Correlation of CRISPR/Cas and Antimicrobial Resistance in Klebsiella pneumoniae Clinical Isolates Recovered from Patients in Egypt Compared to Global Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Clinical Isolates of Klebsiella pneumoniae
2.2. Molecular Screening of cas Genes and Selected ARGs
2.3. Antimicrobial Susceptibility Profiling
2.4. Whole Genome Sequencing (WGS) of Representative Isolates Carrying Type I-E and Type I-E* CRISPR/Cas
2.5. Retrieval, Characterization, and CRISPR/Cas Analysis of Published Genomic Sequences of Klebsiella pneumoniae
2.6. Statistical Analyses
2.7. Data Availability
3. Results
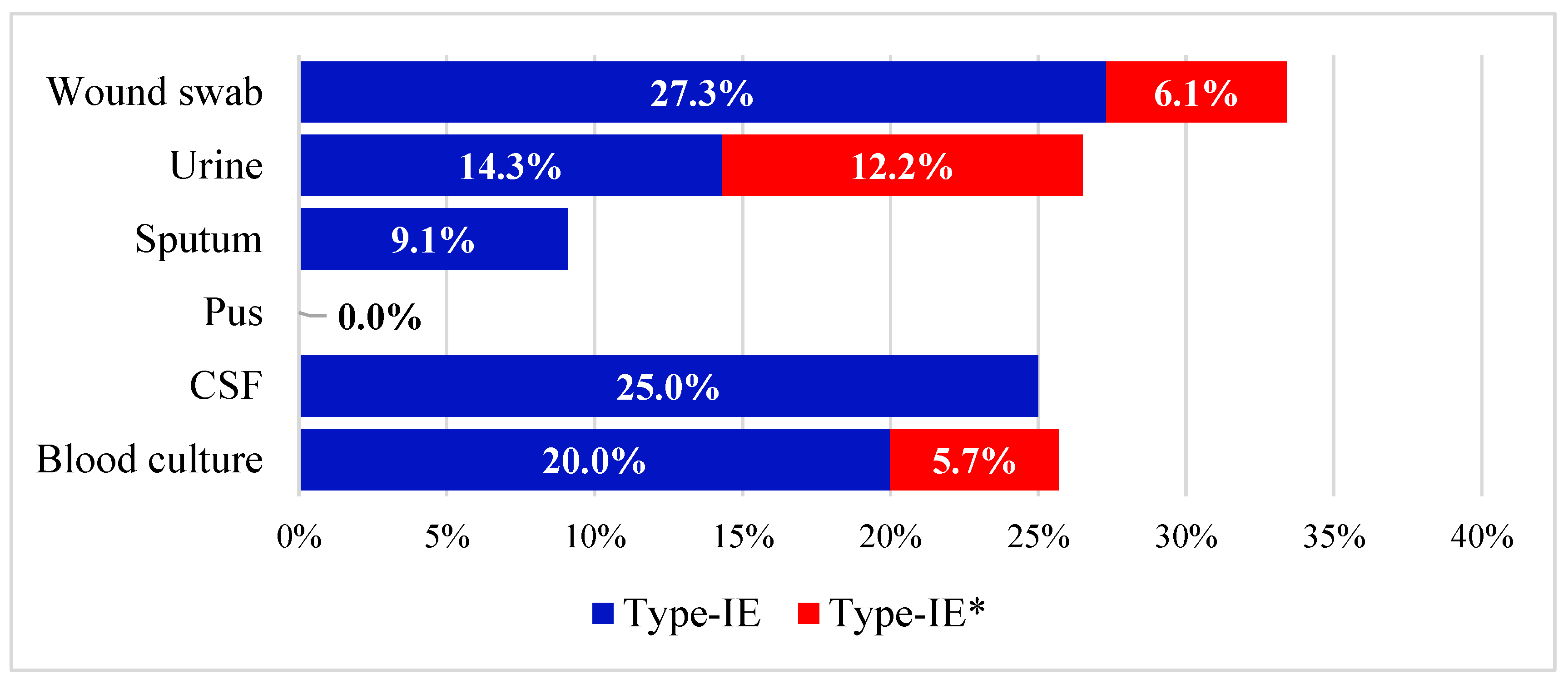
3.1. Prevalence of cas Genes in the Collected Isolates
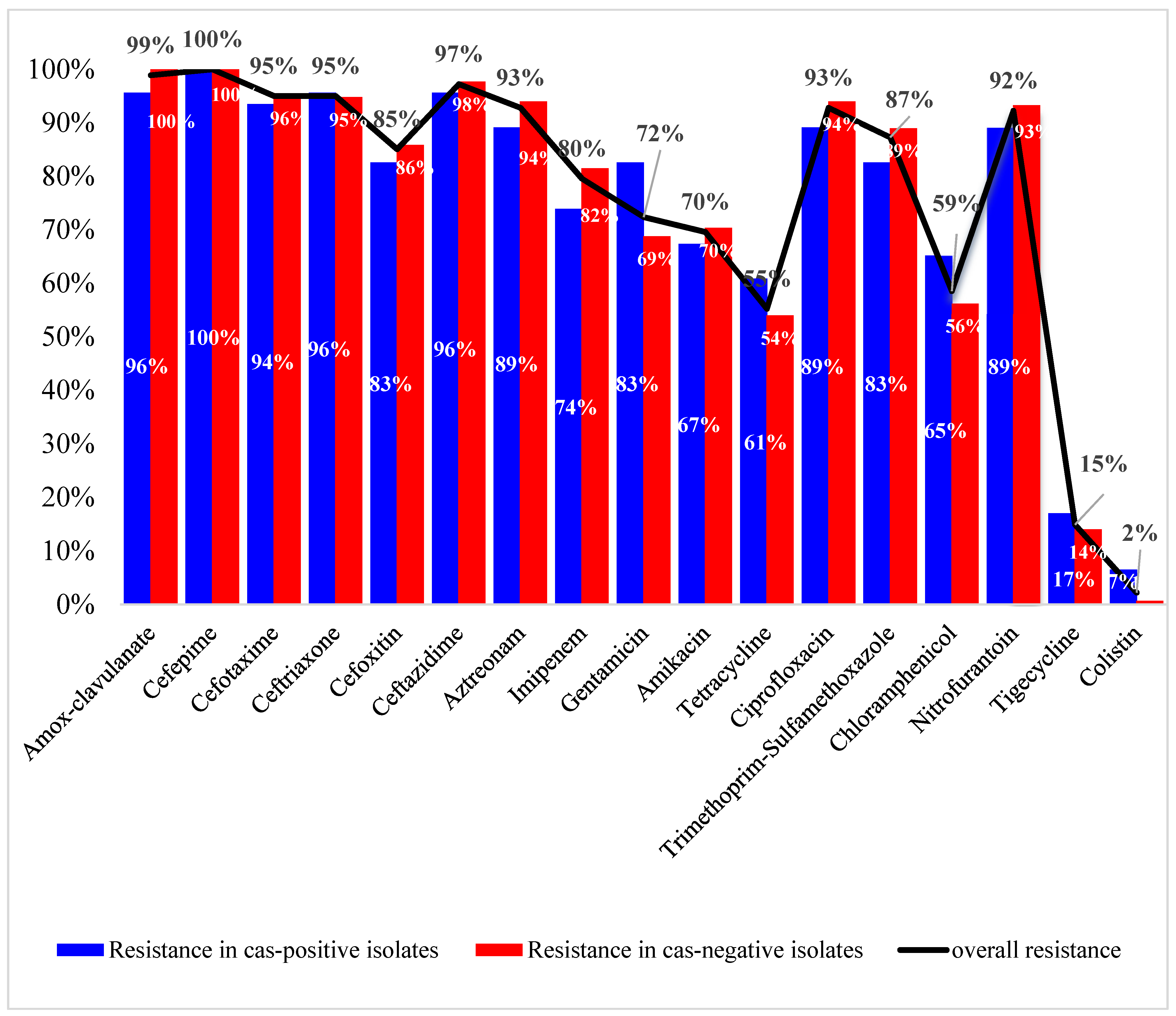
3.2. Antibiotic Susceptibility Profiles and Correlation to cas Genes
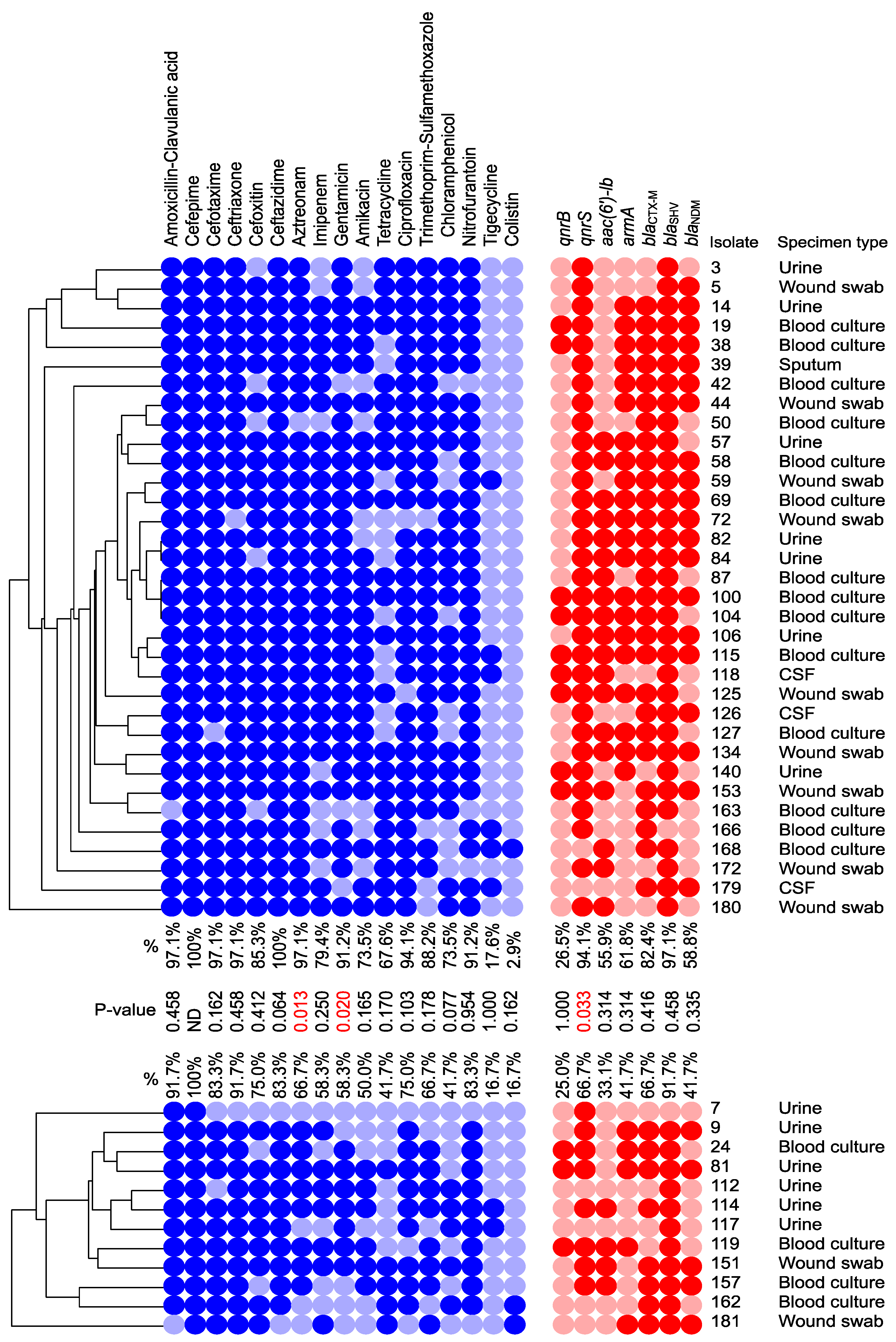
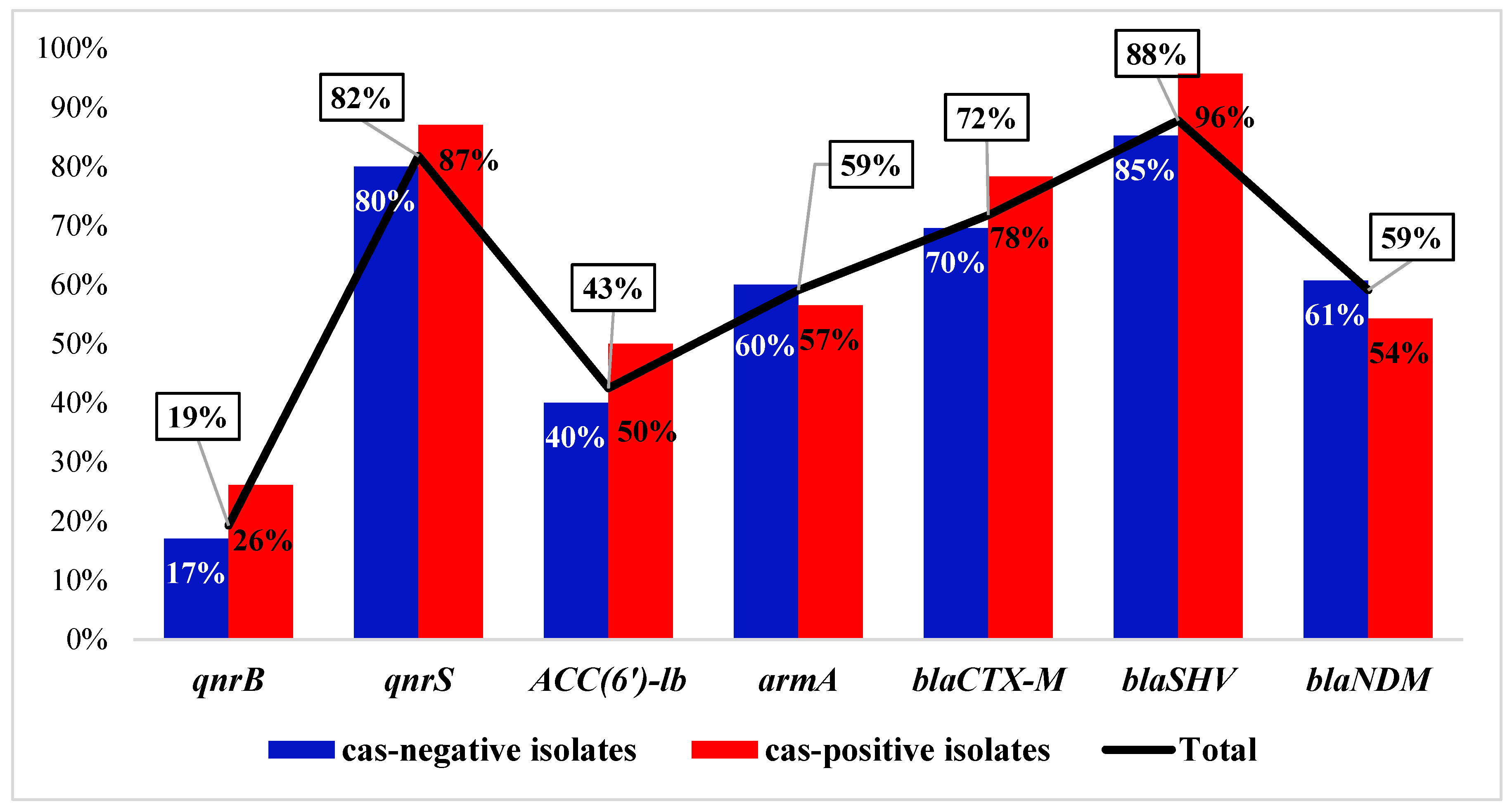
3.3. Distribution of the ARGs and Correlation to cas Genes
3.4. Draft Genome Analysis of Representative Isolates Carrying I-E and I-E* CRISPR/Cas
3.5. Features of the CRISPR/Cas Systems of the Published K. pneumoniae Genomes
3.5.1. ST Distribution of the CRISPR/Cas-Positive Strains
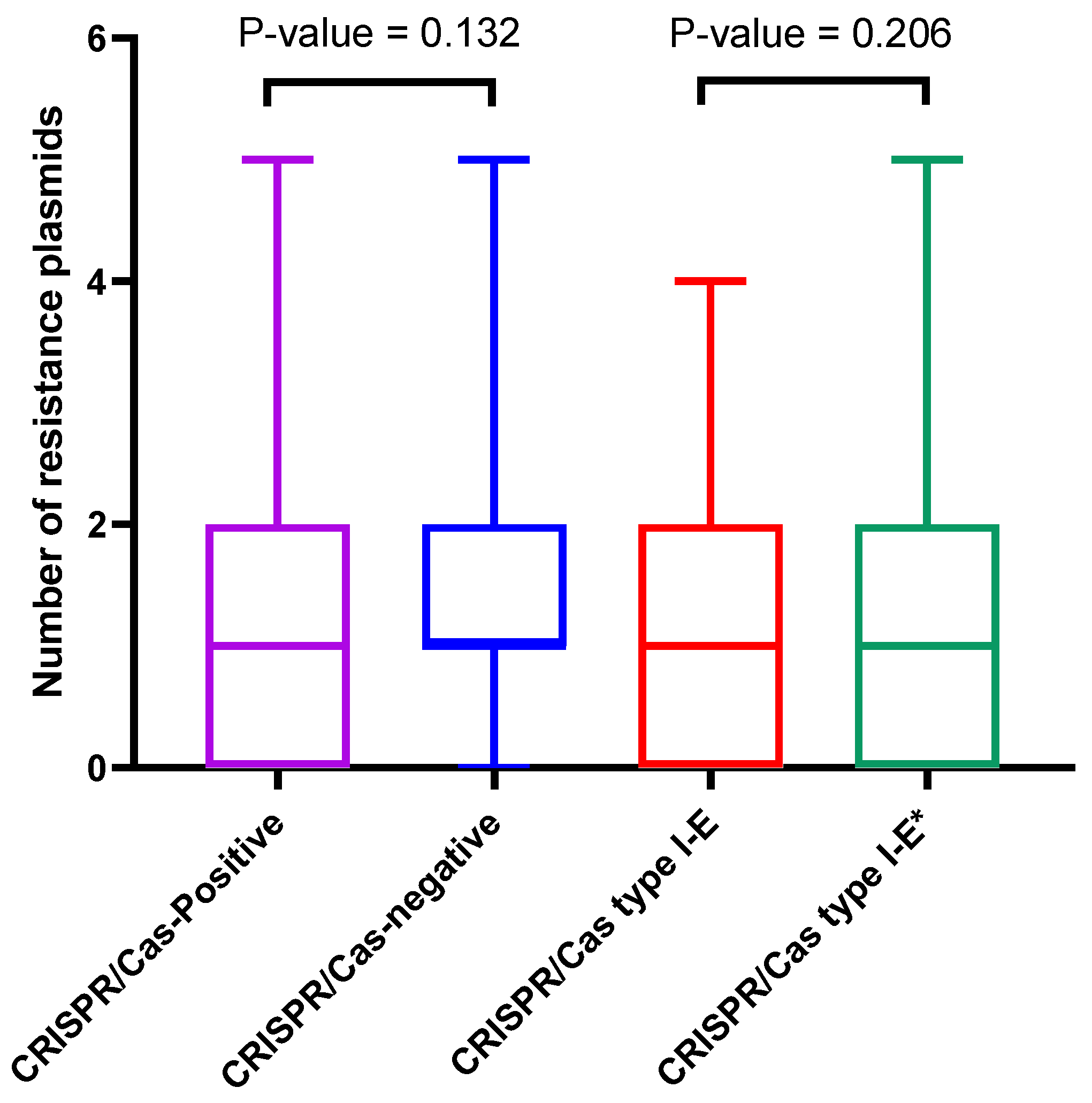
3.5.2. Correlation between CRISPR/Cas Systems and Plasmids Distribution
3.5.3. Correlation between CRISPR/Cas and ARGs
3.5.4. CRISPR/Cas Spacers and Correlation to Resistance Plasmids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Medina-Aparicio, L.; Davila, S.; Rebollar-Flores, J.E.; Calva, E.; Hernandez-Lucas, I. The CRISPR-Cas system in Enterobacteriaceae. Pathog. Dis. 2018, 76, fty002. [Google Scholar] [CrossRef] [PubMed]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [PubMed]
- Ishino, Y.; Krupovic, M.; Forterre, P. History of CRISPR-Cas from Encounter with a Mysterious Repeated Sequence to Genome Editing Technology. J. Bacteriol. 2018, 200, e00580-17. [Google Scholar] [CrossRef] [PubMed]
- Rath, D.; Amlinger, L.; Rath, A.; Lundgren, M. The CRISPR-Cas immune system: Biology, mechanisms and applications. Biochimie 2015, 117, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Garneau, J.E.; Dupuis, M.E.; Villion, M.; Romero, D.A.; Barrangou, R.; Boyaval, P.; Fremaux, C.; Horvath, P.; Magadan, A.H.; Moineau, S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010, 468, 67–71. [Google Scholar] [CrossRef]
- Barrangou, R. CRISPR-Cas systems and RNA-guided interference. Wiley Interdiscip. Rev. RNA 2013, 4, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Koonin, E.V. Annotation and Classification of CRISPR-Cas Systems. Methods Mol. Biol. 2015, 1311, 47–75. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR-Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef]
- Wu, Y.; Battalapalli, D.; Hakeem, M.J.; Selamneni, V.; Zhang, P.; Draz, M.S.; Ruan, Z. Engineered CRISPR-Cas systems for the detection and control of antibiotic-resistant infections. J. Nanobiotechnol. 2021, 19, 401. [Google Scholar] [CrossRef]
- Abdelaziz, S.M.; Aboshanab, K.M.; Yahia, I.S.; Yassien, M.A.; Hassouna, N.A. Correlation between the Antibiotic Resistance Genes and Susceptibility to Antibiotics among the Carbapenem-Resistant Gram-Negative Pathogens. Antibiotics 2021, 10, 255. [Google Scholar] [CrossRef]
- Xu, Z.; Li, M.; Li, Y.; Cao, H.; Miao, L.; Xu, Z.; Higuchi, Y.; Yamasaki, S.; Nishino, K.; Woo, P.C.Y.; et al. Native CRISPR-Cas-Mediated Genome Editing Enables Dissecting and Sensitizing Clinical Multidrug-Resistant P. aeruginosa. Cell Rep. 2019, 29, 1707–1717.e3. [Google Scholar] [CrossRef] [PubMed]
- Touchon, M.; Charpentier, S.; Pognard, D.; Picard, B.; Arlet, G.; Rocha, E.P.; Denamur, E.; Branger, C. Antibiotic resistance plasmids spread among natural isolates of Escherichia coli in spite of CRISPR elements. Microbiology 2012, 158, 2997–3004. [Google Scholar] [CrossRef]
- Shabbir, M.A.; Wu, Q.; Shabbir, M.Z.; Sajid, A.; Ahmed, S.; Sattar, A.; Tang, Y.; Li, J.; Maan, M.K.; Hao, H.; et al. The CRISPR-cas system promotes antimicrobial resistance in Campylobacter jejuni. Future Microbiol. 2018, 13, 1757–1774. [Google Scholar] [CrossRef] [PubMed]
- McDonald, N.D.; Regmi, A.; Morreale, D.P.; Borowski, J.D.; Boyd, E.F. CRISPR-Cas systems are present predominantly on mobile genetic elements in Vibrio species. BMC Genom. 2019, 20, 105. [Google Scholar] [CrossRef]
- Mohd Asri, N.A.; Ahmad, S.; Mohamud, R.; Mohd Hanafi, N.; Mohd Zaidi, N.F.; Irekeola, A.A.; Shueb, R.H.; Yee, L.C.; Mohd Noor, N.; Mustafa, F.H.; et al. Global Prevalence of Nosocomial Multidrug-Resistant Klebsiella pneumoniae: A Systematic Review and Meta-Analysis. Antibiotics 2021, 10, 1508. [Google Scholar] [CrossRef]
- Wyres, K.L.; Lam, M.M.C.; Holt, K.E. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020, 18, 344–359. [Google Scholar] [CrossRef] [PubMed]
- Ostria-Hernandez, M.L.; Sanchez-Vallejo, C.J.; Ibarra, J.A.; Castro-Escarpulli, G. Survey of clustered regularly interspaced short palindromic repeats and their associated Cas proteins (CRISPR/Cas) systems in multiple sequenced strains of Klebsiella pneumoniae. BMC Res. Notes 2015, 8, 332. [Google Scholar] [CrossRef]
- Shen, J.; Lv, L.; Wang, X.; Xiu, Z.; Chen, G. Comparative analysis of CRISPR-Cas systems in Klebsiella genomes. J. Basic. Microbiol. 2017, 57, 325–336. [Google Scholar] [CrossRef]
- Li, H.Y.; Kao, C.Y.; Lin, W.H.; Zheng, P.X.; Yan, J.J.; Wang, M.C.; Teng, C.H.; Tseng, C.C.; Wu, J.J. Characterization of CRISPR-Cas Systems in Clinical Klebsiella pneumoniae Isolates Uncovers Its Potential Association with Antibiotic Susceptibility. Front. Microbiol. 2018, 9, 1595. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; Iredell, J.R. CRISPR-Cas System in Antibiotic Resistance Plasmids in Klebsiella pneumoniae. Front. Microbiol. 2019, 10, 2934. [Google Scholar] [CrossRef]
- Chen, S.; Liu, H.; Liang, W.; Hong, L.; Zhang, B.; Huang, L.; Guo, X.; Duan, G. Insertion sequences in the CRISPR-Cas system regulate horizontal antimicrobial resistance gene transfer in Shigella strains. Int. J. Antimicrob. Agents 2019, 53, 109–115. [Google Scholar] [CrossRef]
- Gholizadeh, P.; Aghazadeh, M.; Ghotaslou, R.; Rezaee, M.A.; Pirzadeh, T.; Cui, L.; Watanabe, S.; Feizi, H.; Kadkhoda, H.; Kafil, H.S. Role of CRISPR-Cas system on antibiotic resistance patterns of Enterococcus faecalis. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 49. [Google Scholar] [CrossRef] [PubMed]
- Alduhaidhawi, A.H.M.; AlHuchaimi, S.N.; Al-Mayah, T.A.; Al-Ouqaili, M.T.S.; Alkafaas, S.S.; Muthupandian, S.; Saki, M. Prevalence of CRISPR-Cas Systems and Their Possible Association with Antibiotic Resistance in Enterococcus faecalis and Enterococcus faecium Collected from Hospital Wastewater. Infect. Drug Resist. 2022, 15, 1143–1154. [Google Scholar] [CrossRef] [PubMed]
- Soliman, M.; Said, H.S.; El-Mowafy, M.; Barwa, R. Novel PCR detection of CRISPR/Cas systems in Pseudomonas aeruginosa and its correlation with antibiotic resistance. Appl. Microbiol. Biotechnol. 2022, 106, 7223–7234. [Google Scholar] [CrossRef]
- Aydin, S.; Personne, Y.; Newire, E.; Laverick, R.; Russell, O.; Roberts, A.P.; Enne, V.I. Presence of Type I-F CRISPR/Cas systems is associated with antimicrobial susceptibility in Escherichia coli. J. Antimicrob. Chemother. 2017, 72, 2213–2218. [Google Scholar] [CrossRef]
- Ren, L.; Deng, L.H.; Zhang, R.P.; Wang, C.D.; Li, D.S.; Xi, L.X.; Chen, Z.R.; Yang, R.; Huang, J.; Zeng, Y.R.; et al. Relationship between drug resistance and the clustered, regularly interspaced, short, palindromic repeat-associated protein genes cas1 and cas2 in Shigella from giant panda dung. Medicine 2017, 96, e5922. [Google Scholar] [CrossRef]
- Mackow, N.A.; Shen, J.; Adnan, M.; Khan, A.S.; Fries, B.C.; Diago-Navarro, E. CRISPR-Cas influences the acquisition of antibiotic resistance in Klebsiella pneumoniae. PLoS ONE 2019, 14, e0225131. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Fu, P.; Zhou, Y.; Xie, Y.; Jin, J.; Wang, B.; Yu, L.; Huang, Y.; Li, G.; Li, M.; et al. Absence of the type I-E CRISPR-Cas system in Klebsiella pneumoniae clonal complex 258 is associated with dissemination of IncF epidemic resistance plasmids in this clonal complex. J. Antimicrob. Chemother. 2020, 75, 890–895. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Song, G.; Xu, Y. Association of CRISPR/Cas System with the Drug Resistance in Klebsiella pneumoniae. Infect. Drug Resist. 2020, 13, 1929–1935. [Google Scholar] [CrossRef]
- Zhou, Y.; Tang, Y.; Fu, P.; Tian, D.; Yu, L.; Huang, Y.; Li, G.; Li, M.; Wang, Y.; Yang, Z.; et al. The type I-E CRISPR-Cas system influences the acquisition of bla(KPC)-IncF plasmid in Klebsiella pneumonia. Emerg. Microbes Infect. 2020, 9, 1011–1022. [Google Scholar] [CrossRef]
- Liu, Y.; Wan, L.G.; Deng, Q.; Cao, X.W.; Yu, Y.; Xu, Q.F. First description of NDM-1-, KPC-2-, VIM-2- and IMP-4-producing Klebsiella pneumoniae strains in a single Chinese teaching hospital. Epidemiol. Infect. 2015, 143, 376–384. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Endimiani, A.; Carias, L.L.; Hujer, A.M.; Bethel, C.R.; Hujer, K.M.; Perez, F.; Hutton, R.A.; Fox, W.R.; Hall, G.S.; Jacobs, M.R.; et al. Presence of plasmid-mediated quinolone resistance in Klebsiella pneumoniae isolates possessing blaKPC in the United States. Antimicrob. Agents Chemother. 2008, 52, 2680–2682. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Ji, S.; Chen, Y.; Zhou, W.; Wei, Z.; Li, L.; Ma, Y. Resistance of strains producing extended-spectrum beta-lactamases and genotype distribution in China. J. Infect. 2007, 54, 53–57. [Google Scholar] [CrossRef] [PubMed]
- EUCAST Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 13.0. Available online: http://www.eucast.org (accessed on 20 January 2022).
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 2018, 7, 1–6. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Moller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Arredondo-Alonso, S.; Rogers, M.R.C.; Braat, J.C.; Verschuuren, T.D.; Top, J.; Corander, J.; Willems, R.J.L.; Schurch, A.C. mlplasmids: A user-friendly tool to predict plasmid- and chromosome-derived sequences for single species. Microb. Genom. 2018, 4, e000224. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, J.; Wang, D.; Guo, Q.; Wang, M. Coexistence of bla (KPC)-IncFII plasmids and type I-E(*) CRISPR-Cas systems in ST15 Klebsiella pneumoniae. Front. Microbiol. 2023, 14, 1125531. [Google Scholar] [CrossRef]
- Liao, W.; Liu, Y.; Chen, C.; Li, J.; Du, F.; Long, D.; Zhang, W. Distribution of CRISPR-Cas Systems in Clinical Carbapenem-Resistant Klebsiella pneumoniae Strains in a Chinese Tertiary Hospital and Its Potential Relationship with Virulence. Microb. Drug Resist. 2020, 26, 630–636. [Google Scholar] [CrossRef]
- Sinkunas, T.; Gasiunas, G.; Fremaux, C.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011, 30, 1335–1342. [Google Scholar] [CrossRef] [PubMed]
- Farhadi, M.; Ahanjan, M.; Goli, H.R.; Haghshenas, M.R.; Gholami, M. High frequency of multidrug-resistant (MDR) Klebsiella pneumoniae harboring several beta-lactamase and integron genes collected from several hospitals in the north of Iran. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 70. [Google Scholar] [CrossRef]
- Nakamura-Silva, R.; Cerdeira, L.; Oliveira-Silva, M.; da Costa, K.R.C.; Sano, E.; Fuga, B.; Moura, Q.; Esposito, F.; Lincopan, N.; Wyres, K.; et al. Multidrug-resistant Klebsiella pneumoniae: A retrospective study in Manaus, Brazil. Arch. Microbiol. 2022, 204, 202. [Google Scholar] [CrossRef] [PubMed]
- Fursova, N.K.; Astashkin, E.I.; Ershova, O.N.; Aleksandrova, I.A.; Savin, I.A.; Novikova, T.S.; Fedyukina, G.N.; Kislichkina, A.A.; Fursov, M.V.; Kuzina, E.S.; et al. Multidrug-Resistant Klebsiella pneumoniae Causing Severe Infections in the Neuro-ICU. Antibiotics 2021, 10, 979. [Google Scholar] [CrossRef] [PubMed]
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef]
- Mostafa, S.H.; Saleh, S.E.; Hamed, S.M.; Aboshanab, K.M. Febrile illness of bacterial etiology in a public fever hospital in Egypt: High burden of multidrug resistance and WHO priority Gram negative pathogens. Germs 2022, 12, 75–85. [Google Scholar] [CrossRef]
- Hassuna, N.A.; AbdelAziz, R.A.; Zakaria, A.; Abdelhakeem, M. Extensively-Drug Resistant Klebsiella pneumoniae Recovered From Neonatal Sepsis Cases From a Major NICU in Egypt. Front. Microbiol. 2020, 11, 1375. [Google Scholar] [CrossRef]
- El-Badawy, M.F.; Tawakol, W.M.; El-Far, S.W.; Maghrabi, I.A.; Al-Ghamdi, S.A.; Mansy, M.S.; Ashour, M.S.; Shohayeb, M.M. Molecular Identification of Aminoglycoside-Modifying Enzymes and Plasmid-Mediated Quinolone Resistance Genes among Klebsiella pneumoniae Clinical Isolates Recovered from Egyptian Patients. Int. J. Microbiol. 2017, 2017, 8050432. [Google Scholar] [CrossRef]
- Hamed, S.M.; Aboshanab, K.M.A.; El-Mahallawy, H.A.; Helmy, M.M.; Ashour, M.S.; Elkhatib, W.F. Plasmid-Mediated Quinolone Resistance in Gram-Negative Pathogens Isolated from Cancer Patients in Egypt. Microb. Drug Resist. 2018, 24, 1316–1325. [Google Scholar] [CrossRef]
- Jlili, N.E.; Rejiba, S.; Smaoui, H.; Guillard, T.; Chau, F.; Kechrid, A.; Cambau, E. Trend of plasmid-mediated quinolone resistance genes at the Children’s Hospital in Tunisia. J. Med. Microbiol. 2014, 63, 195–202. [Google Scholar] [CrossRef]
- Kumar, V.; Sun, P.; Vamathevan, J.; Li, Y.; Ingraham, K.; Palmer, L.; Huang, J.; Brown, J.R. Comparative genomics of Klebsiella pneumoniae strains with different antibiotic resistance profiles. Antimicrob. Agents Chemother. 2011, 55, 4267–4276. [Google Scholar] [CrossRef] [PubMed]
- Bolourchi, N.; Naz, A.; Sohrabi, M.; Badmasti, F. Comparative in silico characterization of Klebsiella pneumoniae hypervirulent plasmids and their antimicrobial resistance genes. Ann. Clin. Microbiol. Antimicrob. 2022, 21, 23. [Google Scholar] [CrossRef] [PubMed]
- David, S.; Reuter, S.; Harris, S.R.; Glasner, C.; Feltwell, T.; Argimon, S.; Abudahab, K.; Goater, R.; Giani, T.; Errico, G.; et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat. Microbiol. 2019, 4, 1919–1929. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhou, J.; Xu, Y.; Xiu, Z. Prophages contribute to genome plasticity of Klebsiella pneumoniae and may involve the chromosomal integration of ARGs in CG258. Genomics 2020, 112, 998–1010. [Google Scholar] [CrossRef]
- Huang, W.; Wang, G.; Sebra, R.; Zhuge, J.; Yin, C.; Aguero-Rosenfeld, M.E.; Schuetz, A.N.; Dimitrova, N.; Fallon, J.T. Emergence and Evolution of Multidrug-Resistant Klebsiella pneumoniae with both bla(KPC) and bla(CTX-M) Integrated in the Chromosome. Antimicrob. Agents Chemother. 2017, 61, e00076-17. [Google Scholar] [CrossRef]
- Bialek-Davenet, S.; Criscuolo, A.; Ailloud, F.; Passet, V.; Jones, L.; Delannoy-Vieillard, A.S.; Garin, B.; Le Hello, S.; Arlet, G.; Nicolas-Chanoine, M.H.; et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg. Infect. Dis. 2014, 20, 1812–1820. [Google Scholar] [CrossRef]
- Stern, A.; Keren, L.; Wurtzel, O.; Amitai, G.; Sorek, R. Self-targeting by CRISPR: Gene regulation or autoimmunity? Trends Genet. 2010, 26, 335–340. [Google Scholar] [CrossRef]
- Enany, S.; Zakeer, S.; Diab, A.A.; Bakry, U.; Sayed, A.A. Whole genome sequencing of Klebsiella pneumoniae clinical isolates sequence type 627 isolated from Egyptian patients. PLoS ONE 2022, 17, e0265884. [Google Scholar] [CrossRef]
- Palmer, K.L.; Gilmore, M.S. Multidrug-resistant enterococci lack CRISPR-cas. mBio 2010, 1, e00227-10. [Google Scholar] [CrossRef]
- van Belkum, A.; Soriaga, L.B.; LaFave, M.C.; Akella, S.; Veyrieras, J.B.; Barbu, E.M.; Shortridge, D.; Blanc, B.; Hannum, G.; Zambardi, G.; et al. Phylogenetic Distribution of CRISPR-Cas Systems in Antibiotic-Resistant Pseudomonas aeruginosa. mBio 2015, 6, e01796-15. [Google Scholar] [CrossRef]
- Hatoum-Aslan, A.; Marraffini, L.A. Impact of CRISPR immunity on the emergence and virulence of bacterial pathogens. Curr. Opin. Microbiol. 2014, 17, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Bikard, D.; Hatoum-Aslan, A.; Mucida, D.; Marraffini, L.A. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe 2012, 12, 177–186. [Google Scholar] [CrossRef]
- Sontheimer, E.J.; Davidson, A.R. Inhibition of CRISPR-Cas systems by mobile genetic elements. Curr. Opin. Microbiol. 2017, 37, 120–127. [Google Scholar] [CrossRef]
- Marino, N.D.; Pinilla-Redondo, R.; Csorgo, B.; Bondy-Denomy, J. Anti-CRISPR protein applications: Natural brakes for CRISPR-Cas technologies. Nat. Methods 2020, 17, 471–479. [Google Scholar] [CrossRef]
- Davidson, A.R.; Lu, W.T.; Stanley, S.Y.; Wang, J.; Mejdani, M.; Trost, C.N.; Hicks, B.T.; Lee, J.; Sontheimer, E.J. Anti-CRISPRs: Protein Inhibitors of CRISPR-Cas Systems. Annu. Rev. Biochem. 2020, 89, 309–332. [Google Scholar] [CrossRef] [PubMed]
- Bondy-Denomy, J.; Pawluk, A.; Maxwell, K.L.; Davidson, A.R. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 2013, 493, 429–432. [Google Scholar] [CrossRef] [PubMed]
- Pawluk, A.; Bondy-Denomy, J.; Cheung, V.H.; Maxwell, K.L.; Davidson, A.R. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. mBio 2014, 5, e00896. [Google Scholar] [CrossRef]
- Watson, B.N.J.; Steens, J.A.; Staals, R.H.J.; Westra, E.R.; van Houte, S. Coevolution between bacterial CRISPR-Cas systems and their bacteriophages. Cell Host Microbe 2021, 29, 715–725. [Google Scholar] [CrossRef]
- Westra, E.R.; Semenova, E.; Datsenko, K.A.; Jackson, R.N.; Wiedenheft, B.; Severinov, K.; Brouns, S.J. Type I-E CRISPR-cas systems discriminate target from non-target DNA through base pairing-independent PAM recognition. PLoS Genet. 2013, 9, e1003742. [Google Scholar] [CrossRef]
- Fu, B.X.; Wainberg, M.; Kundaje, A.; Fire, A.Z. High-Throughput Characterization of Cascade type I-E CRISPR Guide Efficacy Reveals Unexpected PAM Diversity and Target Sequence Preferences. Genetics 2017, 206, 1727–1738. [Google Scholar] [CrossRef]
- Pul, U.; Wurm, R.; Arslan, Z.; Geissen, R.; Hofmann, N.; Wagner, R. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol. Microbiol. 2010, 75, 1495–1512. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.L.; Wick, R.R.; Judd, L.M.; Froumine, R.; Tokolyi, A.; Gorrie, C.L.; Lam, M.M.C.; Duchene, S.; Jenney, A.; Holt, K.E. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 2019, 15, e1008114. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.L.; Pan, Y.J.; Hsieh, P.F.; Hsu, C.R.; Wu, M.C.; Wang, J.T. Imipenem represses CRISPR-Cas interference of DNA acquisition through H-NS stimulation in Klebsiella pneumoniae. Sci. Rep. 2016, 6, 31644. [Google Scholar] [CrossRef]
- Westra, E.R.; Pul, U.; Heidrich, N.; Jore, M.M.; Lundgren, M.; Stratmann, T.; Wurm, R.; Raine, A.; Mescher, M.; Van Heereveld, L.; et al. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol. Microbiol. 2010, 77, 1380–1393. [Google Scholar] [CrossRef] [PubMed]
- Majsec, K.; Bolt, E.L.; Ivancic-Bace, I. Cas3 is a limiting factor for CRISPR-Cas immunity in Escherichia coli cells lacking H-NS. BMC Microbiol. 2016, 16, 28. [Google Scholar] [CrossRef]
- Stoebel, D.M.; Free, A.; Dorman, C.J. Anti-silencing: Overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology 2008, 154, 2533–2545. [Google Scholar] [CrossRef]
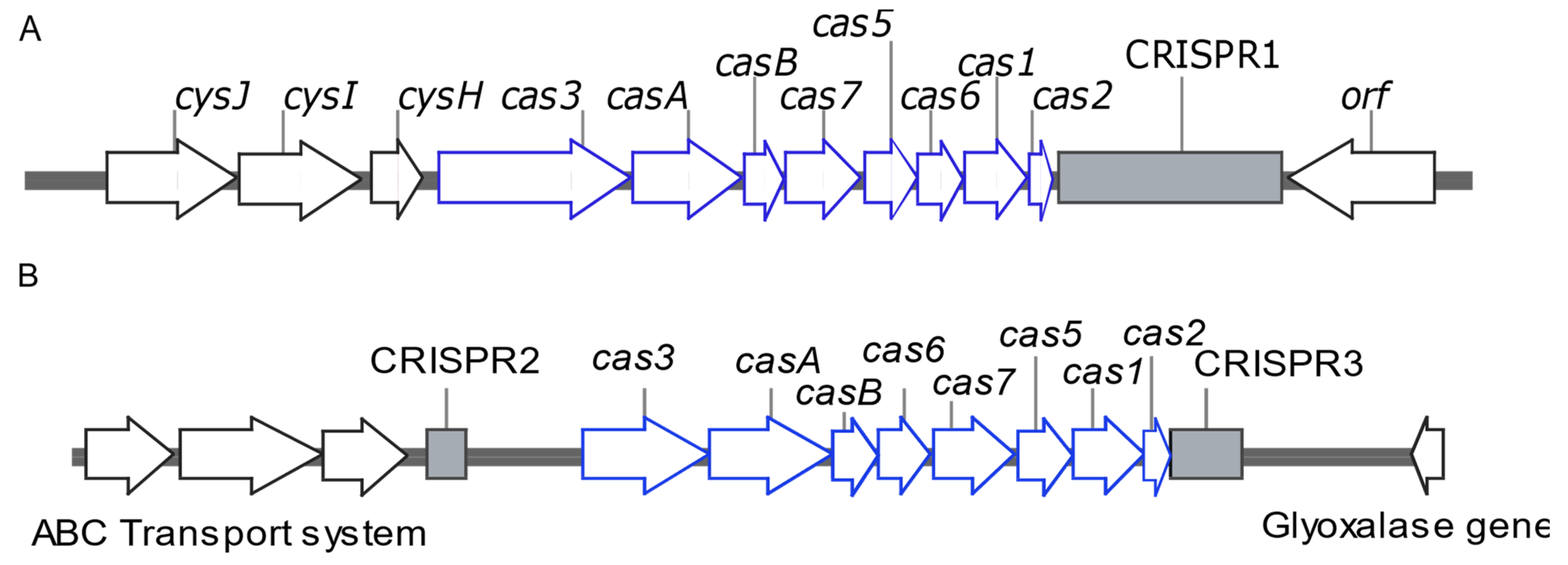


| Isolate No | ST | CRISPR/Cas Type | Total No of Spacers | Plasmid Replicons | ARGs Carried on Plasmids |
|---|---|---|---|---|---|
| K57 | ST1999 | I-E | 50 | IncFIB(K) | blaTEM |
| 117 | ST870 | I-E* | 21 | IncFIB(K) Col440I Col440II IncFIA (HI1) IncFII(pKP91) repB(R1701) | blaDHA-1—tet(D)—aph(6)-Id—dfrA7—aph(3’)-Ia |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alkompoz, A.K.; Hamed, S.M.; Zaid, A.S.A.; Almangour, T.A.; Al-Agamy, M.H.; Aboshanab, K.M. Correlation of CRISPR/Cas and Antimicrobial Resistance in Klebsiella pneumoniae Clinical Isolates Recovered from Patients in Egypt Compared to Global Strains. Microorganisms 2023, 11, 1948. https://doi.org/10.3390/microorganisms11081948
Alkompoz AK, Hamed SM, Zaid ASA, Almangour TA, Al-Agamy MH, Aboshanab KM. Correlation of CRISPR/Cas and Antimicrobial Resistance in Klebsiella pneumoniae Clinical Isolates Recovered from Patients in Egypt Compared to Global Strains. Microorganisms. 2023; 11(8):1948. https://doi.org/10.3390/microorganisms11081948
Chicago/Turabian StyleAlkompoz, Amany K., Samira M. Hamed, Ahmed S. Abu Zaid, Thamer A. Almangour, Mohamed H. Al-Agamy, and Khaled M. Aboshanab. 2023. "Correlation of CRISPR/Cas and Antimicrobial Resistance in Klebsiella pneumoniae Clinical Isolates Recovered from Patients in Egypt Compared to Global Strains" Microorganisms 11, no. 8: 1948. https://doi.org/10.3390/microorganisms11081948
APA StyleAlkompoz, A. K., Hamed, S. M., Zaid, A. S. A., Almangour, T. A., Al-Agamy, M. H., & Aboshanab, K. M. (2023). Correlation of CRISPR/Cas and Antimicrobial Resistance in Klebsiella pneumoniae Clinical Isolates Recovered from Patients in Egypt Compared to Global Strains. Microorganisms, 11(8), 1948. https://doi.org/10.3390/microorganisms11081948







