The Pathogenicity of Fusobacterium nucleatum Modulated by Dietary Fibers—A Possible Missing Link between the Dietary Composition and the Risk of Colorectal Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture
2.2. Cell Lines and Cell Culture
2.3. Cell Viability Assay
2.4. Total RNA Extraction, Reverse Transcription, Quantitative PCR Assay
2.5. GC-Mass Spectrometry Analysis
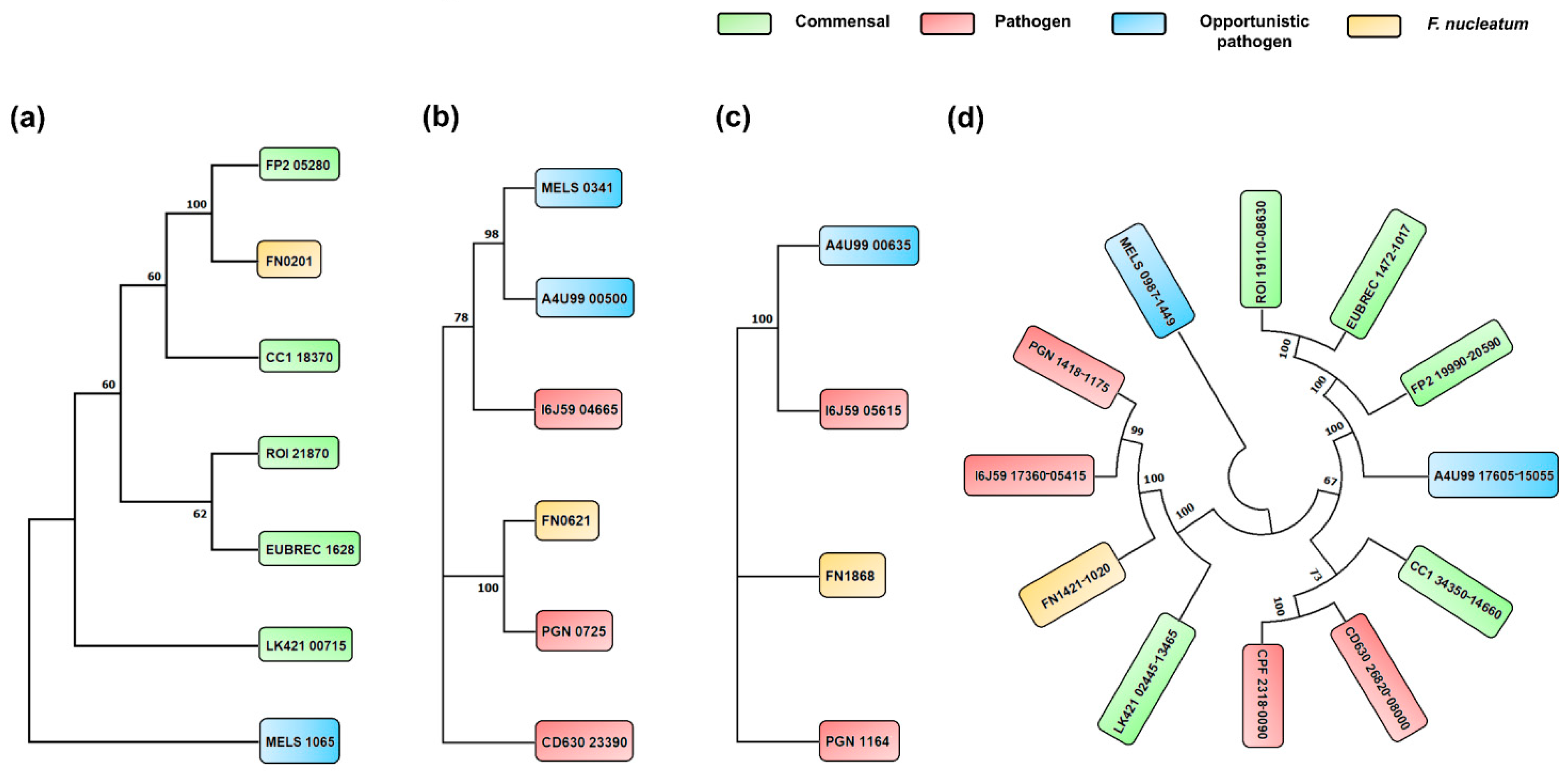
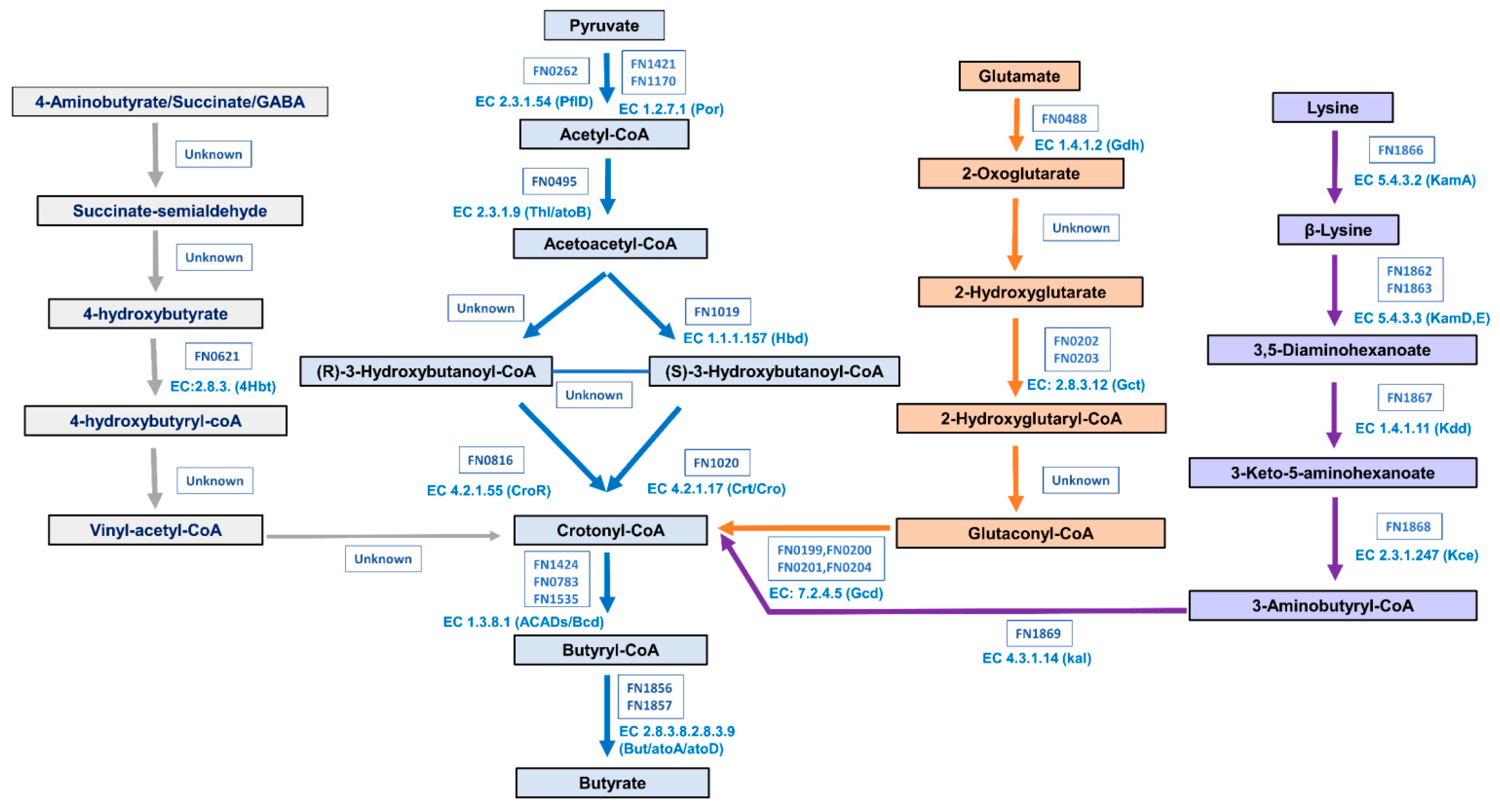
2.6. Phylogenic Analysis of Butyrate Biosynthesis Pathways
2.7. Statistical Analysis
3. Results
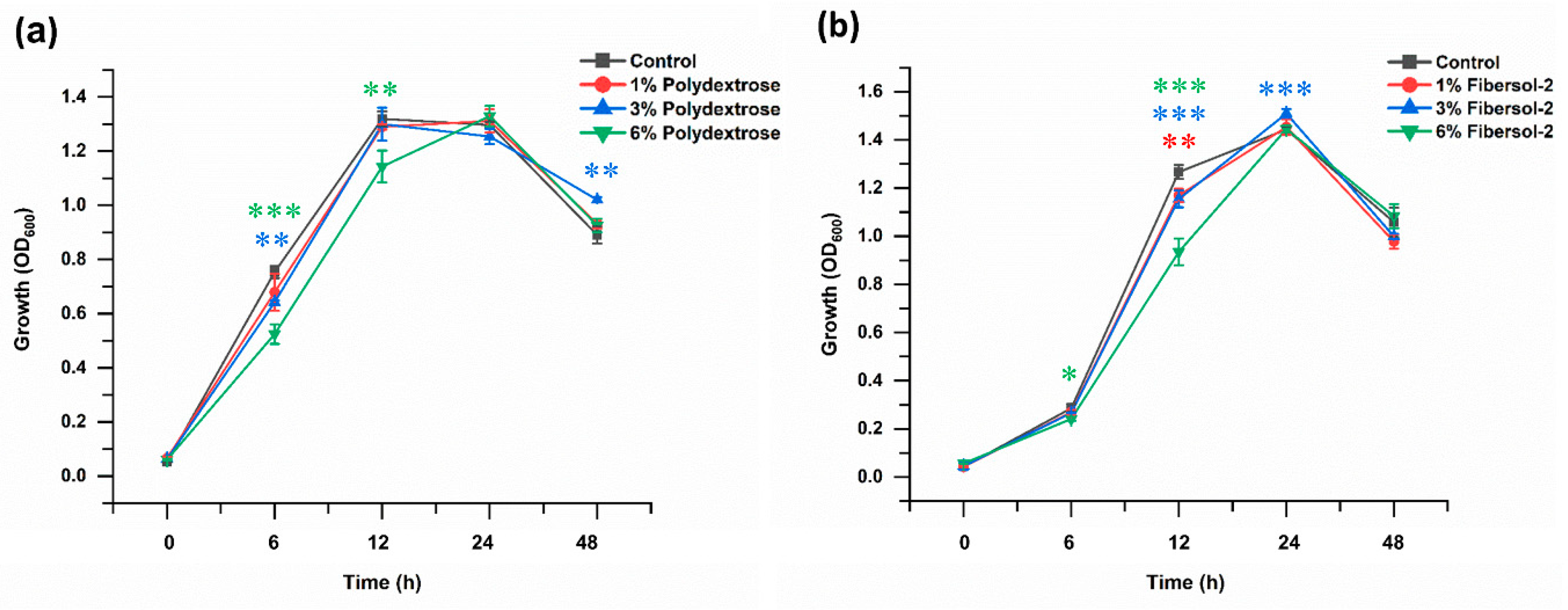
3.1. Polydextrose and Fibersol-2 Did Not Affect the Growth of F. nucleatum Significantly
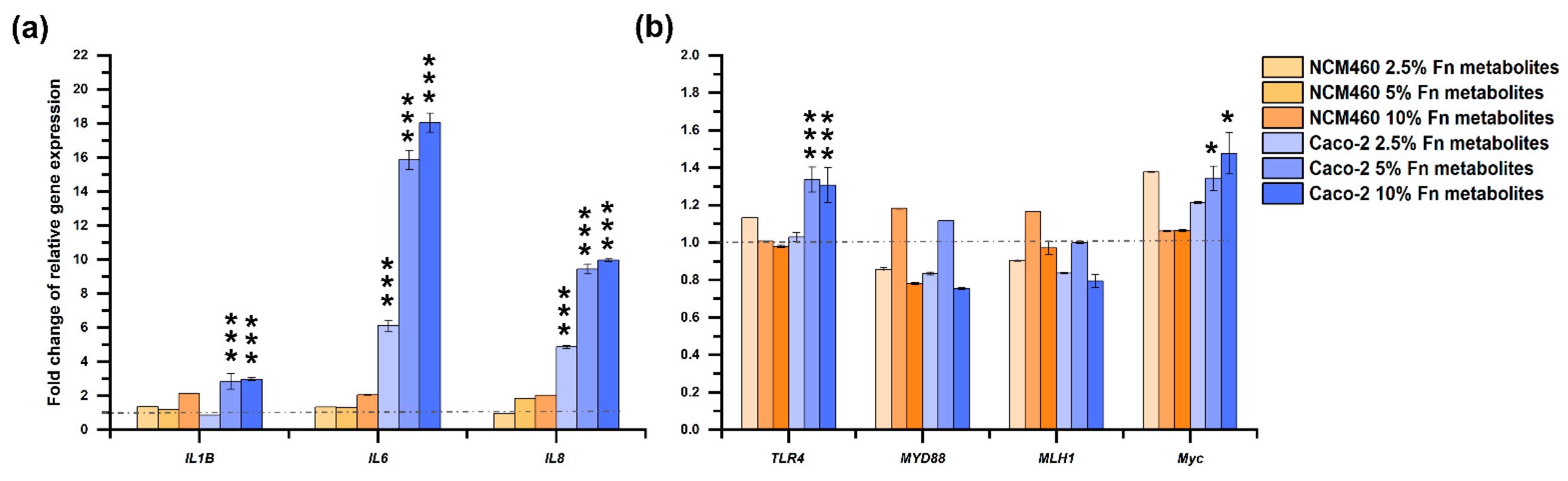
3.2. Fermentation Broth of F. nucleatum Did Not Affect the Growth of Cells but Significantly Up-Regulated Proinflammatory Genes Expression in Caco-2 Cells
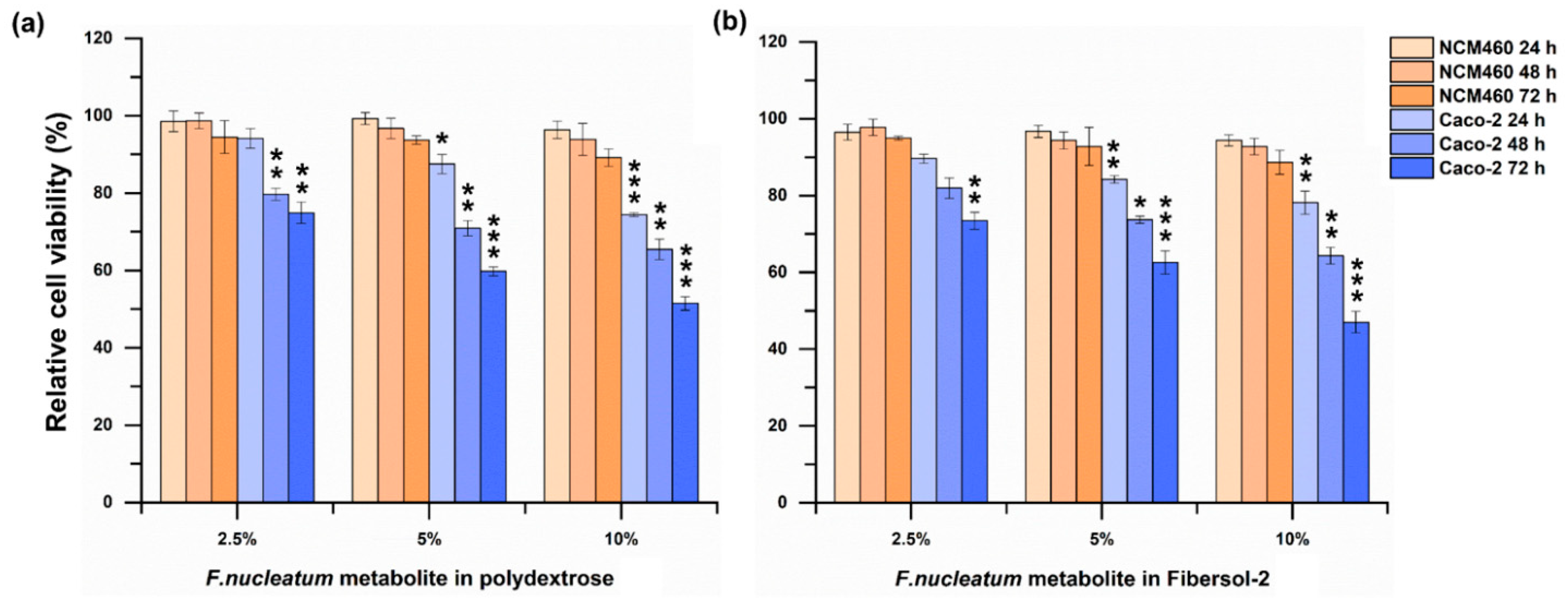
3.3. The Growth of Caco-2 Cells Was Significantly Suppressed by Fermentation Broth of F. nucleatum Cultured in Media Supplemented with Polydextrose or Fibersol-2
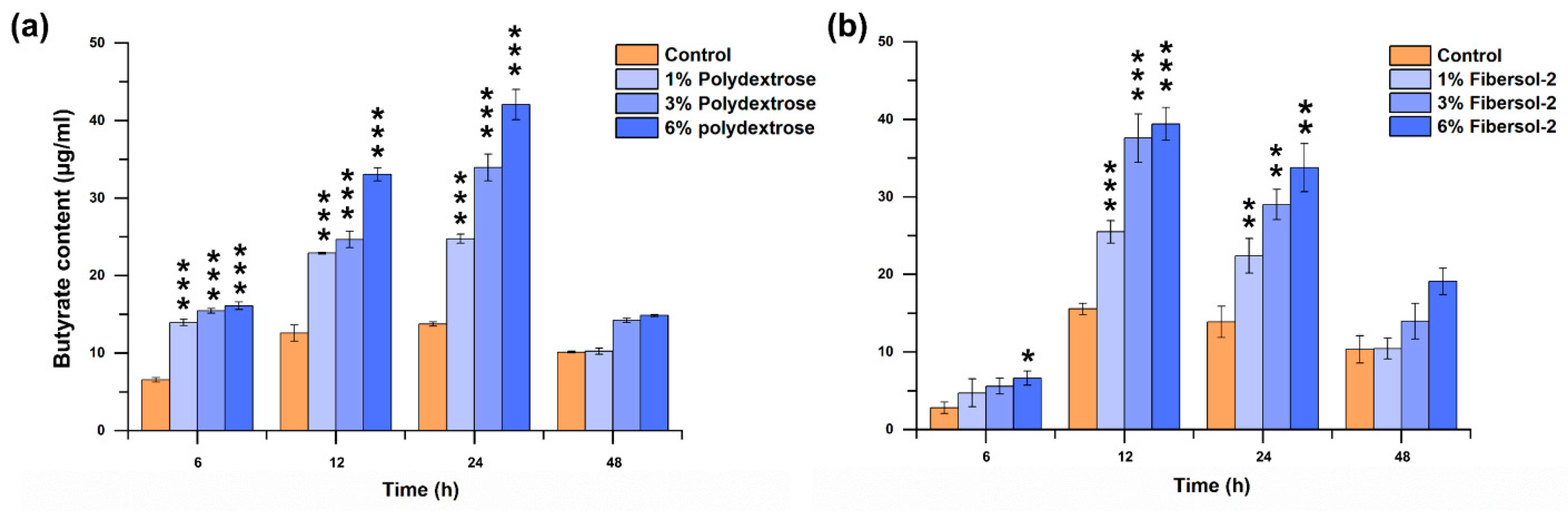
3.4. Both Fibers Significantly Promote F. nucleatum-Producing Butyrate
3.5. Phylogenetic Analysis of Butyrate Biosynthesis Pathways among Selected Species
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AtoB/Thl | Acetyl-CoA acetyltransferase / thiolase |
| Bcd | Butyryl-CoA dehydrogenase |
| But | Butyryl-CoA: acetate CoA transferase |
| But:atoD/atoA | Butyryl-CoA:acetate CoA transferase |
| Cro/Crt | Enoyl-CoA hydratase/crotonase |
| CroR | 3-hydroxybutyryl-CoA dehydratase |
| Gcd | Glutaconyl-CoA decarboxylase |
| Gct | Glutaconate-CoA transferase |
| Gdh | glutamate dehydrogenase |
| 4-Hbt | Butyryl-CoA:4-hydroxybutyrate-CoA transferase |
| Hbd | 3-hydroxybutyryl-CoA dehydrogenase |
| Kal | 3-aminobutyryl-CoA ammonia-lyase |
| Kam D, E | β-lysine-5,6-aminomutase, α/β subunit |
| KamA | Lysine 2,3-aminomutase |
| Kce | 3-keto-5-aminohexanoate cleavage enzymes |
| Kdd | 3,5-diaminohexanoate dehydrogenase |
| PflD | Formate acetyltransferase |
| Por | Pyruvate ferredoxin/flavodoxin oxidoreductase |
References
- Deo, S.; Sharma, J.; Kumar, S. GLOBOCAN 2020 report on global cancer burden: Challenges and opportunities for surgical oncologists. Ann. Surg. Oncol. 2022, 29, 6497–6500. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, A.; Kaushik, A.C.; Wang, Q.; Li, C.-D.; Wei, D.-Q. Bringing structural implications and deep learning-based drug identification for KRAS mutants. J. Chem. Inf. Model. 2021, 61, 571–586. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Dong, X.; Li, H.; Cao, M.; Sun, D.; He, S.; Yang, F.; Yan, X.; Zhang, S.; Li, N. Cancer statistics in China and United States, 2022: Profiles, trends, and determinants. Chin. Med. J. 2022, 135, 584–590. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska, A.; Rudzki, G.; Lewandowski, T.; Stryjkowska-Góra, A.; Rudzki, S. Risk factors for the diagnosis of colorectal cancer. Cancer Control 2022, 29, 10732748211056692. [Google Scholar] [CrossRef]
- Yang, H.; Wang, J. The Changes of Dietary Intake of Chinese Residents from the Year of 1998 to 2012. Food Ind. 2020, 41, 244–248. [Google Scholar]
- Cao, W.; Chen, H.-D.; Yu, Y.-W.; Li, N.; Chen, W.-Q. Changing profiles of cancer burden worldwide and in China: A secondary analysis of the global cancer statistics 2020. Chin. Med. J. 2021, 134, 783–791. [Google Scholar] [CrossRef]
- Sonnenburg, E.D.; Smits, S.A.; Tikhonov, M.; Higginbottom, S.K.; Wingreen, N.S.; Sonnenburg, J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nature 2016, 529, 212–215. [Google Scholar] [CrossRef]
- Parkar, S.G.; Rosendale, D.I.; Stoklosinski, H.M.; Jobsis, C.M.; Hedderley, D.I.; Gopal, P. Complementary food ingredients alter infant gut microbiome composition and metabolism in vitro. Microorganisms 2021, 9, 2089. [Google Scholar] [CrossRef]
- O’Keefe, S.J.; Li, J.V.; Lahti, L.; Ou, J.; Carbonero, F.; Mohammed, K.; Posma, J.M.; Kinross, J.; Wahl, E.; Ruder, E. Fat, fibre and cancer risk in African Americans and rural Africans. Nat. Commun. 2015, 6, 6342. [Google Scholar] [CrossRef]
- Yang, J.; Yu, J. The association of diet, gut microbiota and colorectal cancer: What we eat may imply what we get. Protein Cell 2018, 9, 474–487. [Google Scholar] [CrossRef]
- Vernia, F.; Longo, S.; Stefanelli, G.; Viscido, A.; Latella, G. Dietary factors modulating colorectal carcinogenesis. Nutrients 2021, 13, 143. [Google Scholar] [CrossRef]
- Röytiö, H.; Ouwehand, A. The fermentation of polydextrose in the large intestine and its beneficial effects. Benef. Microbes 2014, 5, 305–313. [Google Scholar] [CrossRef]
- Sorndech, W.; Rodtong, S.; Blennow, A.; Tongta, S. Impact of resistant maltodextrins and resistant starch on human gut microbiota and organic acids production. Starch-Stärke 2019, 71, 1800231. [Google Scholar] [CrossRef]
- Davis, E.C.; Dinsmoor, A.M.; Wang, M.; Donovan, S.M. Microbiome composition in pediatric populations from birth to adolescence: Impact of diet and prebiotic and probiotic interventions. Dig. Dis. Sci. 2020, 65, 706–722. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, J.; He, T.; Becker, S.; Zhang, G.; Li, D.; Ma, X. Butyrate: A double-edged sword for health? Adv. Nutr. 2018, 9, 21–29. [Google Scholar] [CrossRef]
- Do Carmo, M.M.R.; Walker, J.C.L.; Novello, D.; Caselato, V.M.; Sgarbieri, V.C.; Ouwehand, A.C.; Andreollo, N.A.; Hiane, P.A.; Dos Santos, E.F. Polydextrose: Physiological function, and effects on health. Nutrients 2016, 8, 553. [Google Scholar] [CrossRef]
- Sancho, S.C.; Olson, S.L.; Young So, E.; Shimomura, K.; Ouchi, T.; Preuss, F. Fibersol-2 induces apoptosis of Apc-deficient colorectal cancer (SW480) cells and decreases polyp formation in Apc MIN mice. Cancer Biol. Ther. 2016, 17, 657–663. [Google Scholar] [CrossRef]
- Jacobs, L.R. Fiber and colon cancer. Gastroenterol. Clin. N. Am. 1988, 17, 747–760. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, J.; Zhang, S. Dietary fiber—A double-edged sword for balanced nutrition supply and environment sustainability in swine industry: A meta-analysis and systematic review. J. Clean. Prod. 2021, 315, 128130. [Google Scholar] [CrossRef]
- Bennett, G.N.; Rudolph, F.B. The central metabolic pathway from acetyl-CoA to butyryl-CoA in Clostridium acetobutylicum. FEMS Microbiol. Rev. 1995, 17, 241–249. [Google Scholar] [CrossRef]
- Duncan, S.H.; Barcenilla, A.; Stewart, C.S.; Pryde, S.E.; Flint, H.J. Acetate utilization and butyryl coenzyme A (CoA): Acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl. Environ. Microbiol. 2002, 68, 5186–5190. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Flint, H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 2009, 294, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.T.; Duncan, S.H.; Stams, A.J.; Van Dijl, J.M.; Flint, H.J.; Harmsen, H.J. The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic–anoxic interphases. ISME J. 2012, 6, 1578–1585. [Google Scholar] [CrossRef]
- Matthies, C.; Mayer, F.; Schink, B. Fermentative degradation of putrescine by new strictly anaerobic bacteria. Arch. Microbiol. 1989, 151, 498–505. [Google Scholar] [CrossRef]
- Bui, T.P.N.; Ritari, J.; Boeren, S.; De Waard, P.; Plugge, C.M.; De Vos, W.M. Production of butyrate from lysine and the Amadori product fructoselysine by a human gut commensal. Nat. Commun. 2015, 6, 10062. [Google Scholar] [CrossRef]
- Yoshida, Y. Analysis of the butyrate-producing pathway in Porphyromonas gingivalis. Periodontal Pathog. Methods Protoc. 2021, 2210, 167–172. [Google Scholar]
- Shetty, S.A.; Marathe, N.P.; Lanjekar, V.; Ranade, D.; Shouche, Y.S. Comparative genome analysis of Megasphaera sp. reveals niche specialization and its potential role in the human gut. PLoS ONE 2013, 8, e79353. [Google Scholar] [CrossRef]
- Ferreyra, J.A.; Wu, K.J.; Hryckowian, A.J.; Bouley, D.M.; Weimer, B.C.; Sonnenburg, J.L. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 2014, 16, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Buckel, W.; Barker, H. Two pathways of glutamate fermentation by anaerobic bacteria. J. Bacteriol. 1974, 117, 1248–1260. [Google Scholar] [CrossRef] [PubMed]
- Vital, M.; Howe, A.; Tiedje, J. Revealing the bacterial butyrate synthesis pathways by analyzing (meta) genomic data. mBio 2014, 5, e00889-14. [Google Scholar] [CrossRef] [PubMed]
- Oliphant, K.; Allen-Vercoe, E. Macronutrient metabolism by the human gut microbiome: Major fermentation by-products and their impact on host health. Microbiome 2019, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.A.; Macfarlane, G. Dissimilatory amino acid metabolism in human colonic bacteria. Anaerobe 1997, 3, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Mima, K.; Nishihara, R.; Qian, Z.R.; Cao, Y.; Sukawa, Y.; Nowak, J.A.; Yang, J.; Dou, R.; Masugi, Y.; Song, M. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016, 65, 1973–1980. [Google Scholar] [CrossRef]
- Palmieri, O.; Castellana, S.; Latiano, A.; Latiano, T.; Gentile, A.; Panza, A.; Nardella, M.; Ciardiello, D.; Latiano, T.P.; Corritore, G. Mucosal Microbiota from Colorectal Cancer, Adenoma and Normal Epithelium Reveals the Imprint of Fusobacterium nucleatum in Cancerogenesis. Microorganisms 2023, 11, 1147. [Google Scholar] [CrossRef]
- Parhi, L.; Alon-Maimon, T.; Sol, A.; Nejman, D.; Shhadeh, A.; Fainsod-Levi, T.; Yajuk, O.; Isaacson, B.; Abed, J.; Maalouf, N. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat. Commun. 2020, 11, 3259. [Google Scholar] [CrossRef]
- Chu, S.; Cheng, Z.; Yin, Z.; Xu, J.; Wu, F.; Jin, Y.; Yang, G. Airway fusobacterium is associated with poor response to immunotherapy in lung cancer. OncoTargets Ther. 2022, 15, 201–213. [Google Scholar] [CrossRef]
- Alexander, J.L.; Scott, A.J.; Pouncey, A.L.; Marchesi, J.; Kinross, J.; Teare, J. Colorectal carcinogenesis: An archetype of gut microbiota–host interaction. Ecancermedicalscience 2018, 12, 865. [Google Scholar] [CrossRef]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 2017, 170, 548–563.e16. [Google Scholar] [CrossRef]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Sakanaka, A.; Kuboniwa, M.; Shimma, S.; Alghamdi, S.A.; Mayumi, S.; Lamont, R.J.; Fukusaki, E.; Amano, A. Fusobacterium nucleatum metabolically integrates commensals and pathogens in oral biofilms. Msystems 2022, 7, e00170-22. [Google Scholar] [CrossRef] [PubMed]
- Eckburg, P.; Bik, E.; Bernstein, C.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef]
- Han, Y.W. Fusobacterium nucleatum: A commensal-turned pathogen. Curr. Opin. Microbiol. 2015, 23, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Barker, H.; Kahn, J.M.; Hedrick, L. Pathway of lysine degradation in Fusobacterium nucleatum. J. Bacteriol. 1982, 152, 201–207. [Google Scholar] [CrossRef]
- Bolstad, A.; Jensen, H.B.; Bakken, V. Taxonomy, biology, and periodontal aspects of Fusobacterium nucleatum. Clin. Microbiol. Rev. 1996, 9, 55–71. [Google Scholar] [CrossRef]
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012, 22, 299–306. [Google Scholar] [CrossRef]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef]
- Gharbia, S.E.; Shah, H.N. Pathways of glutamate catabolism among Fusobacterium species. Microbiology 1991, 137, 1201–1206. [Google Scholar] [CrossRef]
- Anand, S.; Kaur, H.; Mande, S.S. Comparative in silico analysis of butyrate production pathways in gut commensals and pathogens. Front. Microbiol. 2016, 7, 1945. [Google Scholar] [CrossRef]
- Guan, X.; Li, W.; Meng, H. A double-edged sword: Role of butyrate in the oral cavity and the gut. Mol. Oral Microbiol. 2021, 36, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Stokowa-Sołtys, K.; Wojtkowiak, K.; Jagiełło, K. Fusobacterium nucleatum–Friend or foe? J. Inorg. Biochem. 2021, 224, 111586. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Boppana, S. Characterization of Short Chain Fatty Acids in Microbial Cultures by DART-MS and GC-MS; Eastern Michigan University: Ypsilanti, MI, USA, 2013. [Google Scholar]
- Harrison, C. Quantification of Short-Chain Fatty Acids in Cecal Material by Gas Chromatography-Mass Spectrometry. Master’s Thesis, Eastern Michigan University, Ypsilanti, MI, USA, 2010. [Google Scholar]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki-Kinoshita, K.F.; Itoh, M.; Kawashima, S.; Katayama, T.; Araki, M.; Hirakawa, M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006, 34, D354–D357. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Hillman, E.T.; Kozik, A.J.; Hooker, C.A.; Burnett, J.L.; Heo, Y.; Kiesel, V.A.; Nevins, C.J.; Oshiro, J.M.; Robins, M.M.; Thakkar, R.D. Comparative genomics of the genus Roseburia reveals divergent biosynthetic pathways that may influence colonic competition among species. Microb. Genom. 2020, 6, mgen000399. [Google Scholar] [CrossRef]
- Cockburn, D.W.; Orlovsky, N.I.; Foley, M.H.; Kwiatkowski, K.J.; Bahr, C.M.; Maynard, M.; Demeler, B.; Koropatkin, N.M. Molecular details of a starch utilization pathway in the human gut symbiont E ubacterium rectale. Mol. Microbiol. 2015, 95, 209–230. [Google Scholar] [CrossRef]
- Nogal, A.; Louca, P.; Zhang, X.; Wells, P.M.; Steves, C.J.; Spector, T.D.; Falchi, M.; Valdes, A.M.; Menni, C. Circulating levels of the short-chain fatty acid acetate mediate the effect of the gut microbiome on visceral fat. Front. Microbiol. 2021, 12, 711359. [Google Scholar] [CrossRef]
- Wallace, R.J. Catabolism of amino acids by Megasphaera elsdenii LC1. Appl. Environ. Microbiol. 1986, 51, 1141–1143. [Google Scholar] [CrossRef] [PubMed]
- Borrelli, L.; Coretti, L.; Dipineto, L.; Bovera, F.; Menna, F.; Chiariotti, L.; Nizza, A.; Lembo, F.; Fioretti, A. Insect-based diet, a promising nutritional source, modulates gut microbiota composition and SCFAs production in laying hens. Sci. Rep. 2017, 7, 16269. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.T.; Fujimura, Y.; Mimura, I.; Fujii, Y.; Nguyen, N.L.; Arakawa, K.; Morita, H. Cultivable butyrate-producing bacteria of elderly Japanese diagnosed with Alzheimer’s disease. J. Microbiol. 2018, 56, 760–771. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xu, L.; Wang, Y.; Mao, S. Metagenomic sequencing reveals that high-grain feeding alters the composition and metabolism of cecal microbiota and induces cecal mucosal injury in sheep. Msystems 2021, 6, e00915-21. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D. Human gut microbiome: Hopes, threats and promises. Gut 2018, 67, 1716–1725. [Google Scholar] [CrossRef] [PubMed]
- Oliver, A.; Chase, A.B.; Weihe, C.; Orchanian, S.B.; Riedel, S.F.; Hendrickson, C.L.; Lay, M.; Sewall, J.M.; Martiny, J.B.; Whiteson, K. High-fiber, whole-food dietary intervention alters the human gut microbiome but not fecal short-chain fatty acids. Msystems 2021, 6, e00115-21. [Google Scholar] [CrossRef]
- Jiang, F.; Du, C.; Jiang, W.; Wang, L.; Du, S.-K. The preparation, formation, fermentability, and applications of resistant starch. Int. J. Biol. Macromol. 2020, 150, 1155–1161. [Google Scholar] [CrossRef]
- Wang, S.; Liu, Y.; Li, J.; Zhao, L.; Yan, W.; Lin, B.; Guo, X.; Wei, Y. Fusobacterium nucleatum acts as a pro-carcinogenic bacterium in colorectal cancer: From association to causality. Front. Cell Dev. Biol. 2021, 9, 710165. [Google Scholar] [CrossRef]
- Mehta, R.S.; Nishihara, R.; Cao, Y.; Song, M.; Mima, K.; Qian, Z.R.; Nowak, J.A.; Kosumi, K.; Hamada, T.; Masugi, Y. Association of dietary patterns with risk of colorectal cancer subtypes classified by Fusobacterium nucleatum in tumor tissue. JAMA Oncol. 2017, 3, 921–927. [Google Scholar] [CrossRef]
- Engevik, M.A.; Danhof, H.A.; Ruan, W.; Engevik, A.C.; Chang-Graham, A.L.; Engevik, K.A.; Shi, Z.; Zhao, Y.; Brand, C.K.; Krystofiak, E.S. Fusobacterium nucleatum secretes outer membrane vesicles and promotes intestinal inflammation. mBio 2021, 12, e02706-20. [Google Scholar] [CrossRef]
- Bachrach, G.; Rosen, G.; Bellalou, M.; Naor, R.; Sela, M. Identification of a Fusobacterium nucleatum 65 kDa serine protease. Oral Microbiol. Immunol. 2004, 19, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Curto, E.; Milligan, G. Metabolism meets immunity: The role of free fatty acid receptors in the immune system. Biochem. Pharmacol. 2016, 114, 3–13. [Google Scholar] [CrossRef]
- Zilm, P.S.; Gully, N.J.; Rogers, A.H. Growth pH and transient increases in amino acid availability influence polyglucose synthesis by Fusobacterium nucleatum grown in continuous culture. FEMS Microbiol. Lett. 2002, 215, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [PubMed]
- Bach Knudsen, K.E.; Lærke, H.N.; Hedemann, M.S.; Nielsen, T.S.; Ingerslev, A.K.; Gundelund Nielsen, D.S.; Theil, P.K.; Purup, S.; Hald, S.; Schioldan, A.G. Impact of diet-modulated butyrate production on intestinal barrier function and inflammation. Nutrients 2018, 10, 1499. [Google Scholar] [CrossRef]
- Signat, B.; Roques, C.; Poulet, P.; Duffaut, D. Role of Fusobacterium nucleatum in periodontal health and disease. Curr. Issues Mol. Biol. 2011, 13, 25–36. [Google Scholar] [PubMed]
- Thurnheer, T.; Karygianni, L.; Flury, M.; Belibasakis, G.N. Fusobacterium species and subspecies differentially affect the composition and architecture of supra-and subgingival biofilms models. Front. Microbiol. 2019, 10, 1716. [Google Scholar] [CrossRef]
- Dessì, A.; Bosco, A.; Pintus, R.; Orrù, G.; Fanos, V. Fusobacterium nucleatum and alteration of the oral microbiome: From pregnancy to SARS-CoV-2 infection. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 4579–4596. [Google Scholar]
| Gene | Direction | Sequence (5′→3′) |
|---|---|---|
| IL1B | Forward | 5′-AAACAGATGAAGTGCTCCTTCCAGG-3′ |
| Reverse | 5′-TGGAGAACACCACTTGTTGCTCCA-3′ | |
| IL6 | Forward | 5′-CCCAGGAGAAGATTCCAAAGATGTA-3′ |
| Reverse | 5′-GTCGAGGATGTACCGAATTTGTTTG-3′ | |
| IL8 | Forward | 5′-GAGAGTGATTGAGAGTGGACCAC-3′ |
| Reverse | 5′-CACAACCCTCTGCACCCAGTTT-3′ | |
| IL10 | Forward | 5′-CCTGACCACGCTTTCTAGCTGTT-3′ |
| Reverse | 5′-GGCTCCCTGGTTTCTCTTCCTAAG-3′ | |
| TLR4 | Forward | 5′-CATATCAGAGCCTAAGCCACCTCTC-3′ |
| Reverse | 5′-AGCCACCAGCTTCTGTAAACTTGAT-3′ | |
| MYD88 | Forward | 5′-GAGGCTGAGAAGCCTTTACAGG-3′ |
| Reverse | 5′-GCAGATGAAGGCATCGAAACGC-3′ | |
| MLH1 | Forward | 5′-TTGTTTTTATTGGTTGGATATT-3′ |
| Reverse | 5′-AAATACCAATCAAATTTCTCAA-3′ | |
| Myc | Forward | 5′-GAGGATCTCTGGGAGGAATGCTACT-3′ |
| Reverse | 5′-CTGCTTTGTATTATCTGCGTGGCTA-3′ | |
| GAPDH | Forward | 5′-TCCTGCACCACCAACTGCTT-3′ |
| Reverse | 5′-GGGGCCATCCACAGTCTTCT-3′ |
| Species | Pathway | Representative Gene (Accession Number) | Description | References |
|---|---|---|---|---|
| F. nucleatum ATCC 25586 | Glutarate | FN02019 (AAL94407) | Glutaconyl-COA decarboxylase beta subunit (GcdB) | [49] |
| 4- Aminobutyrate | FN0621 (AAL94817) | 4-hydroxybutyrate coenzyme A transferase (4Hbt/ACH1) | [31] | |
| Lysine | FN1868 (AAL93967) | 3-keto-5-aminohexanoate cleavage enzyme (kce) | [45] | |
| Pyruvate | FN1421 (AAL95614) FN1020 (AAL95216) | Pyruvate-flavodoxin/ferredoxin oxidoreductase (Por/nifJ) Enoyl-Coenzyme A (CoA) hydratase (Crt) | This study | |
| F. prausnitzii L2-6 | Glutarate | FP2_05280 (CBK98162) | Glutaconyl-CoA decarboxylase beta subunit (GcdB) | This study |
| Pyruvate | FP2_19990 (CBK99399) FP2_20590 (CBK99451) | Pyruvate:ferredoxin (flavodoxin) oxidoreductase, homodimeric (Por), Enoyl-CoA hydratase/carnithine racemase (Crt) | [29] | |
| C. catus GD/7 | Glutarate | CC1_18370 (CBK80577) | Glutaconyl-CoA/methylmalonyl-CoA decarboxylase subunit delta (GcdD) | This study |
| Pyruvate | CC1_34350 (CBK81960) CC1_14660 (CBK80244) | Pyruvate-ferredoxin/flavodoxin oxidoreductase (Por), Enoyl-CoA hydratase (Crt) | [31] | |
| R. intestinalis M50/1 | Glutarate | ROI_21870 (CBL09200) | Glutaconyl-CoA/methylmalonyl-CoA decarboxylase subunit delta (GcdD) | This study |
| Pyruvate | ROI_19110 (CBL08968) ROI_08630 (CBL08094) | Pyruvate:ferredoxin (flavodoxin) oxidoreductase (Por/nifJ) Enoyl-CoA hydratase (Crt) | [60] | |
| E. rectale ATCC 33656 | Glutarate | EUBREC_1628 (ACR75372) | Pyruvate carboxylase subunit B (GcdC) | This study |
| Pyruvate | EUBREC_1472 (ACR75226) EUBREC_1017 (ACR74779) | Pyruvate-ferredoxin/flavodoxin oxidoreductase (Por, nifJ) enoyl-CoA hydratase (Crt) | [61] | |
| C. eutactus ATCC 27759 | Glutarate | LK421_00715 (EDP27590) | Sodium ion-translocating decarboxylase subunit beta (GcdB/OAD_beta) | This study |
| Pyruvate | LK421_02445 (EDP24924) LK421_13465 (EDP26522) | Pyruvate ferredoxin oxidoreductase beta subunit (PorB) Enoyl-CoA hydratase (Crt) | [62] | |
| M. elsdenii DSM 20460 | Glutarate | MELS_1065 (CCC73287) | Putative acetyl-CoA carboxylase biotin carboxyl carrier protein subunit (GcdC) | This study |
| 4- Aminobutyrate | MELS_0341 (CCC72564) | Acetyl-CoA hydrolase/transferase/4-hydroxybutyrate CoA-transferase (ACH1) | [63] | |
| Pyruvate | MELS_0987 (CCC73209) MELS_1449 (CCC73670) | Pyruvate-flavodoxin oxidoreductase (Por/nifJ), 3-hydroxybutyryl-CoA dehydratase (Crt) | [31] | |
| F. plautii YL31 | 4- Aminobutyrate | A4U99_00500 (ANU39622) | 4-hydroxybutyrate CoA-transferase (Cat2, AbfT ACH1) | [50] |
| Lysine | A4U99_00635 (ANU39646) | 3-keto-5-aminohexanoate cleavage protein (Kce) | [29] | |
| Pyruvate | A4U99_17605 (ANU42771) A4U99_15055 (ANU42306) | pyruvate:ferredoxin (flavodoxin) oxidoreductase (Por, nifJ) enoyl-CoA hydratase (Crt) | [64] | |
| C. difficile 630 | 4- Aminobutyrate | CD630_23390 (CAJ69226) | 4-hydroxybutyrate CoA transferase (Cat2, AbfT, ACH1) | [31] |
| Pyruvate | CD630_26820 (CAJ69568) CD630_08000 (CAJ67634) | Pyruvate-ferredoxin oxidoreductase (Pfo/Por), 3-hydroxybutyryl-CoA dehydratase (Crt1) | [31] | |
| C. perfringens ATCC 13124 | Pyruvate | CPF_2318 (ABG84858) CPF_0090 (ABG84755) | Pyruvate-flavodoxin oxidoreductase (Por, nifJ) 3-hydroxybutyryl-CoA dehydratase (Crotonase) (Crt) | [31] |
| B. virosa FDAARGOS 1229 | 4- Aminobutyrate | I6J59_04665 (QRO51906) | Acetyl-CoA hydrolase/transferase family protein (ACH1) | [65] |
| Lysine | I6J59_05615 (QRO51094) | 3-keto-5-aminohexanoate cleavage protein (Kce/BKACE) | [66] | |
| Pyruvate | I6J59_17360 (QRO49639) I6J59_05415 (QRO51056) | Pyruvate:ferredoxin (flavodoxin) oxidoreductase (Por, nifJ), Short-chain-enoyl-CoA hydratase (Crt) | [66] | |
| P. gingivalis ATCC 33277 | 4-Aminobutyrate | PGN_0725 (BAG33244) | 4-hydroxybutyrate CoA-transferase (ACH1) | [50] |
| Lysine | PGN_1164 (BAG33683) | 3-keto-5-aminohexanoate cleavage enzyme (kce) | [50] | |
| Pyruvate | PGN_1418 (BAG33937) PGN_1175 (BAG33694) | Pyruvate-flavodoxin oxidoreductase (Por/nifJ) Putative enoyl-CoA hydratase (Crt) | [31] |
| Species | Pathogenic or Nonpathogenic | Pathway | |||
|---|---|---|---|---|---|
| Pyruvate | 4-Amino Butyrate | Glutarate | Lysine | ||
| F. nucleatum ATCC 25586 | Opportunistic pathogen | + | + | + | + |
| F. prausnitzii L2-6 | Commensal | + | - | + | - |
| R. intestinalis M50/1 | Commensal | + | - | + | - |
| E. rectale ATCC 33656 | Commensal | + | - | + | - |
| C. catus GD/7 | Commensal | + | - | + | - |
| C. eutactus ATCC 27759 | Commensal | + | - | + | - |
| M. elsdenii DSM 20460 | Opportunistic pathogen | + | + | + | - |
| F. plautii YL31 | Opportunistic pathogen | + | + | - | + |
| C. difficile 360 | pathogen | + | + | - | - |
| C. perfringens ATCC 13124 | pathogen | + | + | - | - |
| B. virosa FDAARGOS 1229 | pathogen | + | + | - | + |
| P. gingivalis ATCC 33277 | pathogen | + | + | - | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nawab, S.; Bao, Q.; Ji, L.-H.; Luo, Q.; Fu, X.; Fan, S.; Deng, Z.; Ma, W. The Pathogenicity of Fusobacterium nucleatum Modulated by Dietary Fibers—A Possible Missing Link between the Dietary Composition and the Risk of Colorectal Cancer. Microorganisms 2023, 11, 2004. https://doi.org/10.3390/microorganisms11082004
Nawab S, Bao Q, Ji L-H, Luo Q, Fu X, Fan S, Deng Z, Ma W. The Pathogenicity of Fusobacterium nucleatum Modulated by Dietary Fibers—A Possible Missing Link between the Dietary Composition and the Risk of Colorectal Cancer. Microorganisms. 2023; 11(8):2004. https://doi.org/10.3390/microorganisms11082004
Chicago/Turabian StyleNawab, Sadia, Qelger Bao, Lin-Hua Ji, Qian Luo, Xiang Fu, Shuxuan Fan, Zixin Deng, and Wei Ma. 2023. "The Pathogenicity of Fusobacterium nucleatum Modulated by Dietary Fibers—A Possible Missing Link between the Dietary Composition and the Risk of Colorectal Cancer" Microorganisms 11, no. 8: 2004. https://doi.org/10.3390/microorganisms11082004
APA StyleNawab, S., Bao, Q., Ji, L.-H., Luo, Q., Fu, X., Fan, S., Deng, Z., & Ma, W. (2023). The Pathogenicity of Fusobacterium nucleatum Modulated by Dietary Fibers—A Possible Missing Link between the Dietary Composition and the Risk of Colorectal Cancer. Microorganisms, 11(8), 2004. https://doi.org/10.3390/microorganisms11082004







