Abstract
Forty-four bacterial strains isolated from greenhouse soil and beetroots were tested for their antagonistic activity against the plant-parasitic root-knot nematode (RKN) Meloidogyne incognita, which causes significant yield losses in a number of important crops worldwide. Through a novel combination of in vitro and on planta screening assays, Pseudomonas spp. 105 and 108 were identified as the most promising bacterial isolates. Both strains were evaluated for their potential to control different RKN population densities and as root protectants against nematode infestation. Regardless of the application method, both strains significantly reduced root galling caused by M. incognita. These two strains were subjected to whole genome sequencing and de novo genome assembly as a basis for phylogenetic and future functional characterization. Phylogenetic analysis revealed that both Pseudomonas strains cluster within the Pseudomonas fluorescens clade among previously characterized RKN antagonists and Pseudomonas-based biocontrol agents of plant diseases.
1. Introduction
The plant-parasitic nematode Meloidogyne incognita is one of the most common root-knot nematode (RKN) species worldwide [1,2]. This group of obligate, sedentary plant parasites has a wide host range and can cause significant damage to several important agricultural and greenhouse crops, monocots, and dicots [3,4]. M. incognita infections can cause reduced plant growth, stunting, leaf discoloration, and even complete crop loss. The current estimate of the global yield losses caused by plant-parasitic nematodes places the annual losses in the range of billions of dollars [5], higher than those estimated for insects [6,7,8] and accounting for 14% of the total global crop losses [9].
RKN can reproduce both sexually (amphimixis) and asexually (mitotic and meiotic parthenogenesis). During their life cycle, second-stage juveniles (J2) enter the root tip near the elongation zone and migrate intercellularly to establish a permanent feeding site by inducing the formation of giant cells in the vascular cylinder [10]. While depleting plant nutrients and causing root galling, the juveniles molt and develop a swollen, pear-shaped appearance. Mature female nematodes lay their eggs in a gelatinous matrix outside the root system, allowing the J2 to hatch freely in the soil and find new root tips for propagation. Apart from the adult males, only the infective J2 are mobile and can move in the soil after hatching (pre-parasitic stage). During this exophytic phase, RKN spend a long time in the soil before infecting a host root [11].
The exophytic phase is important for both the synthetic chemical and biological control of RKN. For example, soil fumigation has often been used as a rapid, “reliable” means of controlling RKN in soil [12]. Several nematicides are contact nematicides and are applied outside the vegetative period as they can negatively affect seed germination and plant growth. Due to human health and environmental safety concerns, most chemical nematicides are no longer authorized or are strictly regulated in Switzerland and most other countries worldwide [12,13].
In biological control, J2 nematodes in the soil can be antagonized by predators, endoparasites, egg parasites, hyperparasites, and microorganisms in general [14,15]. Such microorganisms have been described to confer nematode suppressiveness to soils [16] and may consist of one or a complex of antagonistic species [17]. Fungal species (e.g., Dactylellina spp., Drechslerella spp., or Arthrobotrys spp.) [18] and bacterial species (e.g., Arthrobacter spp., Burkholderia spp., Lysobacter spp., Pasteuria spp., Pseudomonas spp., Rhizobium spp., and Streptomyces spp.) [19] have also been described to control plant-parasitic nematodes. The genus Pasteuria, which is known to attach to the cuticles of nematodes using an endospore, has been intensively studied as a potential bacterial antagonist of nematodes [20]. Pseudomonas spp. are also widely exploited as antagonistic microorganisms. Some Pseudomonas species are able to utilize plant root exudates for nutritional purposes and produce protective metabolites against pests and pathogens, as well as antimicrobial compounds known to control plant-parasitic nematodes [21,22]. In vitro and on planta studies have shown that Pseudomonas cell suspensions or supernatants can be successfully used to control RKN such as M. incognita and that their RKN control can be transferred to greenhouse conditions [23,24,25].
Pseudomonas spp. are commonly found in natural environments, colonize soil and plant root systems, and are easily maintained and cultured in the laboratory [26,27].
Screening for additional/alternative RKN antagonistic bacteria in the soil rhizosphere may be a promising method for the integrated management of plant-parasitic nematodes. Evaluating the potential of these isolates as RKN antagonists in a screening system that mimics the natural infection process of nematodes could improve the search for potential antagonists and allow the identification of particularly promising isolates.
Therefore, the objectives of this research were to (1) uncover a rapid and reliable method for the screening of nematicidal bacteria, in this proof-of-principle study for potential antagonists of M. incognita; (2) evaluate the selected strains for their nematicidal activity in the greenhouse; and (3) compare the phylogenetic relationships of promising antagonist strains with previously characterized Meloidogyne spp. antagonists to better select and understand potential candidates for RKN control in future trials.
2. Materials and Methods
2.1. Rearing and Collection of Meloidogyne incognita Second-Stage Juveniles
M. incognita was propagated on tomato (Solanum lycopersicum) cv. Oskar under greenhouse conditions (25 °C/19 °C, 60% humidity, 15/9 h light/dark cycle). Second-stage juveniles (J2) were extracted from heavily galled root systems using a mist chamber (23 °C) [28]. Hatched J2 were collected daily and stored at 6 °C for 3–7 days until used in experiments. Total DNA was extracted periodically for barcoding analysis according to Kiewnick et al. [29], to ensure M. incognita identity.
2.2. Bacterial Isolation and Culture Preparation
Beetroot (Beta vulgaris subsp. vulgaris, variety “Pablo”, grown in the greenhouse at Agroscope-Wädenswil (CH)) was harvested, the soil was removed, and small parts of the roots were resuspended in phosphate buffer (15 mM K2HPO4 and 9 mM KH2PO4, pH 7). After 10-fold serial dilution (up to 10−3), 100 µL of the respective dilution steps were plated on tryptic soy broth (TSB, Oxoid/Thermo Fisher Scientific, Waltham, MA, USA) agar plates (15 g/L Agar-Agar, Kobe I (Carl Roth, Karlsruhe, Germany)). Plates were incubated at 26 °C for 24 h; single colonies were picked and subcultured to ensure pure cultures. Bacterial isolates were identified at the genus level with MALDI-TOF by smearing bacterial cell material from a colony on agar plate onto a MALDI targe for the identification of specific biomarker peptides [30] and 16 S rRNA gene sequencing. Forty-four bacterial isolates (Supplementary Table S1) were then tested for their antagonistic potential against M. incognita.
2.3. Antagonistic Screening against Meloidogyne incognita
The bacterial isolates were freshly prepared on TSB (Oxoid/Thermo Fisher Scientific, Waltham, MA, USA) agar plates (15 g/L Agar-Agar, Kobe I (Carl Roth)). Each bacterial strain was cultured overnight at 26 °C in a 250 mL Erlenmeyer flask containing 100 mL TSB medium. Liquid cultures were centrifuged at 6000 rcf for 10 min at room temperature, supernatant decanted, and bacterial pellet resuspended in autoclaved tap water to a final optical density (OD600nm) of 0.5.
Nematode:bacteria mixtures were prepared by pipetting 6 mL of a water suspension containing 250 M. incognita J2 into a 25 mL plastic cup and adding 4 mL of the freshly prepared bacterial suspension to a final volume of 10 mL. The nematode:bacteria suspensions were incubated for three days at 20 °C in the dark. Thereafter, 20 mL of steamed soil:silver sand mixture (1:3, v/v) was added to the cups containing the nematode:bacteria suspensions. A three-day-old germinated cucumber seedling (Cucumis sativus cv. Sprinter F1) was then planted in each cup and grown in a climate chamber at 24 °C, 60% humidity, and a 16/8 h light/dark cycle and watered as needed (n = 3). After three weeks, cucumber roots were washed free of soil, and root gall formation caused by viable M. incognita J2 was determined according to Zeck [31]. After the initial screening, the same experimental setup was repeated for the pre-selected bacterial strains with nematicidal activity.
2.4. In Vitro Screening of Selected Bacterial Strains against Meloidogyne incognita Second-Stage Juveniles
The ability of the selected bacterial strains and the negative control Pseudomonas orientalis F9 [32] to inhibit M. incognita J2 was tested in vitro using 24-well plates (n = 3). Bacteria were processed as described above but adjusted to a final OD600 of 1.0. Each of the 24 wells was filled with 1 mL of a nematode suspension containing approximately 150 J2 and 1 mL of the OD600 = 1.0 bacterial suspension (OD600 of 0.5).
For the exposure of the J2 to the bacterial supernatant, the supernatant was filtered through a 0.2 μm syringe filter and 1 mL was added to a suspension containing 150 J2. The well plates were kept at 20 °C in the dark. For each well, the motility of the first 100 J2 individuals was scored under a light microscope as normal motility, affected motility, or immotile (elongated shape) [33].
2.5. Testing of Pre-Selected Antagonistic Pseudomonas Strains under Soil Conditions
The nematicidal activity of the bacterial isolates showing promising effects in vitro and on planta, as well as the control P. orientalis F9, was further tested under soil conditions in a greenhouse using cucumber plants as indicator plants. Pots (∅ = 14 cm) filled with 750 mL of steamed soil:silver sand mixture (1:3, v/v) were inoculated with ca. 4000 J2. Three days after nematode inoculation, 25 mL bacterial suspensions adjusted to an OD600 of 0.5 were prepared as described above and added to the nematode-infested soil, except for the bacteria-free controls (n = 8). The pots were then covered with plastic foil and kept moist. Seven days after the application of the bacteria, 3-week-old cucumber plants were planted in the soil. The treatments were arranged in a randomized block design in the greenhouse and kept at 25 °C/19 °C, 60% humidity, and a 15/9 h light/dark cycle. Plants were watered according to their needs. After 4 weeks, the plants were uprooted and washed with tap water to remove soil for root gall rating [31].
The experimental setup was repeated for the selected bacterial strains (n = 7), now using 4-week-old tomato cv. Moneymaker plantlets as indicator plants and testing the bacterial suspensions at OD600 = 0.5 and 1.5.
2.6. Evaluation of the Biocontrol Efficacy of Top Pseudomonas Isolates against Meloidogyne incognita by Application to Mineral Wool Plant Starting Plugs
Grodan SBS mineral wool plugs (SBS 36/77, Grodan, Roermond, The Netherlands) were used to grow cucumber seedlings. After 14 days of growth, the plugs were soaked overnight with 10 mL of selected bacterial suspensions (OD600 of 0.5). Bacteria-soaked cucumber plugs were then planted in 14 cm ∅ pots. Three days earlier, the soil:silver sand mixture of these pots had been inoculated with approximately 4000 M. incognita J2. Each treatment (selected Pseudomonas strains and control, P. orientalis F9) was replicated seven times (n = 7). Plants were arranged in a randomized block design in the greenhouse and grown at 25 °C/19 °C, 60% humidity, and a 15/9 h light/dark cycle. After four weeks, plants were uprooted and washed with tap water to remove substrates for root gall rating [31].
2.7. Evaluation of the Biocontrol Efficacy of Selected Pseudomonas Strains against Different Population Densities of Meloidogyne incognita
The top Pseudomonas strains were tested at low (2000 J2) and high (8000 J2) M. incognita soil concentrations (n = 8 for each treatment and nematode concentration, including 8 untreated control plants). Pots (∅ = 14 cm) were prepared with steamed soil:silver sand mixture (1:3 v/v) inoculated with J2. Bacterial suspensions were prepared as described above, but resuspended to a final OD600 of 1.5. Tomato cv. Moneymaker seedlings grown in transplanting trays for 4 weeks were planted in infected pots and arranged in a randomized block design. Tomato growth was measured weekly. Forty days after planting, root galling was evaluated according to Zeck [31].
2.8. Genomic DNA Extraction, Sequencing, De Novo Assembly, and Annotation
Total DNA was isolated from bacterial cells grown in TSB overnight according to [34]. The quality and quantity of the extracted DNA was assessed on a 0.8% (w/v) agarose gel, followed by the Qubit dsDNA GR assay (Life Technologies, Thermo Fisher Scientific, Waltham, MA, USA).
Both Illumina short-read and Oxford Nanopore Technologies long-read sequencing technologies were used. Illumina paired-end reads were trimmed with Trimmomatic v0.39 [35] (parameters: -phred 33 leading:3 trailing:3 sildingwindow:4:15 minlen:36), using FastQC v0.11.9 [36] to check the read quality before and after trimming. Oxford Nanopore raw sequence reads were filtered according to their quality and length using Filtlong v0.2.0 (https://github.com/rrwick/Filtlong, accessed on 29 July 2019). The filtered reads were de novo assembled using Flye v2.9.1-b1780 [37] with default parameters; the dnaA gene was chosen as the start position of the assembly. Assemblies were first polished with long reads using Medaka (https://github.com/nanoporetech/medaka, accessed on 7 October 2021) and then with trimmed Illumina reads and Freebayes v.1.3.2 [38] (minimum alternate fraction, 0.5; minimum alternate count, 5). Variants were manually inspected in Integrated Genome Viewer (IGV) [39] and subsequently corrected using BCFtools v.1.10.2 [40] to remove any potentially remaining sequencing errors. PlasmidSpades v3.13.1 [41] was run to assemble the Illumina short reads to detect short plasmids that may have been missed in the long-read-based assemblies. To verify the circularity and completeness of the de novo assemblies, long and short reads were mapped to each assembly using Minimap2 [42] and BWA-MEM v0.7.17 [43], respectively. Alignments were manually inspected using IGV. The mapping quality of reads was assessed using Qualimap v.2.2.1 [44]. The completeness of the final assembly was further assessed using BUSCO v5.0.0 [45]. As a final QC step, an in-house prototype for the detection of repeats was run to identify large repeats, some of which could indicate misassembled regions [46].
Finally, the finished genomes were annotated using the NCBI’s Prokaryotic Genome Annotation Pipeline (PGAP) [47].
2.9. Phylogenetic Analysis
A phylogenetic tree based on 70 Pseudomonas strains, including the two de novo assembled Pseudomonas strains 105 and 108, was calculated based on 16 core genes (rpsJ, glnK, rplK, infA, rpsE, rplV, rplP, rpsG, rpsC, rpsR, rpsL, mreB, ihfA, rpsK, rplN, rpsU) identified by Page et al. [48]. Multi-sequence alignment of all genes was performed with the ClustalW algorithm [49] using MEGA-CC [50], and a phylogenetic tree was inferred using the Maximum Likelihood method with MEGA-CC and 100 bootstrapping iterations. The 68 strains were selected based on a literature search of available Pseudomonas genomes with putative activity against nematodes, a previous study of Pseudomonas isolates with biocontrol potential that placed them in the context of known Pseudomonas subclades [51], and the five closest genomes of the two isolates reported here (based on sequence identity as estimated by Mash [52] of all publicly available strains on NCBI). All sequences were downloaded from the NCBI database.
2.10. Data Analysis
Statistical analyses were performed using the SPSS software, data were tested for homogeneity of variances (Levene’s test), and treatments were distinguished by one-way ANOVA with Tukey’s Honestly Significant Difference (HSD) post-hoc test (p ≤ 0.05). Results of the in vitro assay using cells and supernatant were expressed as percentages. Significant differences were calculated by comparing data from J2 with normal motility.
3. Results and Discussion
3.1. Identification of Bacteria with Antagonistic Activity against Meloidogyne incognita
Screening of Bacterial Strains with Potential Antagonistic Activity against Root-Knot Nematodes
The antagonistic potential of 44 bacterial strains (Supplementary Table S1) against M. incognita was tested with a newly developed method using a combined “in vitro” on-plant cucumber assay (Figure 1). The screening method was designed to ensure that the J2 were exposed to the bacteria in an aqueous solution and also had the opportunity to infect a host plant. Therefore, in this method, an inhibitory effect of the tested bacteria on the J2 nematodes results in a reduction in or even the absence of root gall formation in the indicator plants. Thus, the effect of the bacteria on J2 plant infectivity can be assessed even if the J2 are visually unaffected. This is an advantage over studies with RKN antagonists, which rely solely on a visual assessment of the viability of the nematodes [53,54,55].
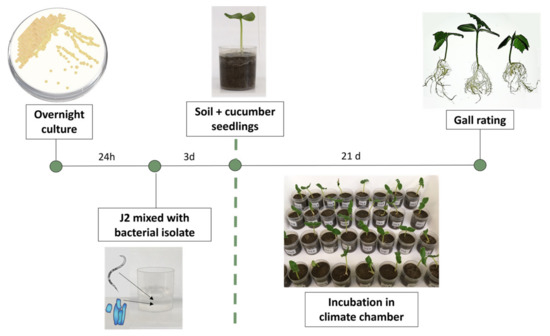
Figure 1.
Schematic representation of the bacterial screening system against M. incognita second-stage juveniles (J2) using cucumber seedlings. Suspensions of the potential bacterial antagonists were prepared and mixed in a cup with 250 J2 in an aqueous solution. Nematodes and potential antagonists were incubated for 3 days before soil was added to the cup and a pre-germinated cucumber seedling was planted. After 21 days, cucumber roots were washed free of substrate and root galls were scored according to Zeck’s [31] 0–10 scale, where 0 indicates no galls and 10 indicates dead roots.
Preliminary characterization revealed that most of the bacterial isolates tested belonged to the genus Pseudomonas (Supplementary Table S1). P. orientalis F9 was included in the study to assess its activity against nematodes, as this strain is known to have antagonistic potential against the fire blight pathogen Erwinia amylovora and also against the oomycete Pythium ultimum [32].
The antagonistic activity of the selected bacteria was determined by the root gall index recorded on cucumber roots caused by M. incognita J2 exposed to the selected bacterial strains and compared to an infection control, in which no bacterial antagonists were added to the J2-infected soil (Figure 2). The most promising antagonists of M. incognita, i.e., strains 102, 105, 108, 112, 119, and 157, caused a twofold or greater reduction in the galling index.
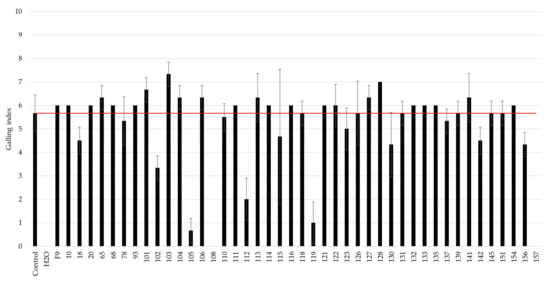
Figure 2.
Screening of bacterial strains against M. incognita using an in vitro + cucumber bioassay. The galling index was scored according to Zeck’s [31] 0–10 scale, where 0 indicates no galls and 10 indicates dead roots, caused by M. incognita. The red line indicates root galling caused by M. incognita-untreated control, J2-infected soil without addition of bacterial strains. H20 represents the negative control where no nematodes or bacteria were added to the cucumber seedlings. Error bars represent the standard deviations of the three replicates (n = 3) per treatment.
Pseudomonas strains (102, 105, 108, 112, 119, and 157) were retested for their ability to antagonize RKN (Figure 3). Based on the results of the initial screening, P. orientalis F9 and isolate 113 were added as negative controls for bacterial antagonism since neither strain showed inhibitory effects on RKN-induced galling (Figure 2).
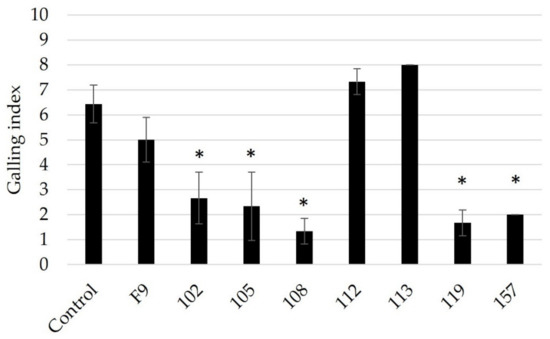
Figure 3.
Testing of pre-selected Pseudomonas strains 102, 105, 108, 112, 119, and 157 against M. incognita using an in vitro + cucumber bioassay (n = 3). Infection control, only nematodes. Negative control for bacterial antagonism, P. orientalis F9 and isolate 113. Root gall index was scored according to Zeck’s [31] 0–10 scale, where 0 indicates no galls and 10 indicates dead roots. Error bars represent standard deviations of three replicates (n = 3) per treatment. Significant differences are indicated by an asterisk, calculated by a one-way ANOVA with post-hoc Tukey HSD test.
With the exception of strain 112, strains 102, 105, 108, 119, and 157 repeatedly showed RKN antagonistic activity, while P. orientalis F9 and isolate 113 again had no significant effect on J2 (Figure 3). The results confirmed the reproducibility of the data obtained in the cucumber assay.
Bacterial cells and sterile filtered supernatants of Pseudomonas strains 102, 105, 108, 112, 119, and 157 were then tested against J2 in a pure in vitro assay to compare the antagonistic effects that occurred with the results obtained from the cucumber experiments. The 7-day in vitro assay showed that, for all strains, bacterial cells added to a J2 suspension significantly inhibited the nematodes after 1 day (Figure 4). However, 4 days (strains F9 and 113) or 7 days (strains F9, 112, and 113) after the application of the cell suspension, the J2 recovered and showed no significant differences compared to the water control. These results were even more pronounced when the supernatant of these strains was tested (Figure 4). Accordingly, the results support the notion that visual screening at a too early time point may give a misleading indication of RKN control in soil, as was observed with P. orientalis F9 and isolates 112 and 113. These negative controls in the cucumber assay significantly inhibited M. incognita J2 after one day when applied either as a cell suspension or its corresponding supernatant (Figure 4). In the literature, similar RKN in vitro assays using Pseudomonas spp. are typically evaluated after 12, 24, 48, and/or 72 h [56,57], and therefore some important information may be overlooked compared to the newly developed screening assay outlined in Figure 1.
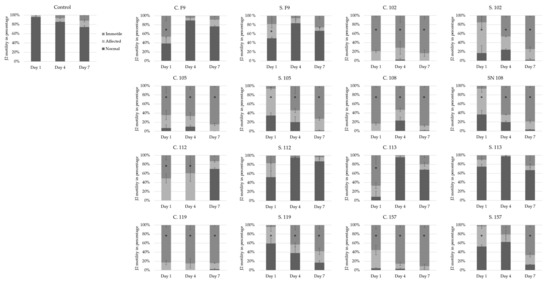
Figure 4.
In vitro effect of different Pseudomonas strains applied as cells (C) or supernatant (S) on the motility of M. incognita second-stage juveniles (J2) after 1, 4, and 7 days. J2 motility was recorded according to normal motility, affected, or immotile nematodes [33]. Error bars represent standard deviations of replicates. * Significantly affected or immotile nematodes in percent (%) relative to the control, calculated using a one-way ANOVA with post-hoc Tukey HSD test (n = 3).
3.2. Testing of Promising Meloidogyne incognita Bacterial Antagonists under Greenhouse Conditions
Pseudomonas strains 102, 105, 108, 119, and 157 were evaluated for their potential to control RKN under soil conditions. P. orientalis F9 was selected as a negative control for the soil experiment. The bacterial cultures were applied to M. incognita-infested soil and cucumbers were planted seven days after application.
Only cucumber roots grown for four weeks in nematode-infested soil and treated with the 105 or 108 strains showed a significant reduction in root galling compared to cucumbers grown in untreated soil or with other Pseudomonas strains (Figure 5). The results showed no significant effect on root gall formation for bacterial strains 102, 112, 119, and 157. It may be that the application method used reduced the RKN control performance of the strains tested, as suggested in previous publications [58].
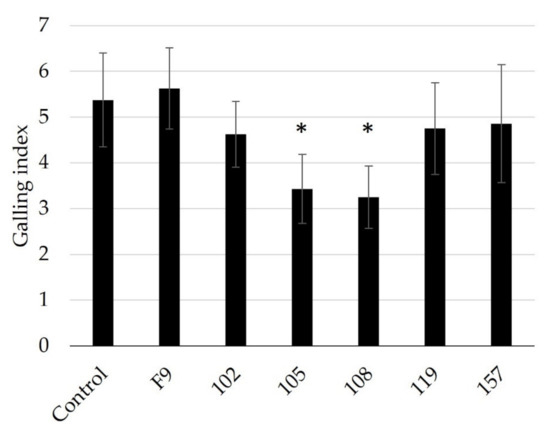
Figure 5.
Testing of selected Pseudomonas strains against the root-knot nematode M. incognita (4000 J2/pot) using cucumber seedlings grown in test soil for four weeks (n = 8). Root gall index was scored according to Zeck’s [31] 0–10 scale, where 0 indicates no galls and 10 indicates dead roots. Error bars represent standard deviations of replicates. Significant differences are indicated by an asterisk, calculated by one-way ANOVA with post-hoc Tukey HSD test.
The selected Pseudomonas strains 102, 105, 108, 119, and 157 were further tested as a root bale treatment. Cucumber seedlings grown on mineral wool plugs (Figure 6) were planted in M. incognita-infested soil. Prior to planting, the mineral wool was inoculated with the selected strains.
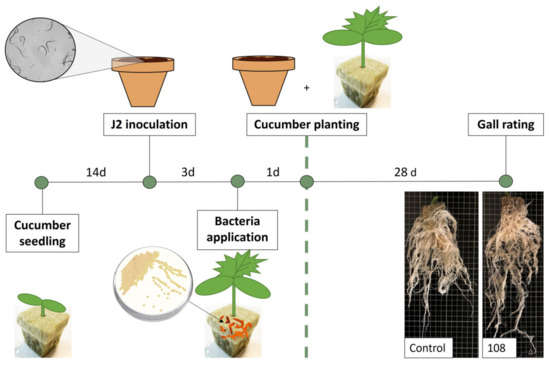
Figure 6.
Selected Pseudomonas strains were tested as root protectants against M. incognita J2 by applying bacterial suspensions to cucumber seedlings grown in mineral wool one day before planting in nematode-infested soil (4000 J2/pot). Cucumber plants were grown for 28 days and root galls were rated according to Zeck’s [31] 0–10 scale, where 0 indicates no galls and 10 indicates dead roots.
Pseudomonas strains 102, 105, 108, and 119 showed a significant reduction in root galling compared to control plants (Figure 7). As in the previous experiments, Pseudomonas strains 105 and 108 showed the strongest reduction in the root galling index. Pseudomonas spp. are known to colonize plant roots [26,57]; therefore, the bacterial treatment of the mineral wool may have promoted the bacterial colonization of the cucumber roots, resulting in protection against M. incognita infection.
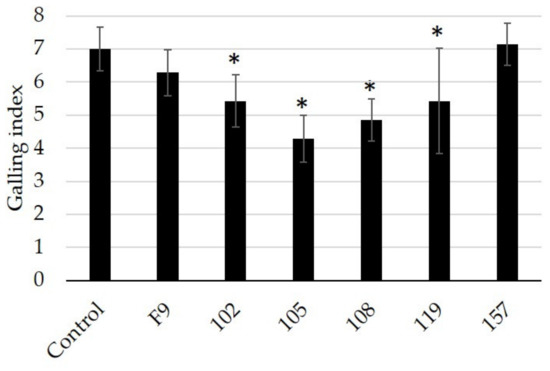
Figure 7.
Testing of selected Pseudomonas strains as protectants of cucumber roots (grown in mineral wool) against the root-knot nematode M. incognita (n = 8). Root galls were scored according to Zeck’s [31] 0–10 scale, where 0 indicates no galls and 10 indicates dead roots. Error bars represent standard deviations of replicates. Significant differences are indicated by an asterisk according to a one-way ANOVA with post-hoc Tukey HSD test.
The ability to control M. incognita J2 using tomato as an indicator plant was tested with the most promising isolates, Pseudomonas strains 105 and 108, and the negative control, P. orientalis F9. Two suspensions with an OD600 of 0.5 or 1.5 were compared (Table 1).

Table 1.
Evaluation of tomato root gall rating 4 weeks after transplanting into soil containing 4000 M. incognita J2/pot and treated with Pseudomonas strain F9, 105, or 108 at OD600 of 0.5 or 1.5.
Root gall formation in tomatoes treated with strain 105 or 108 was reduced regardless of the cell concentration, but a significant reduction in root galling was only achieved with the OD600 = 1.5 bacterial suspension.
Previous studies have shown that Pseudomonas fluorescens strain CHA0 responds specifically to plant species, age, and genotype when tested against M. incognita [59]. Therefore, we hypothesized that Pseudomonas strains 105 and 108 may share a similar trait as CHA0 and act somewhat differently on different crop plants.
To evaluate whether Pseudomonas strains 105 and 108 maintained their control effect at different nematode population densities, their effect was tested in pots inoculated with 2000 or 8000 J2/pot. Both strains showed control of RKN in soil. However, in soil with a higher nematode population density (8000 J2), only strain 105 significantly reduced root gall formation in tomato (Figure 8). Despite the fact that RKN control was significant in our experiments, the control effect was not as strong as, for example, that obtained for Pseudomonas simiae strain MB751. This strain promoted up to an 80% reduction in root galling from 6.3 to 1.2 [60]. However, when compared to the recent study by Zhao et al. [57], the control effect of strains 105 and 108 was stronger than that observed for Pseudomonas protegens strains Sneb1997 or Sneb2001.
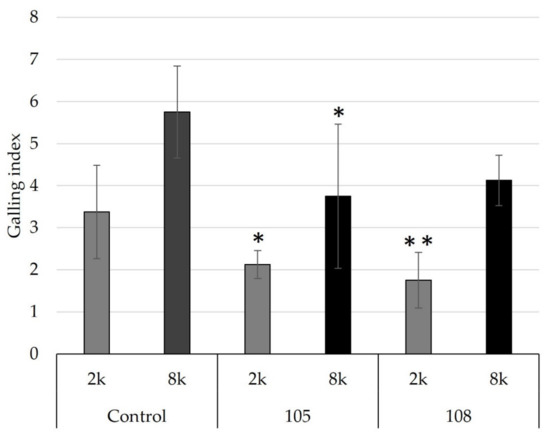
Figure 8.
Control efficacy of Pseudomonas strains 105 and 108 at low or high M. incognita population densities. Low (2k: 2000 J2/pot) or high (8k: 8000 J2/pot) RKN densities were tested, using tomato plants as indicators. The root gall index was determined according to Zeck’s [31] 0–10 scale, where 0 indicates no galls and 10 indicates dead roots. Error bars represent standard deviations of replicates. Significant differences are indicated by an asterisk according to a one-way ANOVA with post-hoc Tukey HSD test (** p < 0.01, * p < 0.05).
The beneficial effect of RKN control by Pseudomonas 105 and 108 strains was also observed on tomato plant height development under soils infested with low (2000 J2/pot) and high (8000 J2/pot) population densities of M. incognita (Supplementary Figure S1). The tomato plant height was lower for plants grown in M. incognita-infested soil than for plants grown in M. incognita-infested soil but treated with Pseudomonas 105 or 108 strains. However, plants performed best when the soil was not inoculated with M. incognita. Similar results were observed when the biological nematicide BioAct or the chemical nematicide fluopyram were applied, as neither nematicide was able to restore the same yield and plant growth as M. incognita-free soil [61]. Based on our experimental design, we cannot conclude that Pseudomonas strains 105 or 108 support plant growth and/or seed germination in the absence of nematodes, as reported for P. protegens [57].
3.3. Sequencing and De Novo Assembly of the Complete Genome of Pseudomonas 105 and 108
To create an optimal basis for the phylogenetic placement of the strains (see below) and for future functional genomics studies to uncover potential mechanisms of action, the complete genomes of both strains were sequenced and de novo assembled. Motivated by recent studies that have demonstrated that Illumina short-read-based genome assemblies can lack important genes that may underlie antagonistic activity, such as non-ribosomal peptide synthetases, phenazine biosynthesis genes, and type six secretion system effectors [62], a combination of long reads (Oxford Nanopore Technologies) and short reads (Illumina) was used (see Methods). Illumina reads were mainly used for polishing and to assemble potential plasmids. The complete genomes of the strains consisted of one chromosome and one plasmid, respectively, and were subsequently annotated with a local installation of the NCBI’s PGAP software (see Methods; Supplementary Table S2). The analysis of the longest repeats indicated that both strains can be classified as difficult to assemble class III genomes [46]; Illumina reads alone would not have allowed us to assemble a complete genome. Finally, an analysis with AntiSmash v6.0.1 [63] predicted several biosynthetic gene clusters of potential relevance (see Supplementary Figure S2A,B), including a lokisin NRP biosynthetic gene cluster, which was shown to exert anti-fungal activity [64]. Moreover, a lokisin derivative was produced in a multispecies bacterial community where the gene and metabolite expression changed depending on the composition [65], underlining the relevance of multispecies consortia.
3.4. Phylogenetic Analysis of Pseudomonas Strains 105 and 108
Phylogenetic analysis using 68 Pseudomonas reference strains available in the NCBI database confirmed that both bacterial isolates, 105 and 108, belonged to the genus Pseudomonas. Both isolates clustered within the P. fluorescence subgroup together with previously identified Pseudomonas isolates with potential for nematode biocontrol (Figure 9, Supplementary Table S3). However, strains 105 and 108 were placed in separate subclades of the P. fluorescence group.
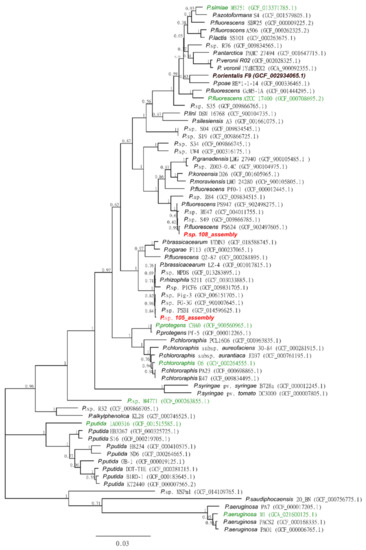
Figure 9.
Phylogenetic tree based on 16 core genes of Pseudomonas strains for the taxonomic placement of the studied Pseudomonas strains 105 and 108 (marked in red) inferred by the Maximum Likelihood method (see Methods). The bootstrap values shown for each node were obtained from 100 bootstrap runs. The genomes sequenced for Pseudomonas strains previously reported to contain nematicidal properties are marked in green (Supplementary Table S3). P. orientalis F9, used as a negative control for the experiments, is marked in brown. The scale of the branch length units at the bottom of the tree indicates the number of nucleotide substitutions per site.
Strain 105 was closely grouped with the characterized Pseudomonas brassicacearum (GCA_001017815) and Pseudomonas rhizophila (GCA_003033885), while strain 108 was grouped with characterized P. fluorescence strains (Figure 9). However, with bootstrap support of 0.63, the branching of both clades was considered low. The majority of the knots in the tree were between 0.95 and 1, indicating that the phylogenetic tree was robust.
Interestingly, this phylogenetic analysis showed that the Pseudomonas isolates used in the present study clustered together with Pseudomonas strains previously tested for potential RKN control (Figure 9, Supplementary Table S3 [56,59,66,67,68,69,70,71,72]). For example, P. simiae MB751, P. fluorescens ATCC-17400, P. protegens CHA0, Pseudomonas putida 1A00316, and Pseudomonas aeruginosa showed nematicidal properties against the RKN M. incognita and/or Meloidogyne javanica [56,59,60,66,67,68,69,70]. Only Pseudomonas chlororaphis O6 has been successfully tested against a temperate RKN, Meloidogyne hapla [71].
The mode of action has only been investigated in a few studies. For example, P. simiae MB751 produces a cyclic dipeptide with nematicidal properties, which is noteworthy as some fungi have also been reported to produce macrocyclic peptides with nematicidal properties against M. incognita [73]. P. putida 1A00316, isolated from Antarctic soil, has been shown to produce volatile nematicides [56]. However, based on phylogenetic analysis alone, no conclusions can be drawn about the mode of action of the isolates tested in this study.
Based on our experiment, the selected Pseudomonas strains were able to protect cucumber root bales (Figure 8) more efficiently than when applied to soil with a higher OD600 of 1.5 (Table 1). Therefore, we hypothesize that the protective properties of Pseudomonas strains 105 and 108 are related to root colonization.
Overall, the phylogenetic analysis shows that the Pseudomonas strains used in this study cluster not only among the RKN control strains but also among other biocontrol strains, such as the P. fluorescens strain 8GCA_902497605, a potato-pathogen-inhibiting strain, or other Pseudomonas strains isolated from the rhizosphere and phyllosphere of potato plants, R84 and S49, which have been reported to inhibit the growth of Phytophthora infestans mycelia [51].
Therefore, it may be worthwhile to test Pseudomonas strains 105 and 108 as biocontrol agents against other important plant-pathogenic nematodes and agricultural fungal and oomycete pathogens. The release of the complete de-novo-assembled genome sequence will serve as an important basis to identify the mechanism(s) of action against various plant pathogens in future studies. On a broader scale, the assay system developed here should also allow the testing of mixtures of different strains with biocontrol activity for robust and potentially even synergistic effects.
4. Conclusions
This study serves as a proof-of-principle for the identification of potentially promising biocontrol strains against RKN, with the help of this newly developed screening method.
The straightforward and reliable combination of screening assays resulted in the isolation of Pseudomonas strains that controlled M. incognita in vitro and in soil.
Further investigation of Pseudomonas strains 105 and 108 demonstrated their potential to control M. incognita under greenhouse conditions with varying RKN population densities. The application of the Pseudomonas strains is versatile, as demonstrated by their efficacy when being used as either soil or root treatments.
Phylogenetic analyses revealed that both Pseudomonas strains clustered in the Pseudomonas fluorescens group alongside previously described plant pathogens and RKN antagonists, but apart from each other.
Future investigations of the selected Pseudomonas candidates for RKN control will need to demonstrate their antagonistic potential in larger greenhouse trials and/or during field applications, investigating the influence of not only biotic but also abiotic factors on the maintenance of their biocontrol potential. Elucidation of the mechanisms of action is now possible given the availability of complete genome sequences.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11082011/s1, Figure S1: Effect of bacterial strains on the height of tomato plants grown in nematode-infected soil; Table S1: Selected bacterial strains used in a screen to identify antagonistic activity against Meloidogyne incognita; Table S2: Selected genome features of Pseudomonas spp. 105 and 108; Figure S2: Graphical output of the AntiSmash prediction server (v.6.0.1) for the two Pseudomonas isolates; Table S3: List of genome sequences of available for Pseudomonas strains that have been linked to plant-parasitic nematode control.
Author Contributions
Conceptualization, P.D., C.H.A. and C.P.; methodology, P.D., T.S. (Tobias Stucky), M.H., S.M., T.S. (Tina Segessemann), A.C.R. and C.P.; validation, all; formal analysis, all; data curation, T.S. (Tina Segessemann), P.D. and C.H.A.; writing—original draft preparation, P.D., A.C.R., C.P. and C.H.A.; writing—review and editing, C.P., A.C.R., T.S. (Tina Segessemann), C.H.A. and P.D.; visualization, T.S. (Tobias Stucky), T.S. (Tina Segessemann), P.D. and C.H.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Data are contained within the article and appendices. The complete genome sequences of the two Pseudomonas isolates are available from the NCBI under bioproject PRJNA975707 (P105) and PRJNA975710 (P108) and with the accession numbers CP126693-CP126694 (P105) and CP126691-CP126692 (P108).
Acknowledgments
The authors acknowledge the Vegetable-Production Extension at Agroscope for providing the soil and beetroots, and Veronika Zengerer, Vera Dreyfuss, Brigitta Teschner, Eliana Thyda Sy, and Reinhard Eder for the technical support during the laboratory and greenhouse experiments. We would like to thank Florian Freimoser for his insightful comments during the experiments and for proofreading the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wesemael, W.; Viaene, N.; Moens, M. Root-knot nematodes (Meloidogyne spp.) in Europe. Nematology 2011, 13, 3–16. [Google Scholar] [CrossRef]
- Dong, L.; Huang, C.; Huang, L.; Li, X.; Zuo, Y. Screening plants resistant against Meloidogyne incognita and integrated management of plant resources for nematode control. Crop Prot. 2012, 33, 34–39. [Google Scholar] [CrossRef]
- Trudgill, D.L.; Blok, V.C. Apomictic, polyphagous root-knot nematodes: Exceptionally successful and damaging biotrophic root pathogens. Annu. Rev. Phytopathol. 2001, 39, 53–77. [Google Scholar] [CrossRef] [PubMed]
- Subedi, S.; Thapa, B.; Shrestha, J. Root-knot nematode (Meloidogyne incognita) and its management: A review. J. Agric. Nat. Resour. 2020, 3, 21–31. [Google Scholar] [CrossRef]
- Bernard, G.C.; Egnin, M.; Bonsi, C. Chapter 4: The impact of plant-parasitic nematodes on agriculture and methods of control. In Nematology: Concepts, Diagnosis and Control; Sha, M.M., Mahamood, M., Eds.; InTech: Rijeka, Croatia, 2017. [Google Scholar] [CrossRef]
- Gianessi, L.P.; Carpenter, J.E. Agricultural Biotechnology: Insect Control Benefits; National Center for Food and Agricultural Policy: Washington, DC, USA, 1999. [Google Scholar]
- Singh, S.; Singh, B.; Singh, A.P. Nematodes: A threat to sustainability of Agriculture. Procedia Environ. Sci. 2015, 29, 215–216. [Google Scholar] [CrossRef]
- Coyne, D.L.; Cortada, L.; Dalzell, J.J.; Claudius-Cole, A.O.; Haukeland, S.; Luambano, N.; Talwana, H. Plant-parasitic nematodes and food security in sub-Saharan Africa. Annu. Rev. Phytopathol. 2018, 56, 381–403. [Google Scholar] [CrossRef]
- Mesa-Valle, C.M.; Garrido-Cardenas, J.A.; Cebrian-Carmona, J.; Talavera, M.; Manzano-Agugliaro, F. Global research on plant nematodes. Agronomy 2020, 10, 1148. [Google Scholar] [CrossRef]
- Caillaud, M.C.; Dubreuil, G.; Quentin, M.; Perfus-Barbeoch, L.; Lecomte, P.; de Almeida Engler, J.; Abad, P.; Rosso, M.-N.; Favery, B. Root-knot nematodes manipulate plant cell functions during a compatible interaction. J. Plant Physiol. 2008, 165, 104–113. [Google Scholar] [CrossRef]
- Vestergård, M. Trap crops for Meloidogyne hapla management and its integration with supplementary strategies. Appl. Soil Ecol. 2019, 134, 105–110. [Google Scholar] [CrossRef]
- Oka, Y. From old-generation to next-generation nematicides. Agronomy 2020, 10, 1387. [Google Scholar] [CrossRef]
- Sasanelli, N.; Konrat, A.; Migunova, V.; Toderas, I.; Iurcu-Straistaru, E.; Rusu, S.; Bivol, A.; Andoni, C.; Veronico, P. Review on Control Methods against Plant Parasitic Nematodes Applied in Southern Member States (C Zone) of the European Union. Agriculture 2021, 11, 602. [Google Scholar] [CrossRef]
- Pal, K.K.; Mc Spadden, G.B. Biological Control of Plant Pathogens. Plant Health Instr. 2006. [Google Scholar] [CrossRef]
- Francisco, B.G.F.; Ponce, I.M.; Espinosa, M.Á.P.; Moctezuma, A.M.; y López, V.E.L. Advances in the biological control of phytoparasitic nematodes via the use of nematophagous fungi. World J. Microbiol. Biotechnol. 2021, 37, 180. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; Mazzola, M. Soil immune responses. Science 2016, 352, 1392–1393. [Google Scholar] [CrossRef]
- Topalović, O.; Santos, S.S.; Heuer, H.; Nesme, J.; Kanfra, X.; Hallmann, J.; Sørensen, S.J.; Vestergård, M. Deciphering bacteria associated with a pre-parasitic stage of the root-knot nematode Meloidogyne hapla in nemato-suppressive and nemato-conducive soils. Appl. Soil Ecol. 2022, 172, 104344. [Google Scholar] [CrossRef]
- Su, H.; Hao, Y.E.; Yang, X.; Yu, Z.; Deng, J.; Mo, M. A new species of Dactylellina producing adhesive knobs and non-constricting rings to capture nematodes. Mycotaxon 2008, 105, 313. [Google Scholar]
- Topalović, O.; Bredenbruch, S.; Schleker, A.S.S.; Heuer, H. Microbes attaching to endoparasitic phytonematodes in soil trigger plant defense upon root penetration by the nematode. Front. Plant. Sci. 2020, 11, 138. [Google Scholar] [CrossRef]
- Chen, Z.X.; Dickson, D.W. Review of Pasteuria penetrans: Biology, ecology, and biological control potential. J. Nematol. 1998, 30, 313–340. [Google Scholar] [PubMed]
- Panpatte, D.G.; Jhala, Y.K.; Shelat, H.N.; Vyas, R.V. Pseudomonas fluorescens: A promising biocontrol agent and PGPR for sustainable agriculture. In Microbial Inoculants in Sustainable Agricultural Productivity; Singh, D., Singh, H., Prabha, R., Eds.; Springer: New Delhi, India, 2016; Volume 1, pp. 257–270. [Google Scholar] [CrossRef]
- Bhat, A.A.; Shakeel, A.; Waqar, S.; Handoo, Z.A.; Khan, A.A. Microbes vs. Nematodes: Insights into Biocontrol through Antagonistic Organisms to Control Root-Knot Nematodes. Plants 2023, 12, 451. [Google Scholar] [CrossRef]
- Soliman, G.M.; Ameen, H.H.; Abdel-Aziz, S.M.; El-Sayed, G.M. In vitro evaluation of some isolated bacteria against the plant parasite nematode Meloidogyne incognita. Bull. Natl. Res. Cent. 2019, 43, 171. [Google Scholar] [CrossRef]
- Nishantha, K.; Jayasiri, H.D.C.M.; Herath, H.; Nugaliyadde, M.M.; Dissanayake, M.; Costa, D.M. Antagonistic effect of eight Sri Lankan isolates of Pseudomonas fluorescens on, Meloidogyne incognita in tomato, Lycopersicon esculentum. Int. J. Environ. Agric. Biotechnol. 2018, 3, 266217. [Google Scholar] [CrossRef]
- Khan, M.R.; Mohidin, F.A.; Khan, U.; Ahamad, F. Native Pseudomonas spp. suppressed the root-knot nematode in in vitro and in vivo, and promoted the nodulation and grain yield in the field grown mungbean. Biol. Control 2016, 101, 159–168. [Google Scholar] [CrossRef]
- Mhatre, P.H.; Karthik, C.; Kadirvelu, K.; Divya, K.L.; Venkatasalam, E.P.; Srinivasan, S.; Shanmuganathan, R. Plant growth promoting rhizobacteria (PGPR): A potential alternative tool for nematodes biocontrol. Biocatal. Agric. Biotechnol. 2019, 17, 119–128. [Google Scholar] [CrossRef]
- Migunova, V.D.; Sasanelli, N. Bacteria as biocontrol tool against phytoparasitic nematodes. Plants 2021, 10, 389. [Google Scholar] [CrossRef]
- Sikora, R.A.; Roberts, P.A. Management practices: An overview of integrated nematode management technologie. In Plant Parasitic Nematodes in Subtropical and Tropical Agriculture, 3rd ed.; Sikora, R.A., Coyne, D., Hallmann, J., Timper, P., Eds.; CABI: Oxfordshire, UK, 2018; pp. 795–839. [Google Scholar] [CrossRef]
- Kiewnick, S.; Wolf, S.; Willareth, M.; Frey, J.E. Identification of the tropical root-knot nematode species Meloidogyne incognita, M. javanica and M. arenaria using a multiplex PCR assay. Nematology 2013, 15, 891–894. [Google Scholar] [CrossRef]
- Gekenidis, M.-T.; Studer, P.; Wüthrich, S.; Brunisholz, R.; Drissner, D. Beyond the matrix-assisted laser desorption ionization (MALDI) biotyping workflow: In search of microorganism-specific tryptic peptides enabling discrimination of subspecies. Appl. Environ. Microbiol. 2014, 80, 4234–4241. [Google Scholar] [CrossRef] [PubMed]
- Zeck, W.M. Rating scheme for field evaluation of root-knot nematode infestations. Pflanzenschutz Nachr. Bayer. 1971, 24, 141–144. [Google Scholar]
- Zengerer, V.; Schmid, M.; Bieri, M.; Müller, D.C.; Remus-Emsermann, M.N.P.; Ahrens, C.H.; Pelludat, C. Pseudomonas orientalis F9: A Potent Antagonist against Phytopathogens with Phytotoxic Effect in the Apple Flower. Front. Microbiol. 2018, 9, 145. [Google Scholar] [CrossRef]
- Oldani, E.; Cabianca, A.; Dahlin, P.; Ruthes, A.C. Biogas digestate as potential source for nematicides. Environ. Technol. Innov. 2023, 29, 103025. [Google Scholar] [CrossRef]
- Davis, L.G.; Dibner, M.D.; Battley, J.F. Basic Methods in Molecular Biology; Elsevier: Amsterdam, The Netherlands, 1986. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 12 March 2023).
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Antipov, D.; Hartwick, N.; Shen, M.; Raiko, M.; Lapidus, A.; Pevzner, P.A. PlasmidSPAdes: Assembling plasmids from whole genome sequencing data. Bioinformatics 2016, 32, 3380–3387. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Okonechnikov, K.; Conesa, A.; García-Alcalde, F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness. In Gene Prediction. Methods in Molecular Biology, 1st ed.; Kollmar, M., Ed.; Humana Press: New York, NY, USA, 2019; Volume 1962, pp. 227–245. [Google Scholar] [CrossRef]
- Schmid, M.; Frei, D.; Patrignani, A.; Schlapbach, R.; Frey, J.E.; Remus-Emsermann, M.N.P.; Ahrens, C.H. Pushing the limits of de novo genome assembly for complex prokaryotic genomes harboring very long, near identical repeats. Nucleic Acids Res. Sep. 2018, 46, 8953–8965. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary. Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J.; Clustal, W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Peterson, D.; Tamura, K. MEGA-CC: Computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics 2012, 28, 2685–2686. [Google Scholar] [CrossRef] [PubMed]
- De Vrieze, M.; Varadarajan, A.R.; Schneeberger, K.; Bailly, A.; Rohr, R.P.; Ahrens, C.H.; Weisskopf, L. Linking comparative genomics of nine potato-associated Pseudomonas isolates with their differing biocontrol potential against late blight. Front. Microbiol. 2020, 11, 857. [Google Scholar] [CrossRef]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallone, A.B.; Bergman, N.H.; Koren, S.; Pjillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef]
- Huang, W.K.; Cui, J.K.; Liu, S.M.; Kong, L.A.; Wu, Q.S.; Peng, H.; He, W.T.; Sun, J.H.; Peng, D.L. Testing various biocontrol agents against the root-knot nematode (Meloidogyne incognita) in cucumber plants identifies a combination of Syncephalastrum racemosum and Paecilomyces lilacinus as being most effective. Biol. Control 2016, 92, 31–37. [Google Scholar] [CrossRef]
- Choi, T.G.; Maung, C.E.H.; Lee, D.R.; Henry, A.B.; Lee, Y.S.; Kim, K.Y. Role of bacterial antagonists of fungal pathogens, Bacillus thuringiensis KYC and Bacillus velezensis CE 100 in control of root-knot nematode, Meloidogyne incognita and subsequent growth promotion of tomato. Biocontrol Sci. Technol. 2020, 30, 685–700. [Google Scholar] [CrossRef]
- Liu, G.; Lin, X.; Xu, S.; Liu, G.; Liu, F.; Mu, W. Screening, identification and application of soil bacteria with nematicidal activity against root-knot nematode (Meloidogyne incognita) on tomato. Pest Manag. Sci. 2020, 76, 2217–2224. [Google Scholar] [CrossRef]
- Zhai, Y.; Shao, Z.; Cai, M.; Zheng, L.; Li, G.; Huang, D.; Cheng, W.; Thomashow, L.S.; Weller, D.M.; Yu, Z.; et al. Multiple modes of nematode control by volatiles of Pseudomonas putida 1A00316 from Antarctic soil against Meloidogyne incognita. Front. Microbiol. 2018, 9, 253. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, S.; Zhu, X.; Wang, Y.; Liu, X.; Duan, Y.; Fan, H.; Chen, L. Isolation and characterization of nodules endophytic bacteria Pseudomonas protegens Sneb1997 and Serratia plymuthica Sneb2001 for the biological control of root-knot nematode. Appl. Soil Ecol. 2021, 164, 103924. [Google Scholar] [CrossRef]
- O’Callaghan, M. Microbial inoculation of seed for improved crop performance: Issues and opportunities. Appl. Microbiol. Biotechnol. 2016, 100, 5729–5746. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, I.A.; Shaukat, S.S. Plant species, host age and host genotype effects on Meloidogyne incognita biocontrol by Pseudomonas fluorescens strain CHA0 and its genetically-modified derivatives. J. Phytopathol. 2003, 151, 231–238. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, R.; Ding, M.; Liu, Y.; Li, L. Biocontrol of the root-knot nematode Meloidogyne incognita by a nematicidal bacterium Pseudomonas simiae MB751 with cyclic dipeptide. Pest Manag. Sci. 2021, 77, 4365–4374. [Google Scholar] [CrossRef] [PubMed]
- Dahlin, P.; Eder, R.; Consoli, E.; Krauss, J.; Kiewnick, S. Integrated control of Meloidogyne incognita in tomatoes using fluopyram and Purpureocillium lilacinum strain 251. Crop Prot. 2019, 124, 104874. [Google Scholar] [CrossRef]
- Varadarajan, A.R.; Allan, R.N.; Valentin, J.D.P.; Castañeda Ocampo, O.E.; Somerville, V.; Pietsch, F.; Buhmann, M.T.; West, J.; Skipp, P.J.; van der Mei, H.C.; et al. An integrated model system to gain mechanistic insights into biofilm-associated antimicrobial resistance in Pseudomonas aeruginosa MPAO1. NPJ Biofilms Microbiomes 2020, 6, 46. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; Van Wezel, G.P.; Medema, M.H.; Weber, T. AntiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Gu, Y.; Ma, Y.N.; Wang, J.; Xia, Z.; Wei, H.L. Genomic insights into a plant growth-promoting Pseudomonas koreensis strain with cyclic lipopeptide-mediated antifungal activity. MicrobiologyOpen 2020, 9, e1092. [Google Scholar] [CrossRef]
- Chevrette, M.G.; Thomas, C.S.; Hurley, A.; Rosario-Meléndez, N.; Sankaran, K.; Tu, Y.; Hall, A.; Magesh, S.; Handelsman, J. Microbiome composition modulates secondary metabolism in a multispecies bacterial community. Proc. Natl. Acad. USA 2022, 119, e2212930119. [Google Scholar] [CrossRef]
- Sharma, M.; Saini, I.; Kaushik, P.; Aldawsari, M.M.; Al Balawi, T.; Alam, P. Mycorrhizal fungi and Pseudomonas fluorescens application reduces root-knot nematode (Meloidogyne javanica) infestation in eggplant. Saudi J. Biol. Sci. 2021, 28, 3685–3691. [Google Scholar] [CrossRef]
- Borrajo, M.P.; Mondino, E.A.; Maroniche, G.A.; Fernández, M.; Creus, C.M. Potential of rhizobacteria native to Argentina for the control of Meloidogyne javanica. Rev. Argent. Microbiol. 2022, 54, 21–30. [Google Scholar] [CrossRef]
- Siddiqui, I.A.; Haas, D.; Heeb, S. Extracellular protease of Pseudomonas fluorescens CHA0, a biocontrol factor with activity against the root-knot nematode Meloidogyne incognita. AEM 2005, 71, 5646–5649. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, I.A.; Shaukat, S.S.; Sheikh, I.H.; Khan, A. Role of cyanide production by Pseudomonas fluorescens CHA0 in the suppression of root-knot nematode, Meloidogyne javanica in tomato. World J. Microbiol. Biotechnol. 2006, 22, 641–650. [Google Scholar] [CrossRef]
- Singh, P.; Siddiqui, Z.A. Biocontrol of root-knot nematode Meloidogyne incognita by the isolates of Pseudomonas on tomato. Arch. Phytopathol. Plant Prot. 2010, 43, 1423–1434. [Google Scholar] [CrossRef]
- Kang, B.R.; Anderson, A.J.; Kim, Y.C. Hydrogen cyanide produced by Pseudomonas chlororaphis O6 exhibits nematicidal activity against Meloidogyne hapla. Plant Pathol. J. 2018, 34, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Proença, D.N.; Francisco, R.; Santos, C.V.; Lopes, A.; Fonseca, L.; Abrantes, I.M.; Morais, P.V. Diversity of bacteria associated with Bursaphelenchus xylophilus and other nematodes isolated from Pinus pinaster trees with pine wilt disease. PLoS ONE 2010, 5, e15191. [Google Scholar] [CrossRef] [PubMed]
- Matabaro, E.; Kaspar, H.; Dahlin, P.; Bader, D.L.V.; Murar, C.E.; Staubli, F.; Field, C.M.; Bode, J.W.; Künzler, M. Identification, heterologous production and bioactivity of lentinulin A and dendrothelin A, two natural variants of backbone N-methylated peptide macrocycle omphalotin A. Sci. Rep. 2021, 11, 3541. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).