CD147 rs8259T>A Variant Confers Susceptibility to COVID-19 Infection within the Mexican Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Genetic Analysis
2.3. CD147, miR-492, and TNF mRNA Expression
2.4. Statistical Analysis
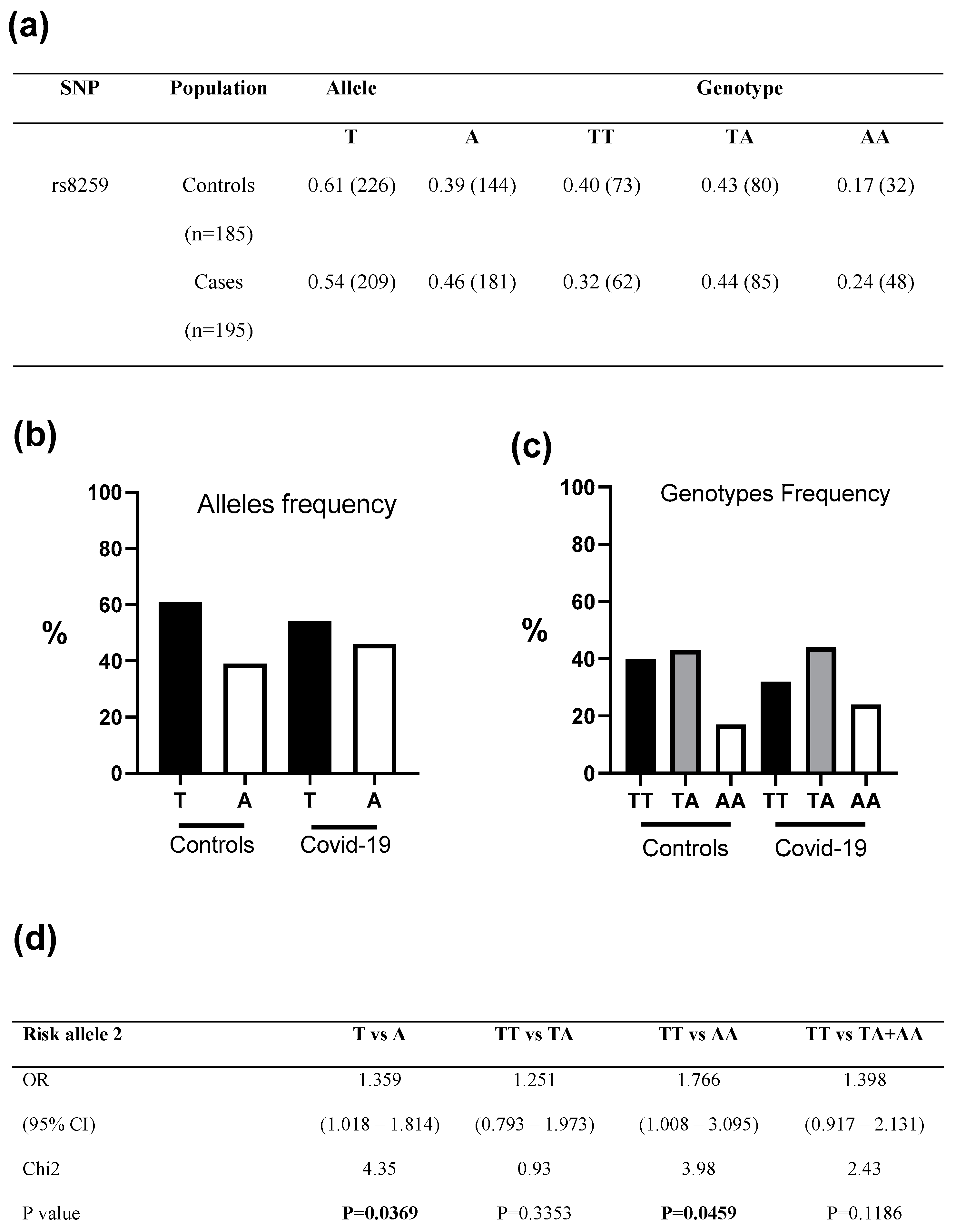
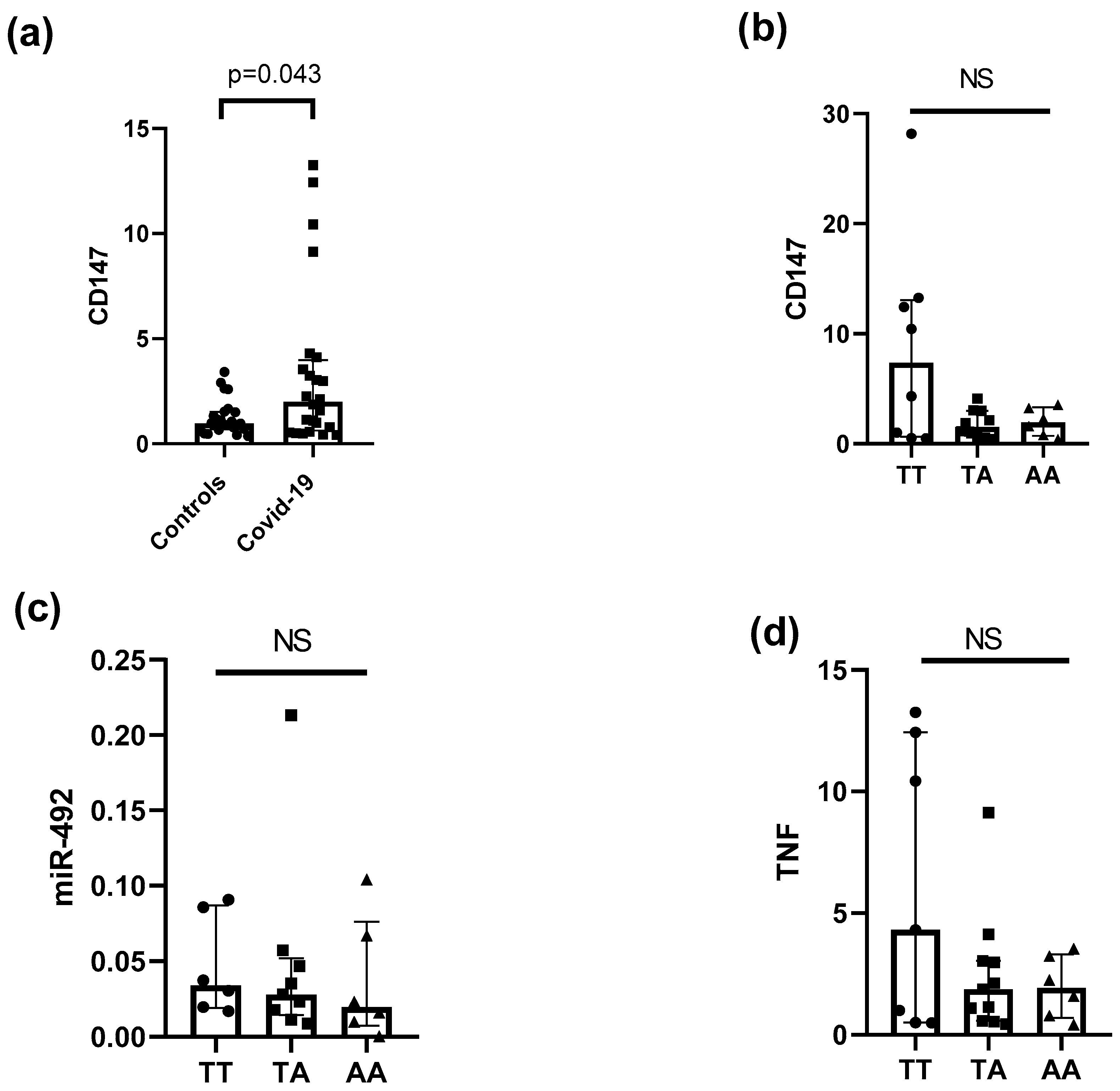
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, H.; Paulson, K.R.; Pease, S.A.; Watson, S.; Comfort, H.; Zheng, P.; Aravkin, A.Y.; Bisignano, C.; Barber, R.M.; Alam, T.; et al. Estimating excess mortality due to the COVID-19 pandemic: A systematic analysis of COVID-19-related mortality, 2020–2021. Lancet 2022, 399, 1513–1536. [Google Scholar] [CrossRef]
- WHO Coronavirus (COVID-19) Dashboard|WHO Coronavirus (COVID-19) Dashboard with Vaccination Data. Available online: https://covid19.who.int/ (accessed on 5 March 2023).
- Fricke-Galindo, I.; Falfán-Valencia, R. Genetics Insight for COVID-19 Susceptibility and Severity: A Review. Front. Immunol. 2021, 12, 622176. [Google Scholar] [CrossRef]
- Zhang, J.; Xiao, T.; Cai, Y.; Chen, B. Structure of SARS-CoV-2 spike protein. Curr. Opin. Virol. 2021, 50, 173–182. Available online: https://pubmed.ncbi.nlm.nih.gov/34534731/ (accessed on 1 May 2023). [CrossRef]
- Scialo, F.; Daniele, A.; Amato, F.; Pastore, L.; Matera, M.G.; Cazzola, M.; Castaldo, G.; Bianco, A. ACE2: The Major Cell Entry Receptor for SARS-CoV-2. Lung 2020, 198, 867–877. Available online: https://pubmed.ncbi.nlm.nih.gov/33170317/ (accessed on 1 May 2023). [CrossRef]
- Radzikowska, U.; Ding, M.; Tan, G.; Zhakparov, D.; Peng, Y.; Wawrzyniak, P.; Wang, M.; Li, S.; Morita, H.; Altunbulakli, C.; et al. Distribution of ACE2, CD147, CD26, and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Allergy 2020, 75, 2829. [Google Scholar] [CrossRef]
- Asgari, R.; Vaisi-Raygani, A.; Aleagha, M.S.E.; Mohammadi, P.; Bakhtiari, M.; Arghiani, N. CD147 and MMPs as key factors in physiological and pathological processes. Biomed. Pharmacother. 2023, 157, 113983. [Google Scholar] [CrossRef]
- Fridley, B.L.; Biernacka, J.M. Gene set analysis of SNP data: Benefits, challenges, and future directions. Eur. J. Hum. Genet. 2011, 19, 837. [Google Scholar] [CrossRef]
- Li, Y.; Wei, L.; He, L.; Sun, J.; Liu, N. Interferon-induced transmembrane protein 3 gene polymorphisms are associated with COVID-19 susceptibility and severity: A meta-analysis. J. Infect. 2022, 84, 825–833. [Google Scholar] [CrossRef]
- Martínez-Gómez, L.E.; Herrera-López, B.; Martinez-Armenta, C.; Ortega-Peña, S.; Camacho-Rea, M.d.C.; Suarez-Ahedo, C.; Vázquez-Cárdenas, P.; Vargas-Alarcón, G.; Rojas-Velasco, G.; Fragoso, J.M.; et al. ACE and ACE2 Gene Variants Are Associated With Severe Outcomes of COVID-19 in Men. Front. Immunol. 2022, 13, 812940. [Google Scholar] [CrossRef]
- Posadas-Sánchez, R.; Fragoso, J.M.; Sánchez-Muñoz, F.; Rojas-Velasco, G.; Ramírez-Bello, J.; López-Reyes, A.; Martínez-Gómez, L.E.; Sierra-Fernández, C.; Rodríguez-Reyna, T.; Regino-Zamarripa, N.E.; et al. Association of the Transmembrane Serine Protease-2 (TMPRSS2) Polymorphisms with COVID-19. Viruses 2022, 14, 1976. [Google Scholar] [CrossRef]
- Kaidashev, I.; Izmailova, O.; Shlykova, O.; Kabaliei, A.; Vatsenko, A.; Ivashchenko, D.; Dudchenko, M.; Volianskyi, A.; Zelinskyy, G.; Koval, T.; et al. Polymorphism of tmprss2 (rs12329760) but not ace2 (rs4240157), tmprss11a (rs353163) and cd147 (rs8259) is associated with the severity of COVID-19 in the Ukrainian population. Acta Bio. Medica. Atenei Parm. 2023, 94, e2023030. [Google Scholar] [CrossRef]
- Behl, T.; Kaur, I.; Aleya, L.; Sehgal, A.; Singh, S.; Sharma, N.; Bhatia, S.; Al-Harrasi, A.; Bungau, S. CD147-spike protein interaction in COVID-19: Get the ball rolling with a novel receptor and therapeutic target. Sci. Total Environ. 2022, 808, 152072–152072. [Google Scholar] [CrossRef] [PubMed]
- Laboratory Testing for Coronavirus Disease (COVID-19) in Suspected Human Cases. Available online: https://apps.who.int/iris/handle/10665/331501 (accessed on 18 May 2023).
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.-X.; Gong, L.; et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020, 5, 283. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Mi, L.; Xu, J.; Yu, J.; Wang, X.; Jiang, J.; Xing, J.; Shang, P.; Qian, A.; Li, Y.; et al. Function of HAb18G/CD147 in Invasion of Host Cells by Severe Acute Respiratory Syndrome Coronavirus. J. Infect. Dis. 2005, 191, 755–760. [Google Scholar] [CrossRef] [PubMed]
- Adimulam, T.; Arumugam, T.; Gokul, A.; Ramsuran, V. Genetic Variants within SARS-CoV-2 Human Receptor Genes May Contribute to Variable Disease Outcomes in Different Ethnicities. Int. J. Mol. Sci. 2023, 24, 8711. Available online: https://www.mdpi.com/1422-0067/24/10/8711/htm (accessed on 18 May 2023). [CrossRef]
- Hashemi, S.M.A.; Thijssen, M.; Hosseini, S.Y.; Tabarraei, A.; Pourkarim, M.R.; Sarvari, J. Human gene polymorphisms and their possible impact on the clinical outcome of SARS-CoV-2 infection. Arch. Virol. 2021, 166, 2089. [Google Scholar]
- Price, A.L.; Patterson, N.; Yu, F.; Cox, D.R.; Waliszewska, A.; McDonald, G.J.; Tandon, A.; Schirmer, C.; Neubauer, J.; Bedoya, G.; et al. A Genomewide Admixture Map for Latino Populations. Am. J. Hum. Genet. 2007, 80, 1024–1036. [Google Scholar] [CrossRef]
- Li, M.-P.; Hu, X.-L.; Yang, Y.-L.; Zhang, Y.-J.; Zhou, J.-P.; Peng, L.-M.; Tang, J.; Chen, X.-P. Basigin rs8259 Polymorphism Confers Decreased Risk of Chronic Heart Failure in a Chinese Population. Int. J. Environ. Res. Public Health 2017, 14, 211. [Google Scholar] [CrossRef]
- Weng, Y.; Chen, T.; Ren, J.; Lu, D.; Liu, X.; Lin, S.; Xu, C.; Lou, J.; Chen, X.; Tang, L. The Association Between Extracellular Matrix Metalloproteinase Inducer Polymorphisms and Coronary Heart Disease: A Potential Way to Predict Disease. DNA Cell Biol. 2020, 39, 244–254. [Google Scholar] [CrossRef]
- Geng, J.; Chen, L.; Yuan, Y.; Wang, K.; Wang, Y.; Qin, C.; Wu, G.; Chen, R.; Zhang, Z.; Wei, D.; et al. CD147 antibody specifically and effectively inhibits infection and cytokine storm of SARS-CoV-2 and its variants delta, alpha, beta, and gamma. Signal Transduct. Target. Ther. 2021, 6, 347. [Google Scholar] [CrossRef]
- Springall, R.; González-Flores, J.; García-Ávila, C.; Juárez-Vicuña, Y.; Hernández-Diazcouder, A.; Márquez-Velasco, R.; Cásares-Alvarado, S.; Sánchez-Muñoz, F.; Basilio-Gálvez, E.; Castillo-Salazar, M.; et al. Elevated Levels of Soluble CD147 are Associated with Hyperinflammation and Disease Severity in COVID-19: A Proof-of-Concept Clinical Study. Arch. Immunol. Ther. Exp. 2022, 70, 18. [Google Scholar] [CrossRef]
- Yan, J.; Mao, Y.; Wang, C.; Wang, Z. Association Study between an SNP in CD147 and Its Expression with Acute Coronary Syndrome in a Jiangsu Chinese Population. Medicine 2015, 94, e1537. [Google Scholar] [CrossRef]
- Wu, L.-S.; Li, F.-F.; Sun, L.-D.; Li, D.; Su, J.; Kuang, Y.-H.; Chen, G.; Chen, X.-P.; Chen, X. A miRNA-492 binding-site polymorphism in BSG (basigin) confers risk to psoriasis in Central South Chinese population. Hum. Genet. 2011, 130, 749–757. [Google Scholar] [CrossRef]
- Mohd Zawawi, Z.; Kalyanasundram, J.; Mohd Zain, R.; Thayan, R.; Basri, D.F.; Yap, W.B. Prospective Roles of Tumor Necrosis Factor-Alpha (TNF-α) in COVID-19: Prognosis, Therapeutic and Management. Int. J. Mol. Sci. 2023, 24, 6142. [Google Scholar] [CrossRef]

| COVID-19 Patients (n = 195) | |
|---|---|
| Age in years, median (IQR) | 55 (46–66) |
| Male sex, n (%) | 122 (62.6) |
| Outcome | |
| 133 (68.2) |
| 62 (31.8) |
| Mechanical Ventilation, n (%) | 60 (30.8) |
| Diabetes Mellitus, n (%) | 69 (35.4) |
| Systemic Hypertension, n (%) | 82 (42.1) |
| Obesity, n (%) | 82 (42.1) |
| Serum creatinine, median (IQR) | 0.87 (0.66–1.35) |
| Ferritin, median (IQR) | 686 (386–1028.7) |
| Lactic Dehydrogenase, median (IQR) | 363.7 (268–471) |
| C-reactive protein, median (IQR) | 8.9 (3.8–23.4) |
| Total bilirubin, median (IQR) | 0.6 (0.43–0.80) |
| ALT, median (IQR) | 47.5 (28–72) |
| AST, median (IQR) | 40 (27–67) |
| TT (n = 62) | TA (n = 85) | AA (n = 48) | p | |
|---|---|---|---|---|
| Mechanical Ventilation (n, %) | 18 (29.0) | 27 (31.8) | 15 (31.3) | 0.787 |
| Diabetes mellitus (n, %) | 24 (38.7) | 27 (31.8) | 18 (37.5) | 0.644 |
| Systemic Hypertension (n, %) | 25 (40.3) | 37 (43.5) | 19 (39.6) | 0.882 |
| Male (n, %) | 39 (62.9) | 54 (63.5) | 29 (60.4) | 0.936 |
| Obesity (n, %) | 29 (46.8) | 34 (40) | 19 (35.6) | 0.659 |
| Creatinine (mg/dL) | 0.84 (0.69–1.28) | 0.88 (0.62–1.32) | 0.96 (0.73–1.42) | 0.31 |
| Ferritin (ng/mL) | 681 (348–1074) | 686 (377–1295) | 693 (396–992) | 0.78 |
| LDH (IU/L) | 366 (256–489) | 362 (275–460) | 350 (269–558) | 0.84 |
| CRP (mg/dL) | 6.38 (3.3–21.1) | 10.20 (4.0–22.2) | 8.32 (4.3–65.2) | 0.60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amezcua-Guerra, L.M.; Guzmán-Martín, C.A.; Montúfar-Robles, I.; Springall, R.; Hernández-Díazcouder, A.; Barbosa-Cobos, R.E.; Sánchez-Muñoz, F.; Ramírez-Bello, J. CD147 rs8259T>A Variant Confers Susceptibility to COVID-19 Infection within the Mexican Population. Microorganisms 2023, 11, 1919. https://doi.org/10.3390/microorganisms11081919
Amezcua-Guerra LM, Guzmán-Martín CA, Montúfar-Robles I, Springall R, Hernández-Díazcouder A, Barbosa-Cobos RE, Sánchez-Muñoz F, Ramírez-Bello J. CD147 rs8259T>A Variant Confers Susceptibility to COVID-19 Infection within the Mexican Population. Microorganisms. 2023; 11(8):1919. https://doi.org/10.3390/microorganisms11081919
Chicago/Turabian StyleAmezcua-Guerra, Luis M., Carlos A. Guzmán-Martín, Isela Montúfar-Robles, Rashidi Springall, Adrián Hernández-Díazcouder, Rosa Elda Barbosa-Cobos, Fausto Sánchez-Muñoz, and Julián Ramírez-Bello. 2023. "CD147 rs8259T>A Variant Confers Susceptibility to COVID-19 Infection within the Mexican Population" Microorganisms 11, no. 8: 1919. https://doi.org/10.3390/microorganisms11081919
APA StyleAmezcua-Guerra, L. M., Guzmán-Martín, C. A., Montúfar-Robles, I., Springall, R., Hernández-Díazcouder, A., Barbosa-Cobos, R. E., Sánchez-Muñoz, F., & Ramírez-Bello, J. (2023). CD147 rs8259T>A Variant Confers Susceptibility to COVID-19 Infection within the Mexican Population. Microorganisms, 11(8), 1919. https://doi.org/10.3390/microorganisms11081919








