SW16-7, a Novel Ackermannviridae Bacteriophage with Highly Effective Lytic Activity Targets Salmonella enterica Serovar Weltevreden
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria Strains and Growth Conditions
2.2. Phage Isolation and Purification
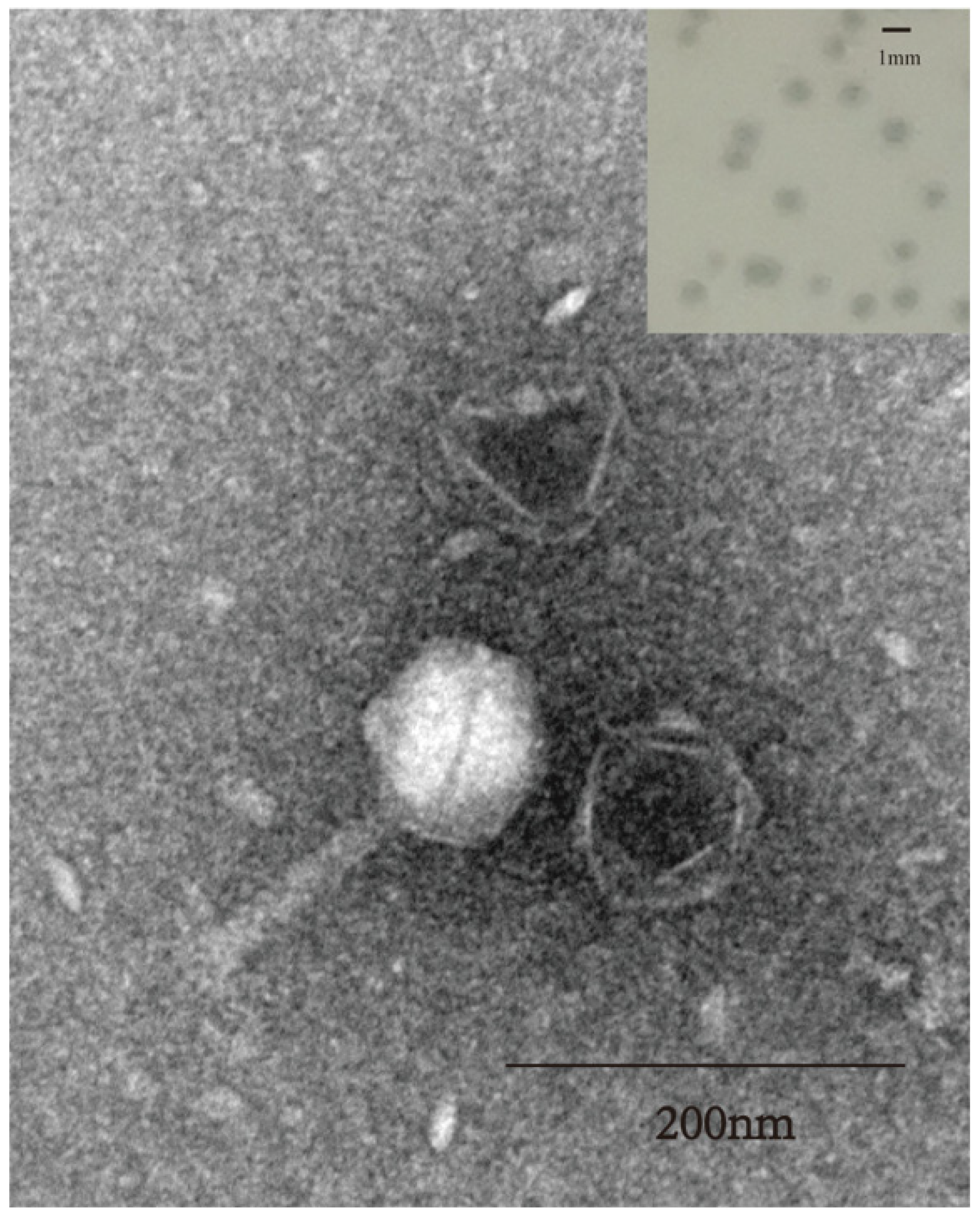
2.3. Transmission Electron Microscopy
2.4. Determination of Host Ranges
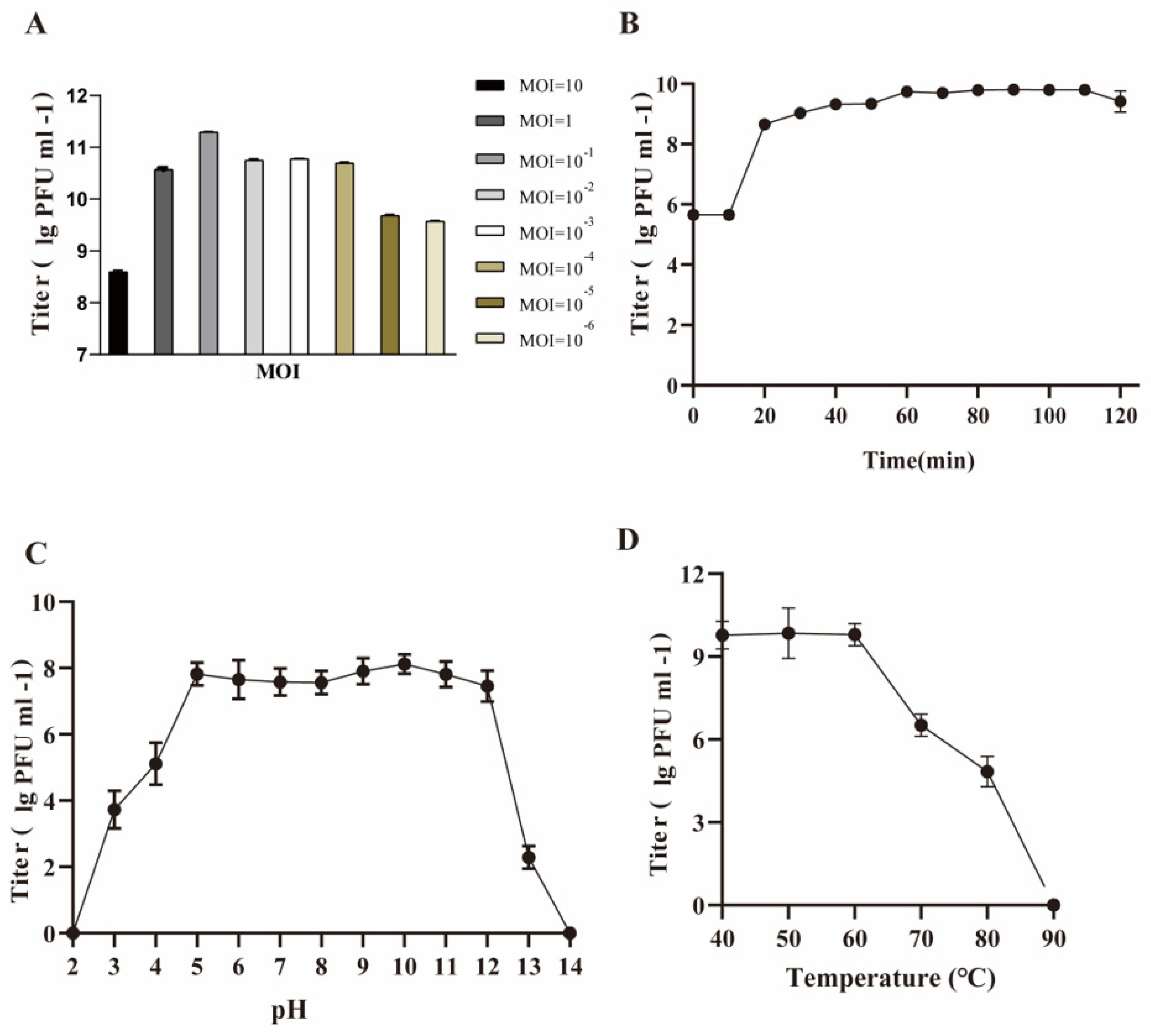
2.5. Thermal and pH Stability Test
2.6. Optimum Multiplicity of Infection (MOI)
2.7. One-Step Growth Curve
2.8. In Vitro Inhibitory Assessment of Phage SW16-7
2.9. Genome Sequencing and Bioinformatics Analysis
2.10. Statistical Analysis
3. Results
3.1. Morphology of Phage SW16-7
3.2. Biological Features of Phage SW16-7
3.3. General Features of the Phage SW16-7 Genome
3.4. Specific Features of the Phage SW16-7 Genome
3.4.1. Morphogenesis-Related Genes
3.4.2. DNA Metabolism-Related Genes
3.4.3. Gene Expression-Related Genes
3.4.4. Auxiliary Metabolism-Related Genes
3.4.5. Lysis-Related Genes
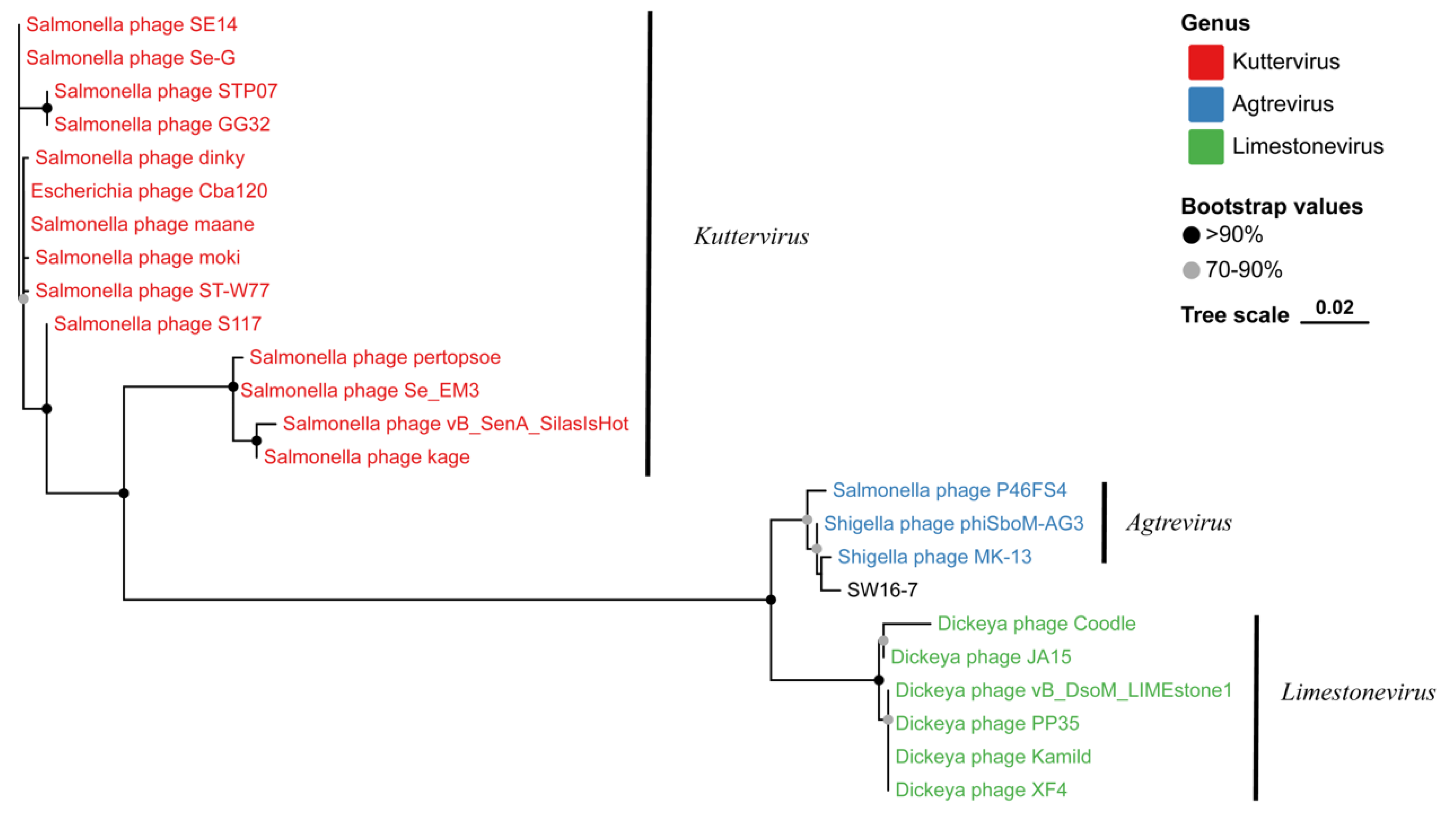
3.5. Phylogenetic Analysis of Phage SW16-7
3.6. Host Range of Phage SW16-7
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ferrari, R.G.; Rosario, D.K.A.; Cunha-Neto, A.; Mano, S.B.; Figueiredo, E.E.S.; Conte-Junior, C.A. Worldwide Epidemiology of Salmonella Serovars in Animal-Based Foods: A Meta-analysis. Appl. Environ. Microbiol. 2019, 85, e00591-19. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Vargas, F.M.; Abu-El-Haija, M.A.; Gómez-Duarte, O.G. Salmonella infections: An update on epidemiology, management, and prevention. Travel Med. Infect. Dis. 2011, 9, 263–277. [Google Scholar] [CrossRef] [PubMed]
- Awang Salleh, N.; Rusul, G.; Hassan, Z.; Reezal, A.; Hajar Isa, S.; Nishibuchi, M.; Radu, S. Incidence of Salmonella spp. in raw vegetables in Selangor, Malaysia. Food Control 2003, 14, 475–479. [Google Scholar] [CrossRef]
- Makendi, C.; Page, A.J.; Wren, B.W.; Le Thi Phuong, T.; Clare, S.; Hale, C.; Goulding, D.; Klemm, E.J.; Pickard, D.; Okoro, C.; et al. A Phylogenetic and Phenotypic Analysis of Salmonella enterica Serovar Weltevreden, an Emerging Agent of Diarrheal Disease in Tropical Regions. PLoS Neglected Trop. Dis. 2016, 10, e0004446. [Google Scholar] [CrossRef]
- Chau, M.L.; Hartantyo, S.H.; Yap, M.; Kang, J.S.; Aung, K.T.; Gutiérrez, R.A.; Ng, L.C.; Tam, C.C.; Barkham, T. Diarrheagenic pathogens in adults attending a hospital in Singapore. BMC Infect. Dis. 2016, 16, 32. [Google Scholar] [CrossRef]
- Brankatschk, K.; Blom, J.; Goesmann, A.; Smits, T.H.; Duffy, B. Genome of a European fresh-vegetable food safety outbreak strain of Salmonella enterica subsp. enterica serovar weltevreden. J. Bacteriol. 2011, 193, 2066. [Google Scholar] [CrossRef]
- Nguyen, T.K.; Bui, H.T.; Truong, T.A.; Lam, D.N.; Ikeuchi, S.; Ly, L.K.T.; Hara-Kudo, Y.; Taniguchi, T.; Hayashidani, H. Retail fresh vegetables as a potential source of Salmonella infection in the Mekong Delta, Vietnam. Int. J. Food Microbiol. 2021, 341, 109049. [Google Scholar] [CrossRef]
- Hounmanou, Y.M.G.; Dalsgaard, A.; Sopacua, T.F.; Uddin, G.M.N.; Leekitcharoenphon, P.; Hendriksen, R.S.; Olsen, J.E.; Larsen, M.H. Molecular Characteristics and Zoonotic Potential of Salmonella Weltevreden from Cultured Shrimp and Tilapia in Vietnam and China. Front. Microbiol. 2020, 11, 1985. [Google Scholar] [CrossRef]
- Li, K.; Petersen, G.; Barco, L.; Hvidtfeldt, K.; Liu, L.; Dalsgaard, A. Salmonella Weltevreden in integrated and non-integrated tilapia aquaculture systems in Guangdong, China. Food Microbiol. 2017, 65, 19–24. [Google Scholar] [CrossRef]
- Noor Uddin, G.M.; Larsen, M.H.; Barco, L.; Minh Phu, T.; Dalsgaard, A. Clonal Occurrence of Salmonella Weltevreden in Cultured Shrimp in the Mekong Delta, Vietnam. PLoS ONE 2015, 10, e0134252. [Google Scholar] [CrossRef]
- Zhao, S.; Datta, A.R.; Ayers, S.; Friedman, S.; Walker, R.D.; White, D.G. Antimicrobial-resistant Salmonella serovars isolated from imported foods. Int. J. Food Microbiol. 2003, 84, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Li, M.; Xu, C.; Cui, K.; Feng, Z.; Fu, Y.; Zhang, J.; Liao, M. Prevalence and molecular characterization of Salmonella enterica isolates throughout an integrated broiler supply chain in China. Epidemiol. Infect. 2016, 144, 2989–2999. [Google Scholar] [CrossRef] [PubMed]
- Minh, D.K.; Hounmanou, Y.M.G.; Mai, H.B.T.; Olsen, J.E.; Dalsgaard, A. Prevalence and genomic characterization of Salmonella Weltevreden in commercial pig feed. Vet. Microbiol. 2020, 246, 108725. [Google Scholar] [CrossRef] [PubMed]
- D’Ortenzio, E.; Weill, F.X.; Ragonneau, S.; Lebon, J.A.; Renault, P.; Pierre, V. First report of a Salmonella enterica serovar Weltevreden outbreak on Reunion Island, France, August 2007. Eurosurveillance 2008, 13, 18949. [Google Scholar] [CrossRef]
- Al-Maqbali, A.A.; Al-Abri, S.S.; Vidyanand, V.; Al-Abaidani, I.; Al-Balushi, A.S.; Bawikar, S.; El Amir, E.; Al-Azri, S.; Kumar, R.; Al-Rashdi, A.; et al. Community Foodborne of Salmonella Weltevreden Outbreak at Northern Governorate, Sultanate of Oman. J. Epidemiol. Glob. Health 2021, 11, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Hassan, R.; Tecle, S.; Adcock, B.; Kellis, M.; Weiss, J.; Saupe, A.; Sorenson, A.; Klos, R.; Blankenship, J.; Blessington, T.; et al. Multistate outbreak of Salmonella Paratyphi B variant L(+) tartrate(+) and Salmonella Weltevreden infections linked to imported frozen raw tuna: USA, March–July 2015. Epidemiol. Infect. 2018, 146, 1461–1467. [Google Scholar] [CrossRef]
- Le Thi Phuong, T.; Rattanavong, S.; Vongsouvath, M.; Davong, V.; Phu Huong Lan, N.; Campbell, J.I.; Darton, T.C.; Thwaites, G.E.; Newton, P.N.; Dance, D.A.B.; et al. Non-typhoidal Salmonella serovars associated with invasive and non-invasive disease in the Lao People’s Democratic Republic. Trans. R. Soc. Trop. Med. Hyg. 2017, 111, 418–424. [Google Scholar] [CrossRef]
- Antony, B.; Dias, M.; Shetty, A.K.; Rekha, B. Food poisoning due to Salmonella enterica serotype weltevreden in Mangalore. Indian J. Med. Microbiol. 2009, 27, 257–258. [Google Scholar] [CrossRef]
- Jain, P.; Nandy, S.; Bharadwaj, R.; Niyogi, S.K.; Dutta, S. Salmonella enterica serovar Weltevreden ST1500 associated foodborne outbreak in Pune, India. Indian J. Med. Res. 2015, 141, 239–241. [Google Scholar] [CrossRef]
- Li, B.; Yang, X.; Tan, H.; Ke, B.; He, D.; Wang, H.; Chen, Q.; Ke, C.; Zhang, Y. Whole genome sequencing analysis of Salmonella enterica serovar Weltevreden isolated from human stool and contaminated food samples collected from the Southern coastal area of China. Int. J. Food Microbiol. 2018, 266, 317–323. [Google Scholar] [CrossRef]
- Zhang, J.; Peng, Z.; Chen, K.; Zhan, Z.; Shen, H.; Feng, S.; Gou, H.; Qu, X.; Ziemann, M.; Layton, D.S.; et al. Genomic Characterization of Salmonella enterica serovar Weltevreden Associated with Human Diarrhea. Microbiol. Spectr. 2023, 11, e0354222. [Google Scholar] [CrossRef]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage Applications for Food Production and Processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef]
- Gambino, M.; Brøndsted, L. Looking into the future of phage-based control of zoonotic pathogens in food and animal production. Curr. Opin. Biotechnol. 2021, 68, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Woolston, J.; Parks, A.R.; Abuladze, T.; Anderson, B.; Li, M.; Carter, C.; Hanna, L.F.; Heyse, S.; Charbonneau, D.; Sulakvelidze, A. Bacteriophages lytic for Salmonella rapidly reduce Salmonella contamination on glass and stainless steel surfaces. Bacteriophage 2013, 3, e25697. [Google Scholar] [CrossRef] [PubMed]
- Endersen, L.; O’Mahony, J.; Hill, C.; Ross, R.P.; McAuliffe, O.; Coffey, A. Phage therapy in the food industry. Annu. Rev. Food Sci. Technol. 2014, 5, 327–349. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Ma, R.; Li, Z.; Mao, X.; Li, Y.; Wu, Y.; Wang, L.; Han, K.; Li, L.; Ma, D.; et al. Broad-Spectrum Salmonella Phages PSE-D1 and PST-H1 Controls Salmonella in Foods. Viruses 2022, 14, 2647. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Chen, H.; Zhao, J.; Tao, Z.; Lan, W.; Zhao, Y.; Sun, X. Characterization of Phage vB_SalM_SPJ41 and the Reduction of Risk of Antibiotic-Resistant Salmonella enterica Contamination in Two Ready-to-Eat Foods. Antibiotics 2023, 12, 364. [Google Scholar] [CrossRef]
- Van Twest, R.; Kropinski, A.M. Bacteriophage enrichment from water and soil. Methods Mol. Biol. 2009, 501, 15–21. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Finn, R.D.; Tate, J.; Mistry, J.; Coggill, P.C.; Sammut, S.J.; Hotz, H.R.; Ceric, G.; Forslund, K.; Eddy, S.R.; Sonnhammer, E.L.; et al. The Pfam protein families database. Nucleic Acids Res. 2008, 36, D281–D288. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Li, H.; Luo, X.; Li, H.; Gao, Q.; Zhang, L.; Teng, Y.; Zhao, Q.; Zuo, Z.; Ren, J. IBS 2.0: An upgraded illustrator for the visualization of biological sequences. Nucleic Acids Res. 2022, 50, W420–W426. [Google Scholar] [CrossRef]
- Lee, I.; Ouk Kim, Y.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef] [PubMed]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363, fnw002. [Google Scholar] [CrossRef]
- Dunne, M.; Hupfeld, M.; Klumpp, J.; Loessner, M.J. Molecular Basis of Bacterial Host Interactions by Gram-Positive Targeting Bacteriophages. Viruses 2018, 10, 397. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Dover, J.A.; Parent, K.N.; Doore, S.M. Host Range Expansion of Shigella Phage Sf6 Evolves through Point Mutations in the Tailspike. J. Virol. 2022, 96, e0092922. [Google Scholar] [CrossRef]
- Baxa, U.; Steinbacher, S.; Weintraub, A.; Huber, R.; Seckler, R. Mutations improving the folding of phage P22 tailspike protein affect its receptor binding activity. J. Mol. Biol. 1999, 293, 693–701. [Google Scholar] [CrossRef]
- Prokhorov, N.S.; Riccio, C.; Zdorovenko, E.L.; Shneider, M.M.; Browning, C.; Knirel, Y.A.; Leiman, P.G.; Letarov, A.V. Function of bacteriophage G7C esterase tailspike in host cell adsorption. Mol. Microbiol. 2017, 105, 385–398. [Google Scholar] [CrossRef]
- Sørensen, A.N.; Woudstra, C.; Sørensen, M.C.H.; Brøndsted, L. Subtypes of tail spike proteins predicts the host range of Ackermannviridae phages. Comput. Struct. Biotechnol. J. 2021, 19, 4854–4867. [Google Scholar] [CrossRef] [PubMed]
- Plattner, M.; Shneider, M.M.; Arbatsky, N.P.; Shashkov, A.S.; Chizhov, A.O.; Nazarov, S.; Prokhorov, N.S.; Taylor, N.M.I.; Buth, S.A.; Gambino, M.; et al. Structure and Function of the Branched Receptor-Binding Complex of Bacteriophage CBA120. J. Mol. Biol. 2019, 431, 3718–3739. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-J.; Dai, N.; Müller, S.I.; Guan, C.; Parker, M.J.; Fraser, M.E.; Walsh, S.E.; Sridar, J.; Mulholland, A.; Nayak, K.; et al. Pathways of thymidine hypermodification. Nucleic Acids Res. 2021, 50, 3001–3017. [Google Scholar] [CrossRef]
- Grimont, P.; Weill, F.-X. Antigenic Formulae of the Salmonella serovars, 9th ed.; WHO Collaborating Centre for Reference and Research on Salmonella: Paris, France, 2007; pp. 1–166. [Google Scholar]
- Sood, L.R.; Basu, S. Bacteriophage typing of Salmonella weltevreden. Antonie Leeuwenhoek 1979, 45, 595–604. [Google Scholar] [CrossRef]
- Sood, L.R.; Basu, S. Sub-classification of phage types of Salmonella weltevreden and typing of strains by an extended phage typing scheme. Antonie Leeuwenhoek 1980, 46, 467–473. [Google Scholar] [CrossRef]
- Sood, L.R.; Basu, S. Phage-typing of Salmonella weltevreden based on lysogeny II. Epidemiological usefulness of the system and geographical distribution of its phage-types. Antonie Leeuwenhoek 1977, 43, 262–268. [Google Scholar] [CrossRef]
- Lekunberri, I.; Subirats, J.; Borrego, C.M.; Balcázar, J.L. Exploring the contribution of bacteriophages to antibiotic resistance. Environ. Pollut. 2017, 220, 981–984. [Google Scholar] [CrossRef]
- Colombo, S.; Arioli, S.; Neri, E.; Scala, G.; Gargari, G.; Mora, D. Viromes As Genetic Reservoir for the Microbial Communities in Aquatic Environments: A Focus on Antimicrobial-Resistance Genes. Front. Microbiol. 2017, 8, 1095. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.-X.; Huang, D.; Ji, J.-H.; Völker, C.; Wurm, F.R. Seawater-Degradable Polymers—Fighting the Marine Plastic Pollution. Adv. Sci. 2021, 8, 2001121. [Google Scholar] [CrossRef]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Oechslin, F. Resistance Development to Bacteriophages Occurring during Bacteriophage Therapy. Viruses 2018, 10, 351. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhang, L.; Feng, C.; Chi, T.; Qi, Y.; Abbas Raza, S.H.; Gao, N.; Jia, K.; Zhang, Y.; Fan, R.; et al. A phage cocktail in controlling phage resistance development in multidrug resistant Aeromonas hydrophila with great therapeutic potential. Microb. Pathog. 2022, 162, 105374. [Google Scholar] [CrossRef] [PubMed]
- Gordillo Altamirano, F.L.; Barr, J.J. Unlocking the next generation of phage therapy: The key is in the receptors. Curr. Opin. Biotechnol. 2021, 68, 115–123. [Google Scholar] [CrossRef]
- Fischer, S.; Kittler, S.; Klein, G.; Glünder, G. Impact of a single phage and a phage cocktail application in broilers on reduction of Campylobacter jejuni and development of resistance. PLoS ONE 2013, 8, e78543. [Google Scholar] [CrossRef]
- Adriaenssens, E.M.; Van Vaerenbergh, J.; Vandenheuvel, D.; Dunon, V.; Ceyssens, P.J.; De Proft, M.; Kropinski, A.M.; Noben, J.P.; Maes, M.; Lavigne, R. T4-related bacteriophage LIMEstone isolates for the control of soft rot on potato caused by ‘Dickeya solani’. PLoS ONE 2012, 7, e33227. [Google Scholar] [CrossRef]
- Anany, H.; Lingohr, E.J.; Villegas, A.; Ackermann, H.W.; She, Y.M.; Griffiths, M.W.; Kropinski, A.M. A Shigella boydii bacteriophage which resembles Salmonella phage ViI. Virol. J. 2011, 8, 242. [Google Scholar] [CrossRef]
- Adriaenssens, E.M.; Ackermann, H.W.; Anany, H.; Blasdel, B.; Connerton, I.F.; Goulding, D.; Griffiths, M.W.; Hooton, S.P.; Kutter, E.M.; Kropinski, A.M.; et al. A suggested new bacteriophage genus: “Viunalikevirus”. Arch. Virol. 2012, 157, 2035–2046. [Google Scholar] [CrossRef]
- Fan, C.; Tie, D.; Sun, Y.; Jiang, J.; Huang, H.; Gong, Y.; Zhao, C. Characterization and Genomic Analysis of Escherichia coli O157:H7 Bacteriophage FEC14, a New Member of Genus Kuttervirus. Curr. Microbiol. 2021, 78, 159–166. [Google Scholar] [CrossRef]
- Walter, M.; Fiedler, C.; Grassl, R.; Biebl, M.; Rachel, R.; Hermo-Parrado, X.L.; Llamas-Saiz, A.L.; Seckler, R.; Miller, S.; van Raaij, M.J. Structure of the receptor-binding protein of bacteriophage det7: A podoviral tail spike in a myovirus. J. Virol. 2008, 82, 2265–2273. [Google Scholar] [CrossRef]
- Steinbacher, S.; Baxa, U.; Miller, S.; Weintraub, A.; Seckler, R.; Huber, R. Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors. Proc. Natl. Acad. Sci. USA 1996, 93, 10584–10588. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Bales, P.; Greenfield, J.; Heselpoth, R.D.; Nelson, D.C.; Herzberg, O. Crystal structure of ORF210 from E. coli O157:H1 phage CBA120 (TSP1), a putative tailspike protein. PLoS ONE 2014, 9, e93156. [Google Scholar] [CrossRef]
- Greenberg, M.; Dunlap, J.; Villafane, R. Identification of the Tailspike Protein from the Salmonella newington Phage ϵ34 and Partial Characterization of Its Phage-Associated Properties. J. Struct. Biol. 1995, 115, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Uetake, H. In vitro interaction between phage and receptor lipopolysaccharide: A novel glycosidase associated with Salmonella phage 15. Virology 1973, 52, 148–159. [Google Scholar] [CrossRef] [PubMed]
- El-Dougdoug, N.K.; Cucic, S.; Abdelhamid, A.G.; Brovko, L.; Kropinski, A.M.; Griffiths, M.W.; Anany, H. Control of Salmonella Newport on cherry tomato using a cocktail of lytic bacteriophages. Int. J. Food Microbiol. 2019, 293, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Petsong, K.; Benjakul, S.; Vongkamjan, K. Evaluation of storage conditions and efficiency of a novel microencapsulated Salmonella phage cocktail for controlling S. enteritidis and S. typhimurium in-vitro and in fresh foods. Food Microbiol. 2019, 83, 167–174. [Google Scholar] [CrossRef]
- Petsong, K.; Benjakul, S.; Chaturongakul, S.; Switt, A.I.M.; Vongkamjan, K. Lysis Profiles of Salmonella Phages on Salmonella Isolates from Various Sources and Efficiency of a Phage Cocktail against S. enteritidis and S. typhimurium. Microorganisms 2019, 7, 100. [Google Scholar] [CrossRef]




| Phages | Genus | OrthoANI |
|---|---|---|
| Salmonella phage P46FS4 | Agtrevirus | 96.04 |
| Shigella phage MK-13 | Agtrevirus | 95.96 |
| Shigella phage Ag3 | Agtrevirus | 95.84 |
| Salmonella phage SKML-39 | Agtrevirus | 95.57 |
| Dickeya phage vB-DsoM-LIMEstone1 | Limestonevirus | 88.24 |
| Dickeya phage Ds23CZ | Limestonevirus | 88.21 |
| Dickeya phage Ds3CZ | Limestonevirus | 88.16 |
| Dickeya phage Ds5CZ | Limestonevirus | 88.16 |
| Dickeya phage PP35 | Limestonevirus | 88.12 |
| Dickeya phage Ds9CZ | Limestonevirus | 88.11 |
| Dickeya phage Ds20CZ | Limestonevirus | 88.10 |
| Dickeya phage XF4 | Limestonevirus | 88.04 |
| Dickeya phage Kamild | Limestonevirus | 88.04 |
| Dickeya phage JA15 | Limestonevirus | 88.02 |
| Dickeya phage Ds16CZ | Limestonevirus | 88.01 |
| Dickeya phage RC-2014 | Limestonevirus | 87.86 |
| Dickeya phage Ds25CZ | Limestonevirus | 87.63 |
| Dickeya phage Coodle | Limestonevirus | 87.16 |
| Salmonella phage SFP10 | Kuttervirus | 78.91 |
| Salmonella phage Sh19 | Kuttervirus | 78.43 |
| Escherichia phage vB_EcoA_4HA11 | Kuttervirus | 76.97 |
| Salmonella phage pertopsoe | Kuttervirus | 76.65 |
| Salmonella phage allotria | Kuttervirus | 76.52 |
| Salmonella phage vB_SenM_SilasIsHot | Kuttervirus | 76.44 |
| Salmonella phage maane | Kuttervirus | 76.22 |
| Salmonella phage aagejoakim | Kuttervirus | 75.47 |
| Salmonella phage Se_EM4 | Kuttervirus | 75.42 |
| Salmonella phage S115 | Kuttervirus | 75.23 |
| Salmonella phage SenASZ3 | Kuttervirus | 75.14 |
| Salmonella phage Se_AO1 | Kuttervirus | 75.13 |
| Salmonella phage SE14 | Kuttervirus | 75.06 |
| Salmonella phage Se-J | Kuttervirus | 75.05 |
| Salmonella phage S118 | Kuttervirus | 75.01 |
| Salmonella phage STP07 | Kuttervirus | 75.00 |
| Salmonella phage dinky | Kuttervirus | 74.98 |
| Escherichia phage FEC14 | Kuttervirus | 74.98 |
| Salmonella phage kage | Kuttervirus | 74.96 |
| Salmonella phage GG32 | Kuttervirus | 74.96 |
| Salmonella phage S117 | Kuttervirus | 74.90 |
| Salmonella phage Se_EM1 | Kuttervirus | 74.86 |
| Salmonella phage vB_SenAc_BPS6 | Kuttervirus | 74.78 |
| Salmonella phage vB-SalM-SJ3 | Kuttervirus | 74.77 |
| Escherichia phage PhaxI | Kuttervirus | 74.76 |
| Salmonella phage SenALZ1 | Kuttervirus | 74.74 |
| Escherichia phage CBA120 | Kuttervirus | 74.68 |
| Salmonella phage SP1 | Kuttervirus | 74.67 |
| Salmonella phage vB_SenAc_BPS3 | Kuttervirus | 74.63 |
| Salmonella phage Se-G | Kuttervirus | 74.61 |
| Salmonella phage ST-W77 | Kuttervirus | 74.59 |
| Salmonella phage ISTP3 | Kuttervirus | 74.56 |
| Salmonella phage Se-B | Kuttervirus | 74.55 |
| Salmonella phage moki | Kuttervirus | 74.54 |
| Salmonella phage Se_EM3 | Kuttervirus | 74.47 |
| Salmonella Serovar | Group | No. of Strains Tested | No. of Lysed Strains |
|---|---|---|---|
| S. Weltevreden | Group O:3,10 | 11 | 11 |
| S. Muenster | Group O:3,10 | 11 | 11 |
| S. London | Group O:3,10 | 11 | 9 |
| S. Meleagridis | Group O:3,10 | 7 | 6 |
| S. Give | Group O:3,10 | 6 | 5 |
| S. Anatum | Group O:3,10 | 5 | 4 |
| S. Derby | Group O:4 | 17 | 2 |
| S. Stanley | Group O:4 | 16 | 1 |
| S. Typhimurium | Group O:4 | 16 | 1 |
| S. Enteritidis | Group O:9 | 15 | 0 |
| S. Rissen | Group O:7 | 16 | 1 |
| S. Thompson | Group O:7 | 3 | 1 |
| S. Goldcoast | Groups O:6,8 | 14 | 2 |
| S. Hadar | Groups O:6,8 | 2 | 1 |
| S. Corvallis | Groups O:8 | 16 | 1 |
| S. Kentucky | Groups O:8 | 15 | 1 |
| S. Hillingdon | Group O:9,46 | 1 | 1 |
| S. Gateshead | Group O:9,46 | 1 | 1 |
| S. Michigan | Group O:17 | 1 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Li, J.; Yan, Y.; Han, P.; Tong, Y.; Li, X. SW16-7, a Novel Ackermannviridae Bacteriophage with Highly Effective Lytic Activity Targets Salmonella enterica Serovar Weltevreden. Microorganisms 2023, 11, 2090. https://doi.org/10.3390/microorganisms11082090
Xu J, Li J, Yan Y, Han P, Tong Y, Li X. SW16-7, a Novel Ackermannviridae Bacteriophage with Highly Effective Lytic Activity Targets Salmonella enterica Serovar Weltevreden. Microorganisms. 2023; 11(8):2090. https://doi.org/10.3390/microorganisms11082090
Chicago/Turabian StyleXu, Jialiang, Jia Li, Yi Yan, Pengjun Han, Yigang Tong, and Xu Li. 2023. "SW16-7, a Novel Ackermannviridae Bacteriophage with Highly Effective Lytic Activity Targets Salmonella enterica Serovar Weltevreden" Microorganisms 11, no. 8: 2090. https://doi.org/10.3390/microorganisms11082090
APA StyleXu, J., Li, J., Yan, Y., Han, P., Tong, Y., & Li, X. (2023). SW16-7, a Novel Ackermannviridae Bacteriophage with Highly Effective Lytic Activity Targets Salmonella enterica Serovar Weltevreden. Microorganisms, 11(8), 2090. https://doi.org/10.3390/microorganisms11082090






