D.; Men’shenin, A.V.; Kokryakov, V.N. Aurelin, a novel antimicrobial peptide from jellyfish Aurelia aurita with structural features of defensins and channel-blocking toxins. Biochem. Biophys. Res. Commun. 2006, 348, 514–523. [Google Scholar] [CrossRef]
- Shenkarev, Z.O.; Panteleev, P.V.; Balandin, S.V.; Gizatullina, A.K.; Altukhov, D.A.; Finkina, E.I.; Kokryakov, V.N.; Arseniev, A.S.; Ovchinnikova, T.V. Recombinant expression and solution structure of antimicrobial peptide aurelin from jellyfish Aurelia aurita. Biochem. Biophys. Res. Commun. 2012, 429, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Pitts, B.; Hamilton, M.A.; Zelver, N.; Stewart, P.S. A microtiter-plate screening method for biofilm disinfection and removal. J. Microbiol. Methods 2003, 54, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Azeredo, J.; Azevedo, N.F.; Briandet, R.; Cerca, N.; Coenye, T.; Costa, A.R.; Desvaux, M.; Di Bonaventura, G.; Hébraud, M.; Jaglic, Z. Critical review on biofilm methods. Crit. Rev. Microbiol. 2017, 43, 313–351. [Google Scholar] [CrossRef] [PubMed]
- Kragh, K.N.; Alhede, M.; Kvich, L.; Bjarnsholt, T. Into the well—A close look at the complex structures of a microtiter biofilm and the crystal violet assay. Biofilm 2019, 1, 100006. [Google Scholar] [CrossRef]
- Kragh, K.N.; Alhede, M.; Rybtke, M.; Stavnsberg, C.; Jensen, P.Ø.; Tolker-Nielsen, T.; Whiteley, M.; Bjarnsholt, T. The inoculation method could impact the outcome of microbiological experiments. Appl. Environ. Microbiol. 2018, 84, e02264-17. [Google Scholar] [CrossRef]
- Wieczorek, M.; Jenssen, H.; Kindrachuk, J.; Scott, W.R.; Elliott, M.; Hilpert, K.; Cheng, J.T.; Hancock, R.E.; Straus, S.K. Structural studies of a peptide with immune modulating and direct antimicrobial activity. Chem. Biol. 2010, 17, 970–980. [Google Scholar] [CrossRef]
- Qin, S.; Xiao, W.; Zhou, C.; Pu, Q.; Deng, X.; Lan, L.; Liang, H.; Song, X.; Wu, M. Pseudomonas aeruginosa: Pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct. Target. Ther. 2022, 7, 199. [Google Scholar] [CrossRef]
- Chen, H.; Wubbolts, R.W.; Haagsman, H.P.; Veldhuizen, E.J. Inhibition and eradication of Pseudomonas aeruginosa biofilms by host defence peptides. Sci. Rep. 2018, 8, 10446. [Google Scholar] [CrossRef]
- Casciaro, B.; Lin, Q.; Afonin, S.; Loffredo, M.R.; de Turris, V.; Middel, V.; Ulrich, A.S.; Di, Y.P.; Mangoni, M.L. Inhibition of Pseudomonas aeruginosa biofilm formation and expression of virulence genes by selective epimerization in the peptide Esculentin-1a (1-21) NH 2. FEBS J. 2019, 286, 3874–3891. [Google Scholar] [CrossRef]
- Ferreira Cespedes, G.; Nicolas Lorenzon, E.; Festozo Vicente, E.; Jose Soares Mendes-Giannini, M.; Fontes, W.; de Souza Castro, M.; Maffud Cilli, E. Mechanism of action and relationship between structure and biological activity of Ctx-Ha: A new ceratotoxin-like peptide from Hypsiboas albopunctatus. Protein Pept. Lett. 2012, 19, 596–603. [Google Scholar] [CrossRef]
- Lorenzón, E.; Cespedes, G.; Vicente, E.; Nogueira, L.; Bauab, T.; Castro, M.; Cilli, E.M. Effects of dimerization on the structure and biological activity of antimicrobial peptide Ctx-Ha. Antimicrob. Agents Chemother. 2012, 56, 3004–3010. [Google Scholar] [CrossRef] [PubMed]
- Yawata, Y.; Nguyen, J.; Stocker, R.; Rusconi, R. Microfluidic studies of biofilm formation in dynamic environments. J. Bacteriol. 2016, 198, 2589–2595. [Google Scholar] [CrossRef]
- Subramanian, S.; Huiszoon, R.C.; Chu, S.; Bentley, W.E.; Ghodssi, R. Microsystems for biofilm characterization and sensing—A review. Biofilm 2020, 2, 100015. [Google Scholar] [CrossRef] [PubMed]
- Yasir, M.; Willcox, M.D.P.; Dutta, D. Action of antimicrobial peptides against bacterial biofilms. Materials 2018, 11, 2468. [Google Scholar] [CrossRef] [PubMed]
- Wimley, W.C. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem. Biol. 2010, 5, 905–917. [Google Scholar] [CrossRef]
- Torrent, M.; Valle, J.; Nogués, M.V.; Boix, E.; Andreu, D. The generation of antimicrobial peptide activity: A trade-off between charge and aggregation? Angew. Chem. 2011, 123, 10874–10877. [Google Scholar] [CrossRef]
- Lorenzon, E.N.; Piccoli, J.P.; Santos-Filho, N.A.; Cilli, E.M. Dimerization of antimicrobial peptides: A promising strategy to enhance antimicrobial peptide activity. Protein Pept. Lett. 2019, 26, 98–107. [Google Scholar] [CrossRef]
- Thamri, A.; Létourneau, M.; Djoboulian, A.; Chatenet, D.; Deziel, E.; Castonguay, A.; Perreault, J. Peptide modification results in the formation of a dimer with a 60-fold enhanced antimicrobial activity. PLoS ONE 2017, 12, e0173783. [Google Scholar] [CrossRef]
- Jiang, Z.; Higgins, M.P.; Whitehurst, J.; Kisich, K.O.; Voskuil, M.I.; Hodges, R.S. Anti-tuberculosis activity of α-helical antimicrobial peptides: De novo designed L-and D-enantiomers versus L-and D-LL37. Protein Pept. Lett. 2011, 18, 241–252. [Google Scholar] [CrossRef]
- Asma, S.T.; Imre, K.; Morar, A.; Herman, V.; Acaroz, U.; Mukhtar, H.; Arslan-Acaroz, D.; Shah, S.R.A.; Gerlach, R. An overview of biofilm formation–combating strategies and mechanisms of action of antibiofilm agents. Life 2022, 12, 1110. [Google Scholar] [CrossRef]
- Krukiewicz, K.; Kazek-Kęsik, A.; Brzychczy-Włoch, M.; Łos, M.J.; Ateba, C.N.; Mehrbod, P.; Ghavami, S.; Shyntum, D.Y. Recent Advances in the Control of Clinically Important Biofilms. Int. J. Mol. Sci. 2022, 23, 9526. [Google Scholar] [CrossRef] [PubMed]
- Rima, M.; Rima, M.; Fajloun, Z.; Sabatier, J.-M.; Bechinger, B.; Naas, T. Antimicrobial peptides: A potent alternative to antibiotics. Antibiotics 2021, 10, 1095. [Google Scholar] [CrossRef] [PubMed]
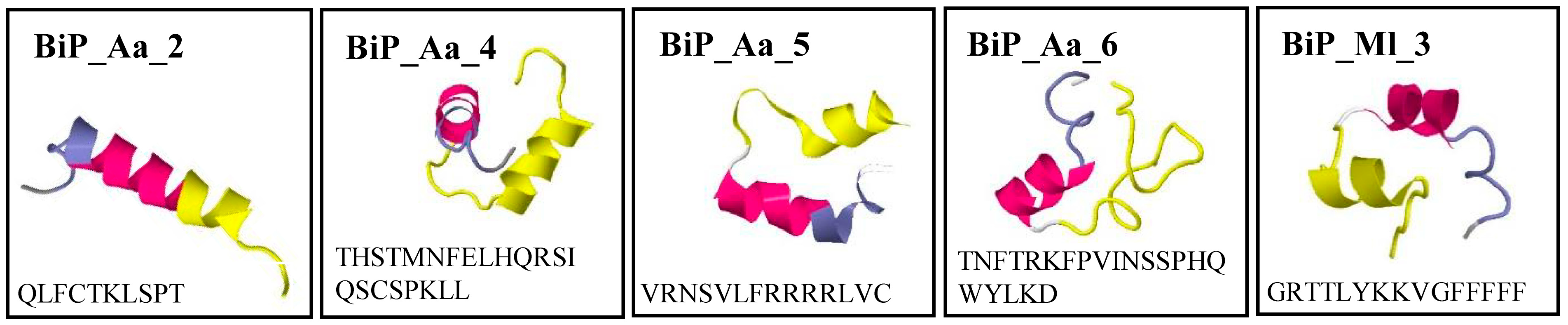
| Peptide Designation | Related EST Clone | Amino Acid Sequence | MWCALC (kDA) | pICALC | Aggregation Propensity [46] |
|---|---|---|---|---|---|
| BiP_Aa_2 | A. aurita 112_6C | QLFCTKLSPT | 1.14 | 8.22 | 0.201 |
| BiP_Aa_4 | A. aurita 127_8E | THSTMNFELHQRSIQSCSPKLL | 2.55 | 7.95 | −0.091 |
| BiP_Aa_5 | A. aurita 127_8F | VRNSVLFRRRRLVC | 1.78 | 12.18 | 0.150 |
| BiP_Aa_6 | A. aurita 127_8H | TNFTRKFPVINSSPHQWYLKD | 2.58 | 9.7 | −0.042 |
| BiP_Ml_3 | M. leidyi 010_9A | GRTTLYKKVGFFFFF | 1.8 | 10.29 | 0.561 |
| control IDR-1018 [47] | VRLIVAVRIWRR | 1.5 | 12.48 | 0.487 | |
| Clone Designation | Biofilm Formation (%) | |||
|---|---|---|---|---|
| K. oxytoca | P. aeruginosa | S. aureus | S. epidermidis | |
| Aa_112_4H | 71 ± 12 | 85 ± 19 | 77 ± 9 | 55 ± 14 |
| Aa_112_6C | 92 ± 14 | 79 ± 24 | 84 ± 13 | 49 ± 9 |
| Aa_127_8A | 22 ± 2 | 100 ± 18 | 100 ± 13 | 77 ±13 |
| Aa_127_8E | 80 ± 3 | 69 ± 17 | 78 ± 13 | 6 ± 1 |
| Aa_127_8F | 28 ± 3 | 75 ± 18 | 84 ± 23 | 95 ± 10 |
| Aa_127_8H | 104 ± 5 | 77 ± 18 | 61 ± 10 | 40 ± 12 |
| Ml_068_11H | 107 ± 13 | 81 ± 13 | 69 ± 7 | 29 ± 3 |
| Ml_011_11H | 27 ± 2 | 100 ± 22 | 67 ± 3 | 77 ± 10 |
| Ml_010_9A | 71 ± 8 | 78 ± 14 | 76 ± 5 | 94 ± 9 |
| Ml_010_9G | 68 ± 14 | 75 ± 18 | 89 ± 17 | 91 ± 7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ladewig, L.; Gloy, L.; Langfeldt, D.; Pinnow, N.; Weiland-Bräuer, N.; Schmitz, R.A. Antimicrobial Peptides Originating from Expression Libraries of Aurelia aurita and Mnemiopsis leidyi Prevent Biofilm Formation of Opportunistic Pathogens. Microorganisms 2023, 11, 2184. https://doi.org/10.3390/microorganisms11092184
Ladewig L, Gloy L, Langfeldt D, Pinnow N, Weiland-Bräuer N, Schmitz RA. Antimicrobial Peptides Originating from Expression Libraries of Aurelia aurita and Mnemiopsis leidyi Prevent Biofilm Formation of Opportunistic Pathogens. Microorganisms. 2023; 11(9):2184. https://doi.org/10.3390/microorganisms11092184
Chicago/Turabian StyleLadewig, Lisa, Leon Gloy, Daniela Langfeldt, Nicole Pinnow, Nancy Weiland-Bräuer, and Ruth A. Schmitz. 2023. "Antimicrobial Peptides Originating from Expression Libraries of Aurelia aurita and Mnemiopsis leidyi Prevent Biofilm Formation of Opportunistic Pathogens" Microorganisms 11, no. 9: 2184. https://doi.org/10.3390/microorganisms11092184
APA StyleLadewig, L., Gloy, L., Langfeldt, D., Pinnow, N., Weiland-Bräuer, N., & Schmitz, R. A. (2023). Antimicrobial Peptides Originating from Expression Libraries of Aurelia aurita and Mnemiopsis leidyi Prevent Biofilm Formation of Opportunistic Pathogens. Microorganisms, 11(9), 2184. https://doi.org/10.3390/microorganisms11092184







