Probiotics as an Alternative to Antibiotics: Genomic and Physiological Characterization of Aerobic Spore Formers from the Human Intestine
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolation and Characterization
2.2. Whole-Genome Sequencing and Bioinformatic Analysis
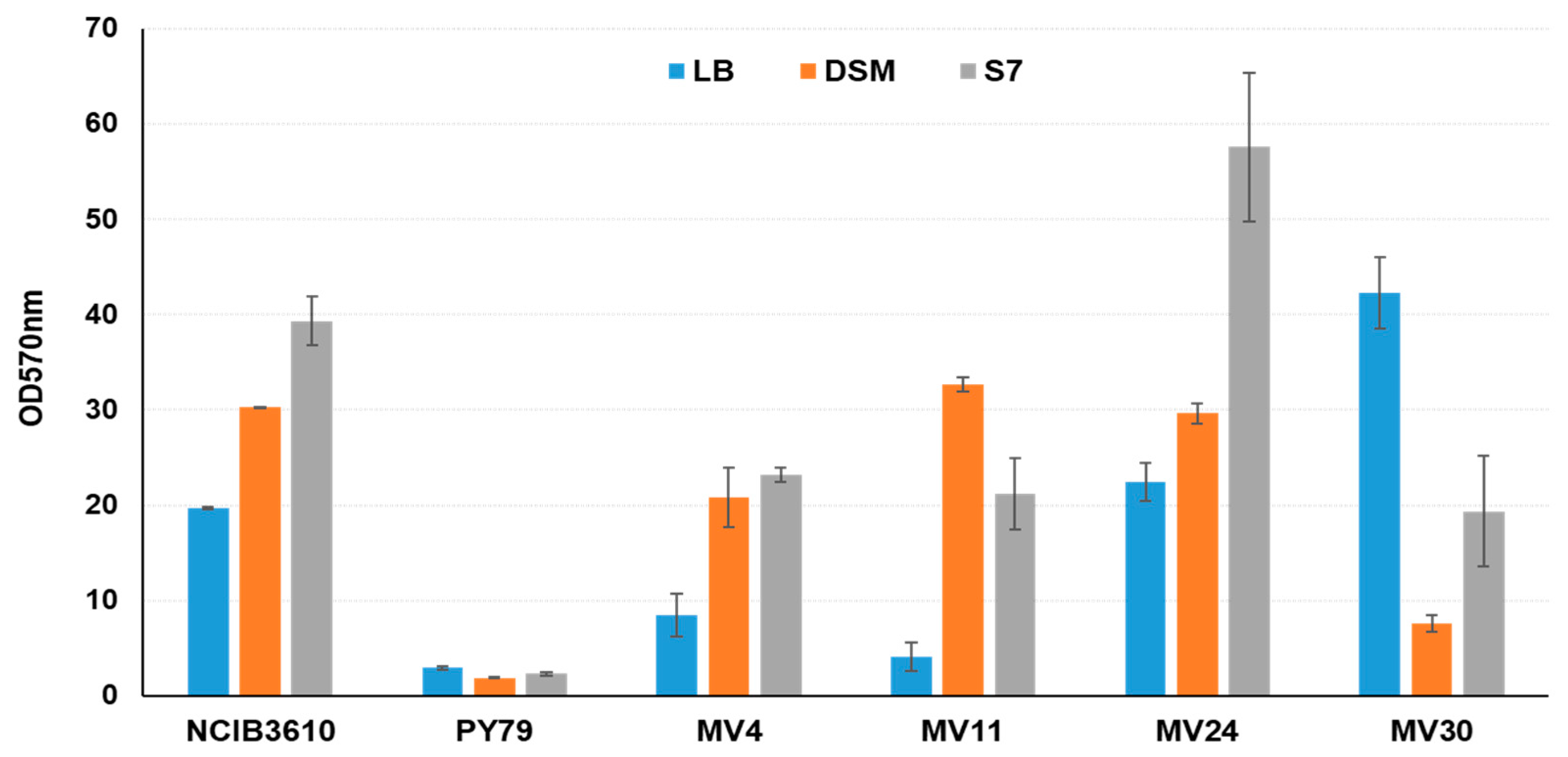
2.3. Antimicrobial and Biofilm Production Assays
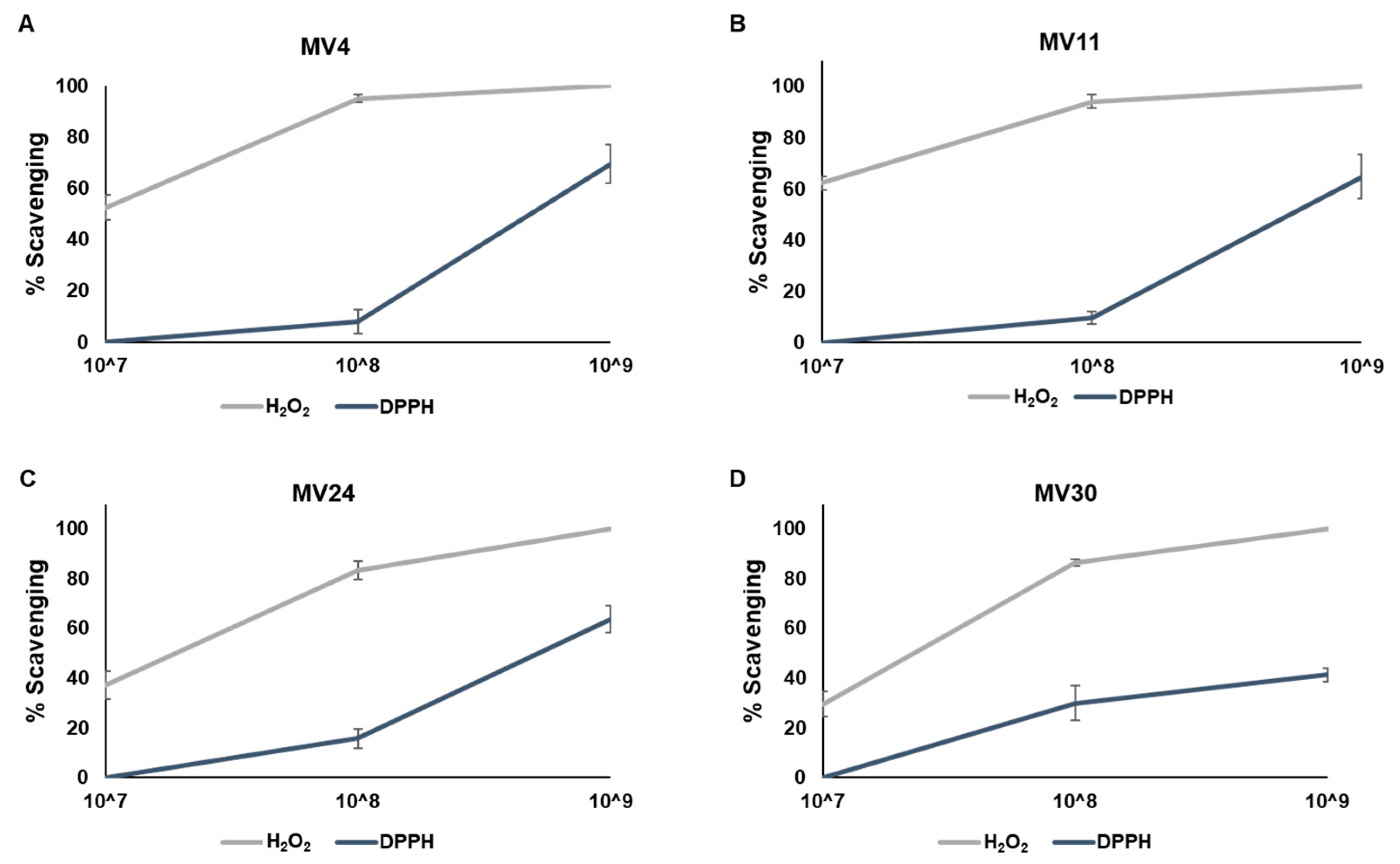
2.4. Antioxidant Activity Tests
2.4.1. Hydrogen Peroxide Scavenging Assay
2.4.2. DPPH Assay
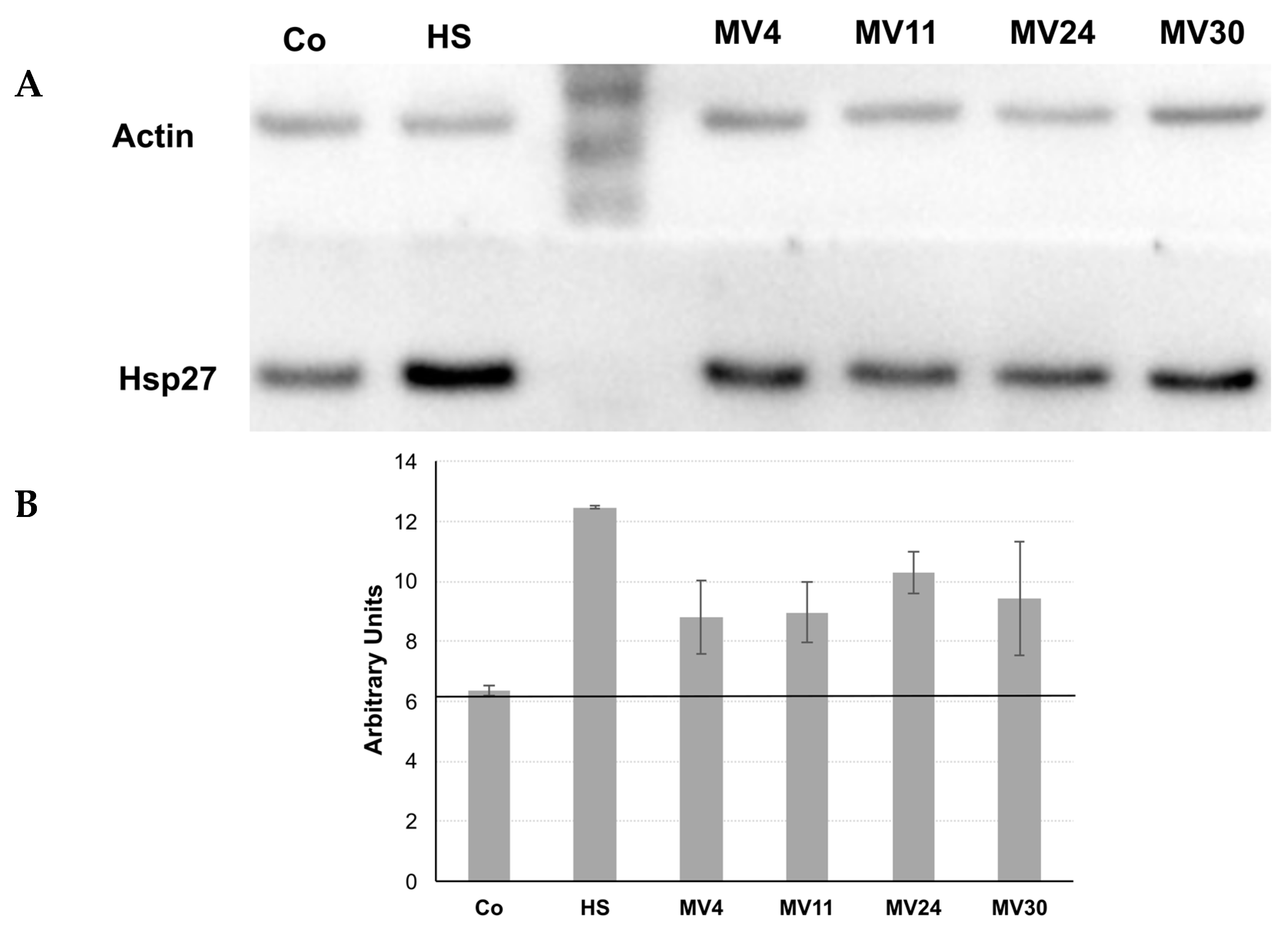
2.5. Growth and Treatment of HT-29 Cells with Bacterial Conditioned Medium
2.6. Protein Extraction, SDS-PAGE, and Western Blot Analysis
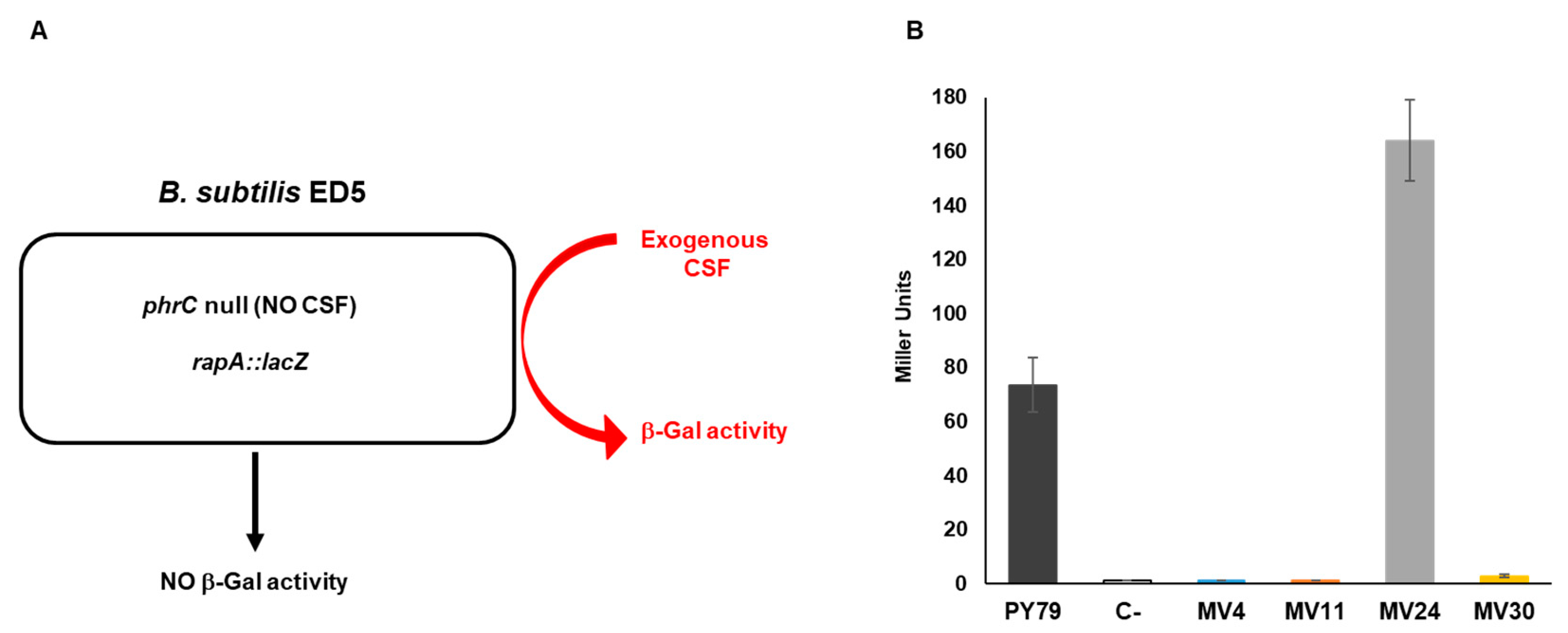
2.7. β-Galactosidase Activity
3. Results
3.1. Preliminary Characterization and Selection of Strains with Probiotic Potentials
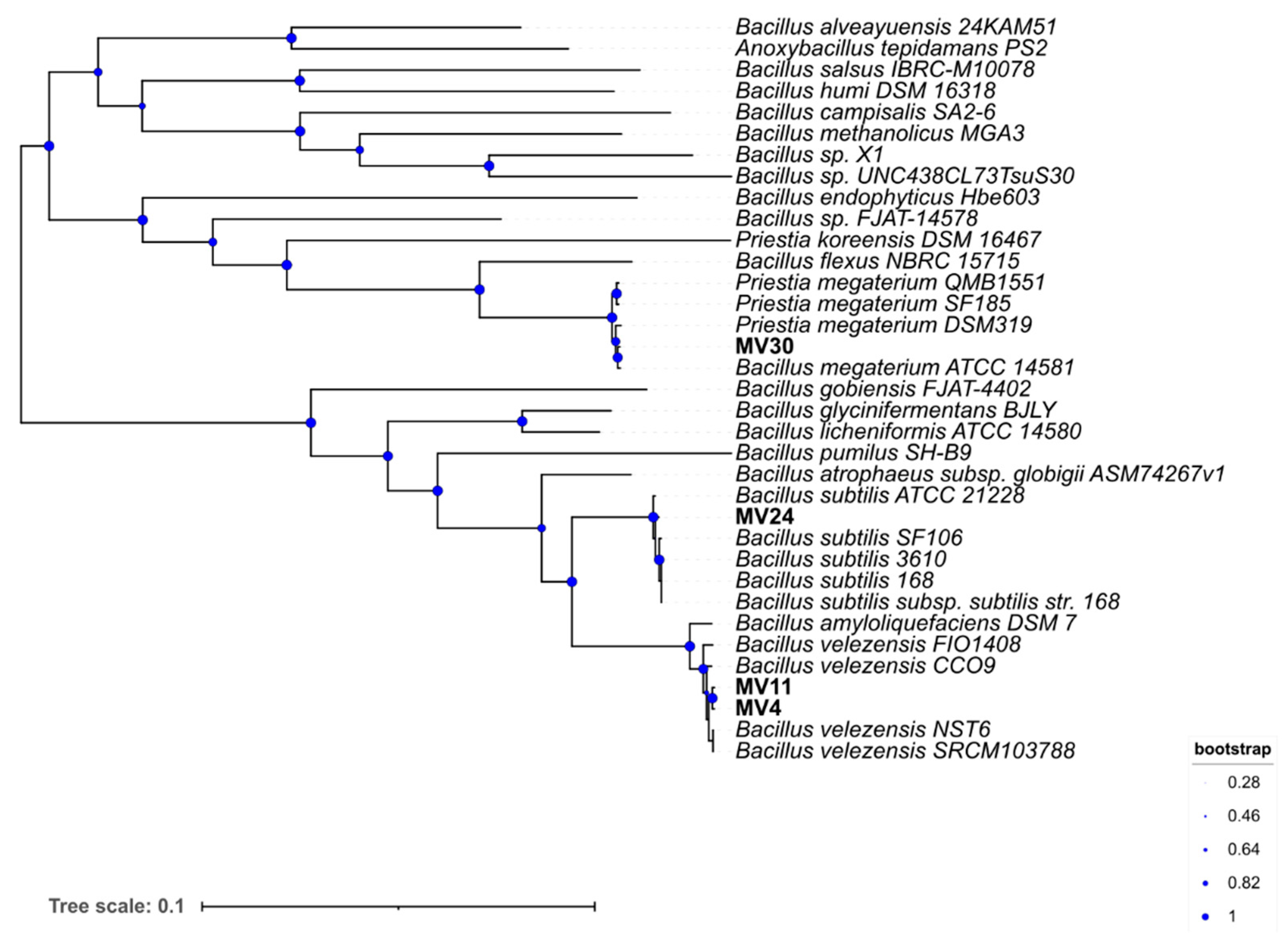
3.2. Species Assignment and Phylogenetic Analysi
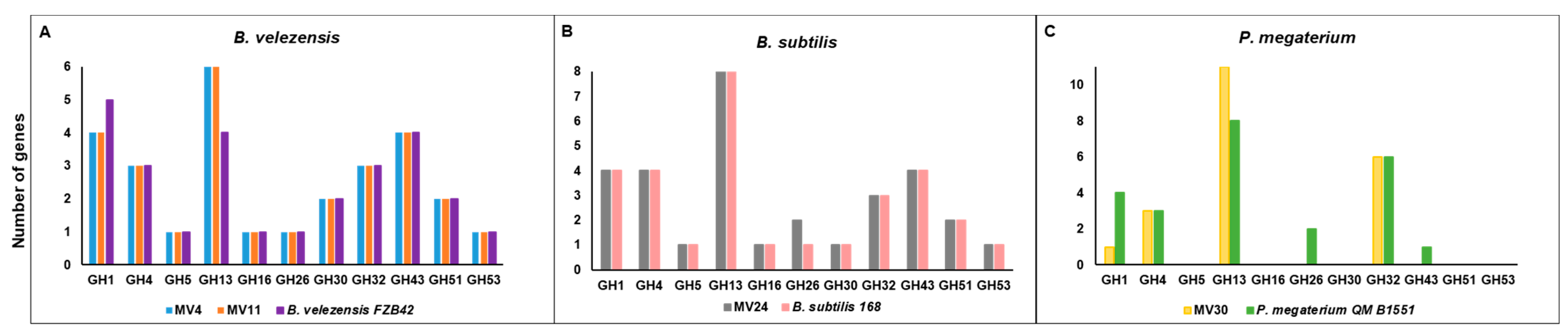
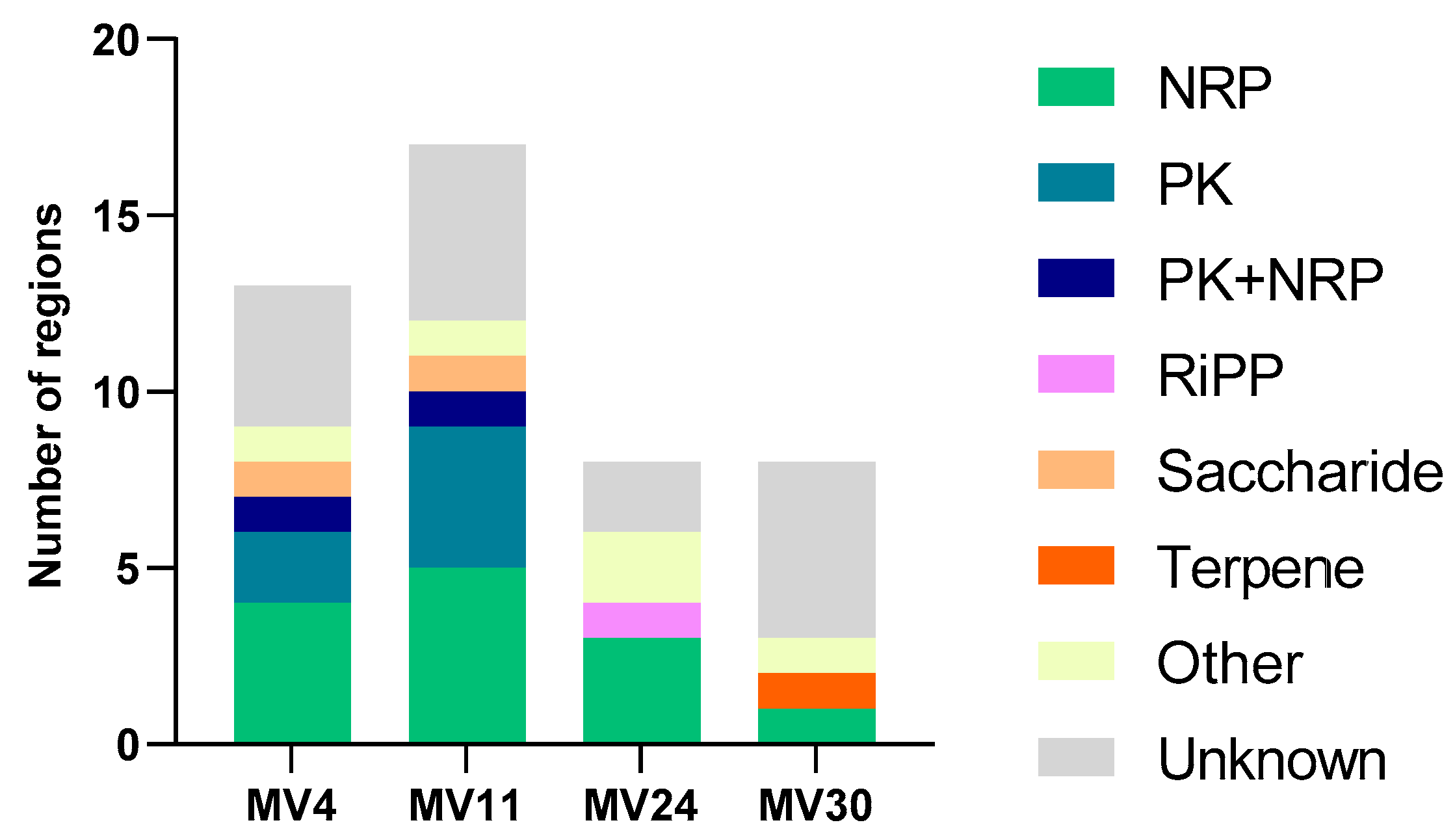
3.3. Genome Analyses
3.4. Antimicrobial and Antioxidant Activities
3.5. Matrix Formation
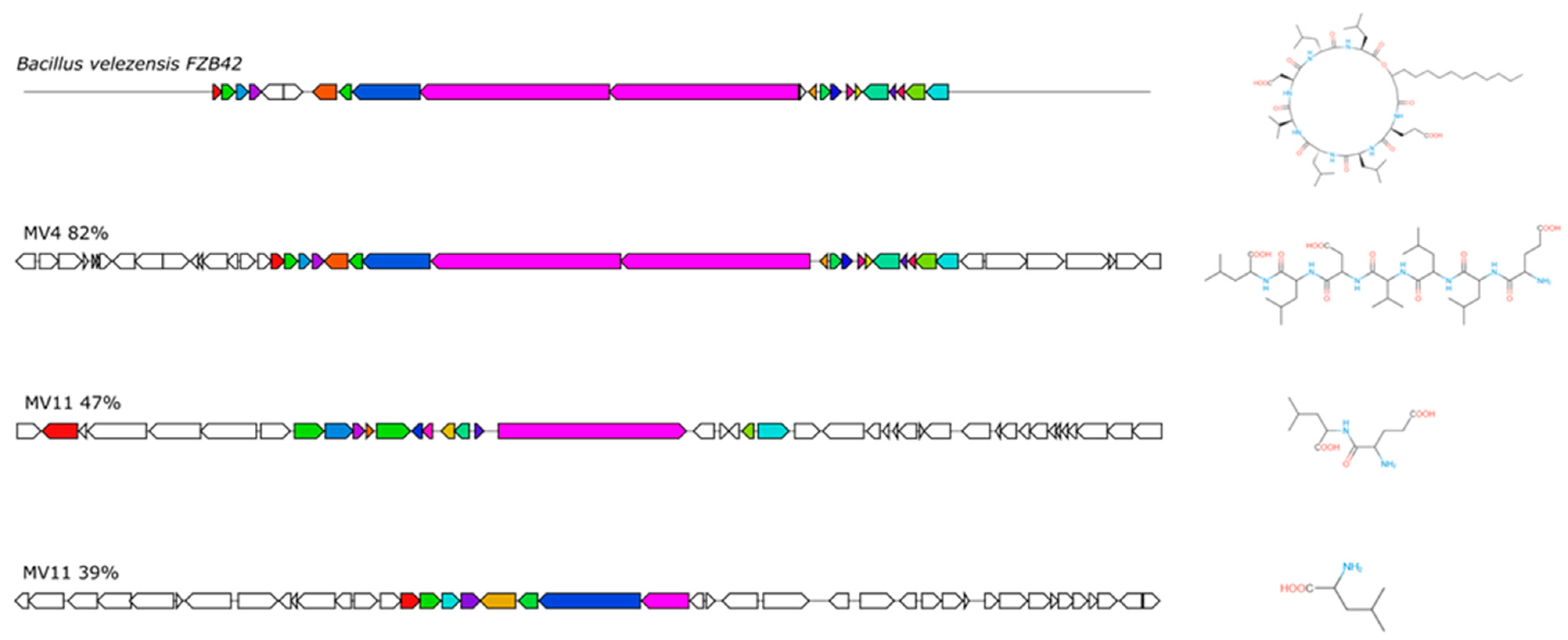
3.6. Production of CSF or CSF-like Peptides
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Browne, K.; Chakraborty, S.; Chen, R.; Willcox, M.D.; Black, D.S.; Walsh, W.R.; Kumar, N. A New era of antibiotics: The clinical potential of antimicrobial peptides. Int. J. Mol. Sci. 2020, 21, 7047. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Xue, Y.; Gao, W.; Li, J.; Zu, X.; Fu, D.; Bai, X.; Zuo, Y.; Hu, Z.; Zhang, F. Bacillaceae-derived peptide antibiotics since 2000. Peptides 2018, 101, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tran, D.Q.; Rhoads, J.M. Probiotics in disease prevention and treatment. J. Clin. Pharmacol. 2018, 58 (Suppl. 10), S164–S179. [Google Scholar] [CrossRef] [PubMed]
- Sanders, M.E.; Merenstein, D.J.; Reid, G.; Gibson, G.R.; Rastall, R.A. Probiotics and prebiotics in intestinal health and disease: From biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef]
- Martín, R.; Miquel, S.; Benevides, L.; Bridonneau, C.; Robert, V.; Hudault, S.; Chain, F.; Berteau, O.; Azevedo, V.; Chatel, J.M.; et al. Functional Characterization of novel Faecalibacterium prausnitzii strains isolated from healthy volunteers: A step forward in the use of F. prausnitzii as a Next-Generation Probiotic. Front. Microbiol. 2017, 8, 1664-302x. [Google Scholar] [CrossRef]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef]
- Nicholson, W.L.; Munakata, N.; Horneck, G.; Melosh, H.J.; Setlow, P. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 2000, 64, 548–572. [Google Scholar] [CrossRef]
- Christie, G.; Setlow, P. Bacillus spore germination: Knowns, unknowns and what we need to learn. Cell Signal 2020, 74, 109729. [Google Scholar] [CrossRef]
- Cutting, S.M. Bacillus probiotics. Food Microbiol. 2011, 28, 214–220. [Google Scholar] [CrossRef]
- Egan, M.; Dempsey, E.; Ryan, C.A.; Ross, R.P.; Stanton, C. The sporobiota of the human gut. Gut Microbes 2021, 13, e1863134. [Google Scholar] [CrossRef]
- Saggese, A.; Baccigalupi, L.; Ricca, E. Spore formers as beneficial microbes for humans and animals. Appl. Microbiol. 2021, 1, 498–509. [Google Scholar] [CrossRef]
- Casula, G.; Cutting, S.M. Bacillus probiotics: Spore germination in the gastrointestinal tract. Appl. Environ. Microbiol. 2002, 68, 2344–2352. [Google Scholar] [CrossRef] [PubMed]
- Bernardeau, M.; Lehtinen, M.J.; Forssten, S.D.; Nurminen, P. Importance of the gastrointestinal life cycle of Bacillus for probiotic functionality. J. Food Sci. Technol. 2017, 54, 2570–2584. [Google Scholar] [CrossRef]
- Tam, N.K.; Uyen, N.Q.; Hong, H.A.; Duc, L.H.; Hoa, T.T.; Serra, C.R.; Henriques, A.O.; Cutting, S.M. The intestinal life cycle of Bacillus subtilis and close relatives. J. Bacteriol. 2006, 188, 2692–2700. [Google Scholar] [CrossRef]
- Thirabunyanon, M.; Thongwittaya, N. Protection activity of a novel probiotic strain of Bacillus subtilis against Salmonella Enteritidis infection. Res. Vet. Sci. 2012, 93, 74–81. [Google Scholar] [CrossRef]
- Paparo, L.; Tripodi, L.; Bruno, C.; Pisapia, L.; Damiano, C.; Pastore, L.; Berni Canani, R. Protective action of Bacillus clausii probiotic strains in an in vitro model of Rotavirus infection. Sci. Rep. 2020, 10, 12636. [Google Scholar]
- Rhee, K.-J.; Sethupathi, P.; Driks, A.; Lanning, D.K.; Knight, K.L. Role of commensal bacteria in development of gut-associated lymphoid tissues and preimmune antibody repertoire. J. Immunol. 2004, 172, 1118. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.M.; La Ragione, R.; Nunez, A.; Cutting, S.M. Immunostimulatory activity of Bacillus spores. FEMS Immunol. Med. Microbiol. 2008, 53, 195–203. [Google Scholar] [CrossRef]
- Kotowicz, N.; Bhardwaj, R.K.; Ferreira, W.T.; Hong, H.A.; Olender, A.; Ramirez, J.; Cutting, S.M. Safety and probiotic evaluation of two Bacillus strains producing antioxidant compounds. Benef. Microbes 2019, 10, 759–771. [Google Scholar] [CrossRef]
- Mazzoli, A.; Donadio, G.; Lanzilli, M.; Saggese, A.; Guarino, A.M.; Rivetti, M.; Crescenzo, R.; Ricca, E.; Ferrandino, I.; Iossa, S.; et al. Bacillus megaterium SF185 spores exert protective effects against oxidative stress in vivo and in vitro. Sci. Rep. 2014, 9, 12082. [Google Scholar] [CrossRef]
- Petruk, G.; Donadio, G.; Lanzilli, M.; Isticato, R.; Monti, D.M. Alternative use of Bacillus subtilis spores: Protection against environmental oxidative stress in human normal keratinocytes. Sci. Rep. 2018, 8, 1745. [Google Scholar] [CrossRef]
- Hou, Q.; Jia, J.; Lin, J.; Zhu, L.; Xie, S.; Yu, Q.; Li, Y. Bacillus subtilis programs the differentiation of intestinal secretory lineages to inhibit Salmonella infection. Cell Rep. 2022, 40, 111416. [Google Scholar] [CrossRef]
- Ji, L.; Zhang, L.; Shen, J.; Zhang, Y.; Lu, L.; Zhang, X.; Ma, X. Bacillus subtilis M6 improves intestinal barrier, antioxidant capacity and gut microbial composition in AA broiler. Front. Nutr. 2022, 9, 965310. [Google Scholar] [CrossRef]
- Marzorati, M.; Van den Abbeele, P.; Bubeck, S.; Bayne, T.; Krishnan, K.; Young, A. Treatment with a spore-based probiotic containing five strains of Bacillus induced changes in the metabolic activity and community composition of the gut microbiota in a SHIME® model of the human gastrointestinal system. Food Res. Intern. 2021, 149, 110676. [Google Scholar] [CrossRef] [PubMed]
- Altaf, F.; Wu, S.; Kasim, V. Role of fibrinolytic enzymes in anti-thrombosis therapy. Front. Mol. Biosci. 2021, 8, 680397. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Irorita Fugaban, J.I.; Holzapfel, W.H.; Todorov, S.D. Selection of beneficial bacterial strains with potential as oral probiotic candidates. Probiotics Antimicrob. Proteins 2022, 14, 1077–1093. [Google Scholar] [CrossRef]
- Mnif, I.; Ghribi, D. Review lipopeptides biosurfactants: Mean classes and new insights for industrial, biomedical, and environmental applications. Biopolymers 2015, 104, 129–147. [Google Scholar] [CrossRef] [PubMed]
- Saggese, A.; De Luca, Y.; Baccigalupi, L.; Ricca, E. An antimicrobial peptide specifically active against Listeria monocytogenes is secreted by Bacillus pumilus SF214. BMC Microbiol. 2022, 22, 3. [Google Scholar] [CrossRef]
- Fujita, M.; Musch, M.W.; Nakagawa, Y.; Hu, S.; Alverdy, J.; Kohgo, Y.; Schneewind, O.; Jabri, B.; Chang, E.B. The Bacillus subtilis quorum-sensing molecule CSF contributes to intestinal homeostasis via OCTN2, a host cell membrane transporter. Cell Host Microbe 2007, 1, 299–308. [Google Scholar] [CrossRef]
- Okamoto, K.; Fujiya, M.; Nata, T.; Ueno, N.; Inaba, Y.; Ishikawa, C.; Ito, T.; Moriichi, K.; Tanabe, H.; Mizukami, Y.; et al. Competence and sporulation factor derived from Bacillus subtilis improves epithelial cell injury in intestinal inflammation via immunomodulation and cytoprotection. Int. J. Colorectal Dis. 2012, 27, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Di Luccia, B.; D’Apuzzo, E.; Varriale, F.; Baccigalupi, L.; Ricca, E.; Pollice, A. Bacillus megaterium SF185 induces stress pathways and affects the cell cycle distribution of human intestinal epithelial cells. Benef. Microbes 2016, 7, 609–620. [Google Scholar] [CrossRef]
- Cogliati, S.; Clementi, V.; Francisco, M.; Crespo, C.; Argañaraz, F.; Grau, R. Bacillus subtilis delays neurodegeneration and behavioral impairment in the Alzheimer’s disease model Caenorhabditis elegans. J. Alzheimers Dis. 2020, 73, 1035–1052. [Google Scholar] [CrossRef]
- Donato, V.; Rodrıguez Ayala, F.; Cogliati, S.; Bauman, C.; Costa, J.C.; Lenini, C.; Grau, R. Bacillus subtilis biofilm extends Caenorhabditis elegans longevity through downregulation of the insulin-like signalling pathway. Nat. Commun. 2017, 8, 14332. [Google Scholar] [CrossRef] [PubMed]
- Rodrıguez Ayala, F.; Bauman, C.; Cogliati, S.; Lenini, C.; Bartolini, M.; Grau, R. Microbial flora, probiotics, Bacillus subtilis and the search for a long and healthy human longevity. Microbial. Cell 2017, 4, 133–136. [Google Scholar] [CrossRef] [PubMed]
- Fakhry, S.; Sorrentini, I.; Ricca, E.; De Felice, M. and Baccigalupi, L. Characterisation of spore forming Bacilli isolated from the human gastrointestinal tract. J. App. Microbiol. 2008, 105, 2178–2186. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP). Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012, 10, 2740. [Google Scholar]
- Saggese, A.; Isticato, R.; Cangiano, G.; Ricca, E.; Baccigalupi, L. CotG-like modular proteins are common among spore-forming Bacilli. J. Bacteriol. 2016, 198, 1513–1520. [Google Scholar] [CrossRef]
- Lugli, G.A.; Milani, C.; Mancabelli, L.; Van Sinderen, D.; Ventura, M. MEGAnnotator: A user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol. Lett. 2016, 363, fnw049. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Saggese, A.; Giglio, R.; D’Anzi, N.; Baccigalupi, L.; Ricca, E. Comparative genomics and physiological characterization of two aerobic spore formers isolated from human ileal samples. Int. J Mol. Sci. 2022, 23, 14946. [Google Scholar] [CrossRef]
- Vittoria, M.; Saggese, A.; Di Gregorio Barletta, G.; Castaldi, S.; Isticato, R.; Baccigalupi, L.; Ricca, E. Sporulation efficiency and spore quality in a human intestinal isolate of Bacillus cereus. Res. Microbiol. 2023, 2, 104030. [Google Scholar] [CrossRef] [PubMed]
- Isticato, R.; Sirec, T.; Vecchione, S.; Crispino, A.; Saggese, A.; Baccigalupi, L.; Notomista, E.; Driks, A.; Ricca, E. The direct interaction between two morphogenetic proteins is essential for spore coat formation in Bacillus subtilis. PLoS ONE 2015, 10, e0141040. [Google Scholar] [CrossRef] [PubMed]
- Zanfardino, A.; Migliardi, A.; D’Alonzo, D.; Lombardi, A.; Varcamonti, M.; Cordone, A. Inactivation of MSMEG_0412 gene drastically affects surface related properties of Mycobacterium smegmatis. BMC Microbiol 2016, 16, 267. [Google Scholar] [CrossRef] [PubMed]
- Di Luccia, B.; Acampora, V.; Saggese, A.; Calabrò, V.; Vivo, M.; Angrisano, T.; Baccigalupi, L.; Ricca, E.; Pollice, A. Modulation of intestinal epithelial cell proliferation and apoptosis by Lactobacillus gasseri SF1183. Sci. Rep. 2022, 12, 20248. [Google Scholar] [CrossRef] [PubMed]
- Guinebretiere, M.H.; Broussolle, V.; Nguyen-The, C. Enterotoxigenic profiles of food-poisoning and food-borne Bacillus cereus strains. J. Clin. Microbiol. 2002, 40, 3053–3056. [Google Scholar] [CrossRef]
- Phelps, R.J.; McKillip, J.L. Enterotoxin production in natural isolates of Bacillaceae outside the Bacillus cereus group. Appl. Environ. Microbiol. 2002, 68, 3147–3151. [Google Scholar] [CrossRef]
- Li, Y.; Lei, L.; Zheng, L.; Xiao, X.; Tang, L.; Luo, C. Genome sequencing of gut symbiotic Bacillus velezensis LC1 for bioethanol production from bamboo shoots. Biotechnol. Biofuels 2020, 13, 34. [Google Scholar] [CrossRef]
- Arnaouteli, S.; Bamford, N.C.; Stanley-Wall, N.R.; Kovacs, A.T. Bacillus subtilis biofilm formation and social interactions. Nat. Rev. Microbiol. 2021, 19, 600–614. [Google Scholar]
- Vilain, S.; Pretorius, J.M.; Theron, J.; Brozel, V.S. DNA as an adhesin: Bacillus cereus requires extracellular DNA to form biofilm. Appl. Environ. Microbiol. 2009, 75, 2861–2868. [Google Scholar] [CrossRef]
- Nijland, R.; Burgess, J.G.; Errington, J.; Veening, J.W. Transfromation of environmental Bacillus subtilis isolates by transiently inducing genetic competence. PLoS ONE 2010, 5, e9724. [Google Scholar] [CrossRef]
- Ero, M.P.; Ng, C.M.; Mihailovski, T.; Harvey, N.R.; Lewis, B.H. A pilot study on the serum pharmacokinetics of nattokinase in humans following a single, oral, daily dose. Altern. Ther. Health Med. 2013, 19, 16–19. [Google Scholar] [PubMed]
| Strain | Van | Gen | Kan | Strep | Ery | Clin | Tet | Chl |
|---|---|---|---|---|---|---|---|---|
| MV4 | 1.500 | 0.500 | 0.750 | 8.000 | 0.094 | 0.750 | 8.000 | 1.000 |
| MV6 | 0.500 | 0.125 | 2.000 | R 1 | 0.125 | 0.190 | 1.000 | 0.250 |
| MV11 | 0.190 | 0.750 | 0.500 | 1.000 | 0.047 | 0.125 | 2.000 | 2.000 |
| MV14 | 3.000 | 0.125 | 0.750 | 8.000 | 0.250 | 4.000 | R 1 | 5.000 |
| MV15 | 0.380 | 0.250 | R 1 | 2.000 | 0.380 | R 1 | 0.250 | R 1 |
| MV17 | 0.047 | 0.500 | R 1 | R 1 | R 1 | 0.750 | 0.750 | R 1 |
| MV20 | 0.190 | 0.250 | 0.500 | 1.000 | 0.125 | R 1 | 1.000 | 0.750 |
| MV24 | 1.500 | 0.500 | 1.500 | 3.000 | 0.380 | 0.380 | 0.094 | 4.000 |
| MV26 | 0.125 | 0.047 | 0.125 | 1.000 | 0.064 | R 1 | 0.250 | 2.000 |
| MV30 | 0.125 | 0.047 | 0.190 | 8.000 | 0.064 | 4.000 | 0.500 | 1.500 |
| EFSA Breakpoints | 4.000 | 4.000 | 8.000 | 8.000 | 4.000 | 4.000 | 8.000 | 8.000 |
| MV4 | MV11 | MV24 | MV30 | |
|---|---|---|---|---|
| Size (bp) | 3,987,695 | 4,035,847 | 4,116,400 | 5,853,282 |
| Number of contigs | 36 | 58 | 112 | 35 |
| Average Coverage | 58 | 23 | 41 | 133 |
| Number of predicted ORFs | 3860 | 3956 | 4380 | 6003 |
| Average GC (percentage) | 46.35 | 46.2 | 43.26 | 37.46 |
| B. velezensis FZB42 | B. subtilis 168 | P. megaterium QM B1551 | |
|---|---|---|---|
| MV4 | 98.36 | 77.3 | 68.77 |
| MV11 | 98.36 | 77.04 | 68.44 |
| MV24 | 77.3 | 98.54 | 68.97 |
| MV30 | 68.31 | 68.58 | 97.42 |
| Species and Strain | GH | GT | PL | CE | CBM | AA | Total |
|---|---|---|---|---|---|---|---|
| B. velezensis MV4 | 47 | 37 | 3 | 11 | 16 | 3 | 117 |
| B. velezensis MV11 | 48 | 37 | 3 | 11 | 16 | 3 | 118 |
| B. velezensis FZB42 | 40 | 35 | 3 | 13 | 14 | 5 | 110 |
| B. subtilis MV24 | 56 | 39 | 7 | 14 | 22 | 3 | 141 |
| B. subtilis 168 | 56 | 34 | 7 | 14 | 23 | 4 | 138 |
| P. megaterium MV30 | 49 | 42 | 1 | 20 | 23 | 5 | 140 |
| P. megaterium QM B1551 | 53 | 42 | 1 | 24 | 21 | 5 | 146 |
| MV4 | MV11 | MV24 | MV30 | |
|---|---|---|---|---|
| Listeria monocytogenes ATCC 7644 | + | + | + | - |
| Bacillus cereus ATCC 10987 | ++ | ++ | + | - |
| Enterococcus faecalis ATCC 29212 | - | - | - | - |
| Streptococcus faecalis ATCC 33186 | +/- | +/- | - | - |
| Escherichia coli ATCC 25922 | +/- | +/- | - | - |
| Salmonella enterica subsp. enterica serovar Typhimurium ATCC 14028 | +/- | +/- | - | - |
| Citrobacter rodensis ATCC 14580 | +/- | +/- | - | - |
| Shigella sonnei ATCC 25931 | - | - | - | - |
| Candida albicans ATCC 14028 | +/- | +/- | - | - |
| Lacticaseibacillus rhamnosus GG 1 | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vittoria, M.; Saggese, A.; Isticato, R.; Baccigalupi, L.; Ricca, E. Probiotics as an Alternative to Antibiotics: Genomic and Physiological Characterization of Aerobic Spore Formers from the Human Intestine. Microorganisms 2023, 11, 1978. https://doi.org/10.3390/microorganisms11081978
Vittoria M, Saggese A, Isticato R, Baccigalupi L, Ricca E. Probiotics as an Alternative to Antibiotics: Genomic and Physiological Characterization of Aerobic Spore Formers from the Human Intestine. Microorganisms. 2023; 11(8):1978. https://doi.org/10.3390/microorganisms11081978
Chicago/Turabian StyleVittoria, Maria, Anella Saggese, Rachele Isticato, Loredana Baccigalupi, and Ezio Ricca. 2023. "Probiotics as an Alternative to Antibiotics: Genomic and Physiological Characterization of Aerobic Spore Formers from the Human Intestine" Microorganisms 11, no. 8: 1978. https://doi.org/10.3390/microorganisms11081978
APA StyleVittoria, M., Saggese, A., Isticato, R., Baccigalupi, L., & Ricca, E. (2023). Probiotics as an Alternative to Antibiotics: Genomic and Physiological Characterization of Aerobic Spore Formers from the Human Intestine. Microorganisms, 11(8), 1978. https://doi.org/10.3390/microorganisms11081978








