Potential Self-Attenuation of Arsenic by Indigenous Microorganisms in the Nakdong River
Abstract
1. Introduction
2. Materials and Methods
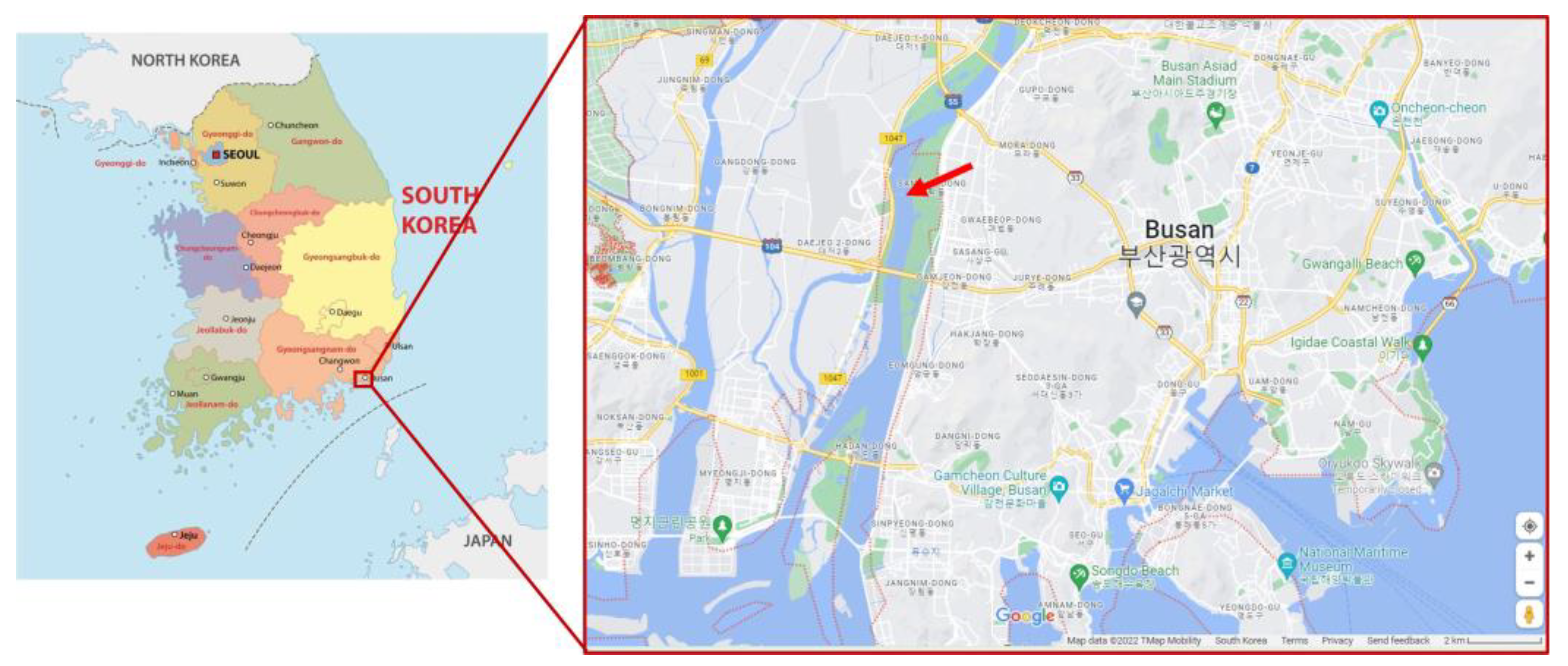
2.1. Sample Collection and Preparation
2.2. As(III) Oxidation by the Indigenous Microcosm
2.3. Bacterial Isolation and Characterization
2.4. As(III) Oxidation by Newly Isolated Bacteria
2.5. Analytical Techniques and Chemicals
2.6. Phylogenetic Study of Newly Isolated Bacteria
3. Results and Discussions
3.1. River Water Characteristics
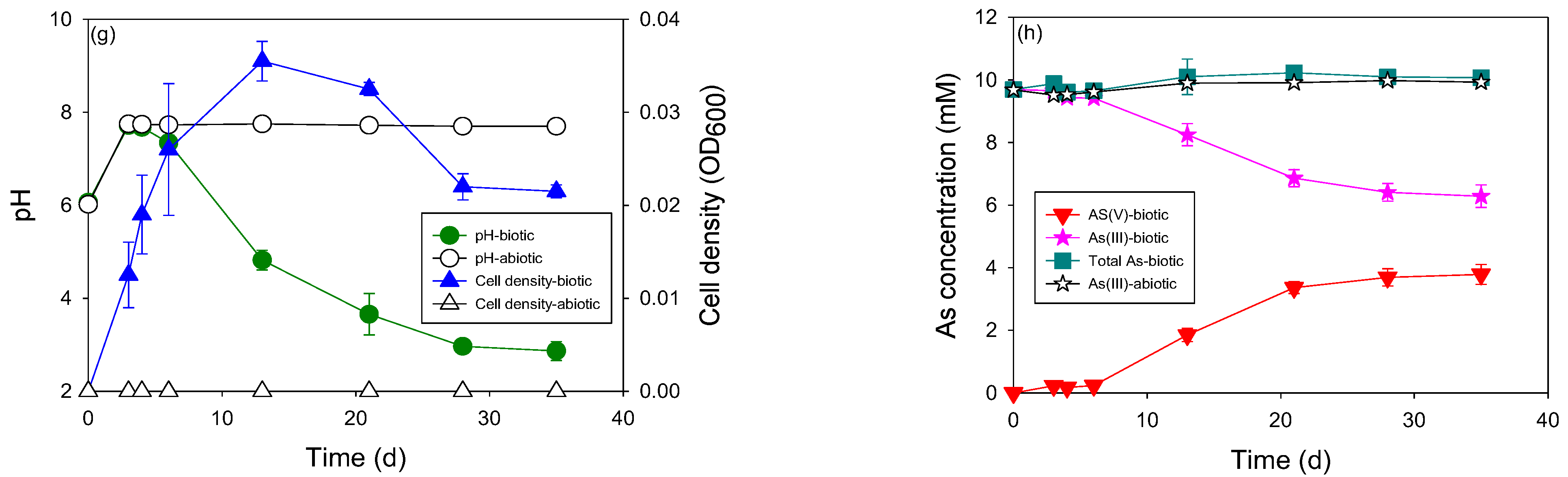
3.2. Heterotrophic and Autotrophic As(III) Oxidation by the Indigenous Microcosm
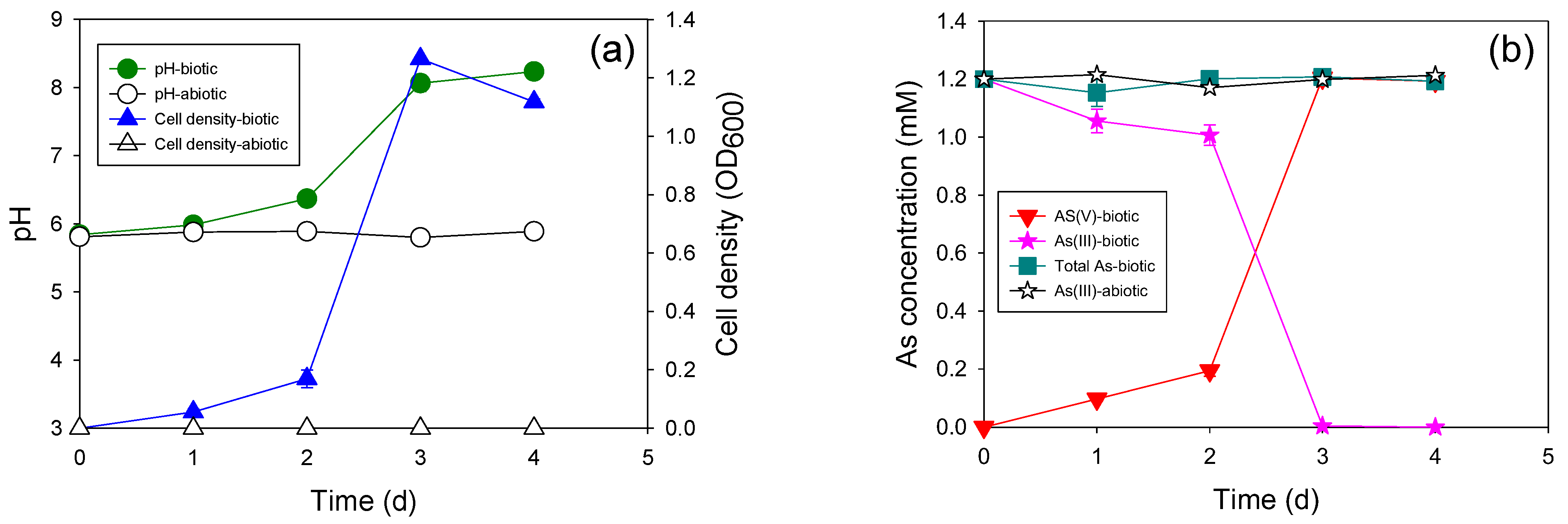
3.3. Heterotrophic and Autotrophic As(III) Oxidation by the Newly Isolated Bacteria
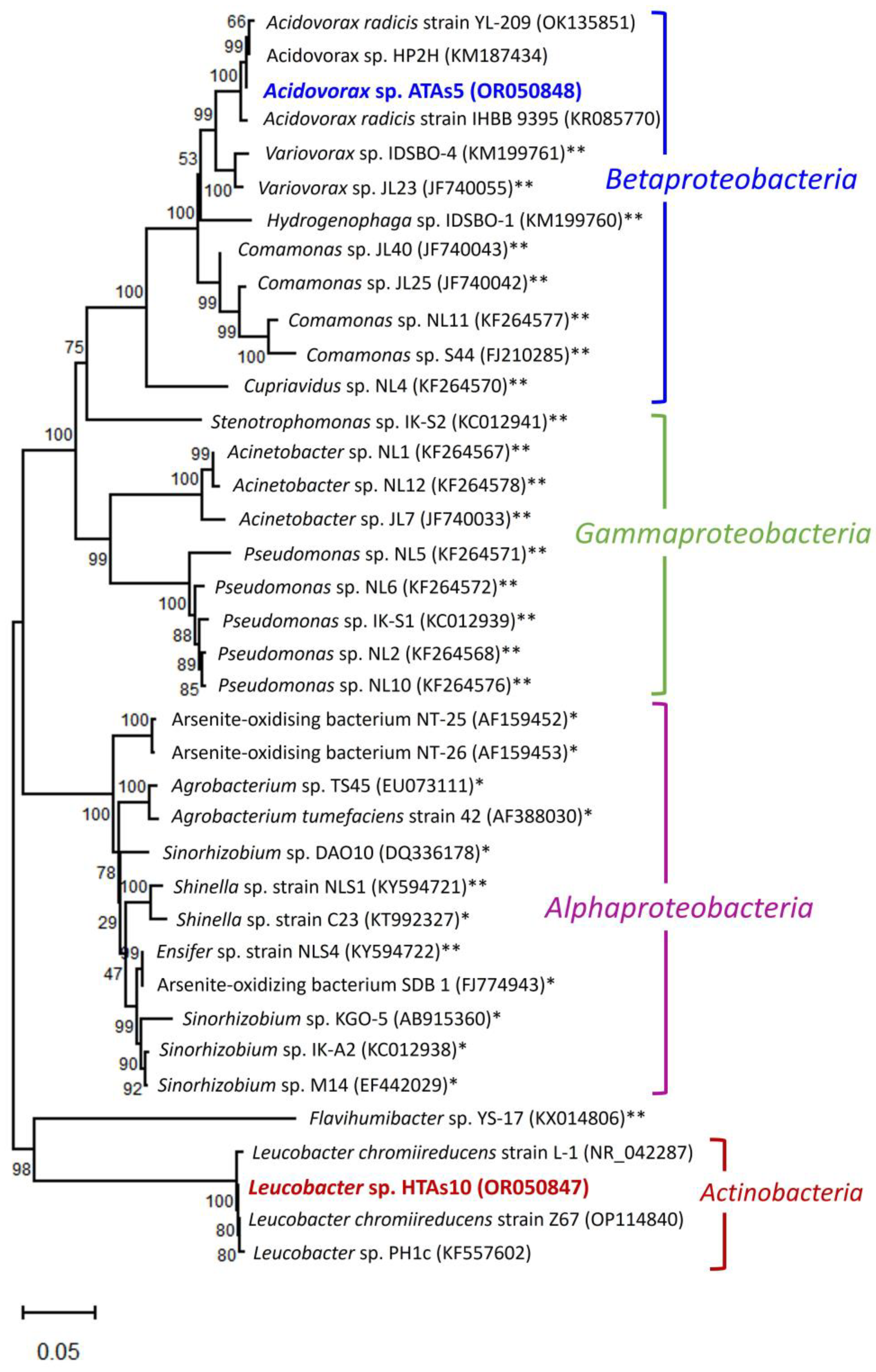
3.4. Evolutionary History of the Newly Isolated Bacteria
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tighe, M.; Ashley, P.; Lockwood, P.; Wilson, S. Soil, water, and pasture enrichment of antimony and arsenic within a coastal floodplain system. Sci. Total Environ. 2005, 347, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Mandal, B.K.; Suzuki, K.T. Arsenic round the world: A review. Talanta 2002, 58, 201–235. [Google Scholar] [CrossRef] [PubMed]
- Jain, C.; Ali, I. Arsenic: Occurrence, toxicity and speciation techniques. Water Res. 2000, 34, 4304–4312. [Google Scholar] [CrossRef]
- Hashim, M.A.; Mukhopadhyay, S.; Sahu, J.N.; Sengupta, B. Remediation technologies for heavy metal contaminated groundwater. J. Environ. Manag. 2011, 92, 2355–2388. [Google Scholar] [CrossRef]
- Inskeep, W.P.; Macur, R.E.; Hamamura, N.; Warelow, T.P.; Ward, S.A.; Santini, J.M. Detection, diversity and expression of aerobic bacterial arsenite oxidase genes. Environ. Microbiol. 2007, 9, 934–943. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, Y.; Liu, C.; Wu, M.; Wang, H. Characterization of the antimonite- and arsenite-oxidizing bacterium Bosea sp. AS-1 and its potential application in arsenic removal. J. Hazard. Mater. 2018, 359, 527–534. [Google Scholar] [CrossRef]
- Terry, L.R.; Kulp, T.R.; Wiatrowski, H.; Miller, L.G.; Oremland, R.S. Microbiological oxidation of antimony(III) with oxygen or nitrate by bacteria isolated from contaminated mine sediments. Appl. Environ. Microbiol. 2015, 81, 8478–8488. [Google Scholar] [CrossRef]
- Santini, J.M.; Stolz, J.F.; Macy, J.M. Isolation of a New Arsenate-Respiring Bacterium-Physiological and Phylogenetic Studies. Geomicrobiol. J. 2002, 19, 41–52. [Google Scholar] [CrossRef]
- Kinegam, S.; Yingprasertchai, T.; Tanasupawat, S.; Leepipatpiboon, N.; Akaracharanya, A.; Kim, K.-W. Isolation and characterization of arsenite-oxidizing bacteria from arsenic-contaminated soils in Thailand. World J. Microbiol. Biotechnol. 2008, 24, 3091–3096. [Google Scholar] [CrossRef]
- Rodríguez-Freire, L.; Sun, W.; Sierra-Alvarez, R.; Field, J.A. Flexible bacterial strains that oxidize arsenite in anoxic or aerobic conditions and utilize hydrogen or acetate as alternative electron donors. Biodegradation 2012, 23, 133–143. [Google Scholar] [CrossRef]
- Ai, L.N.; Sato, A.; Inoue, D.; Sei, K.; Soda, S.; Ike, M. Enrichment of arsenite oxidizing bacteria under autotrophic conditions and the isolation and characterization of facultative chemolithoautotrophic arsenite oxidizing bacteria for removal of arsenic from groundwater. Water Sci. Technol. Water Supply 2012, 12, 707–714. [Google Scholar] [CrossRef]
- Pous, N.; Casentini, B.; Rossetti, S.; Fazi, S.; Puig, S.; Aulenta, F. Anaerobic arsenite oxidation with an electrode serving as the sole electron acceptor: A novel approach to the bioremediation of arsenic-polluted groundwater. J. Hazard. Mater. 2015, 283, 617–622. [Google Scholar] [CrossRef]
- Nguyen, V.K.; Nguyen, D.D.; Ha, M.G.; Kang, H.Y. Potential of versatile bacteria isolated from activated sludge for the bioremediation of arsenic and antimony. J. Water Process Eng. 2021, 39, 101890. [Google Scholar] [CrossRef]
- Nguyen, V.K.; Tran, H.T.; Park, Y.; Yu, J.; Lee, T. Microbial arsenite oxidation with oxygen, nitrate, or an electrode as the sole electron acceptor. J. Ind. Microbiol. Biotechnol. 2017, 44, 857–868. [Google Scholar] [CrossRef]
- Nguyen, V.K.; Park, Y.; Yu, J.; Lee, T. Simultaneous arsenite oxidation and nitrate reduction at the electrodes of bioelectrochemical systems. Environ. Sci. Pollut. Res. 2016, 23, 19978–19988. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Nguyen, V.K.; Choi, W.; Ha, Y.; Gu, Y.; Lee, C.; Park, J.; Jang, G.; Shin, C.; Cho, S. Microbial tellurite reduction and production of elemental tellurium nanoparticles by novel bacteria isolated from wastewater. J. Ind. Eng. Chem. 2019, 78, 246–256. [Google Scholar] [CrossRef]
- Lund, P.; Tramonti, A.; De Biase, D. Coping with low pH: Molecular strategies in neutralophilic bacteria. FEMS Microbiol. Rev. 2014, 38, 1091–1125. [Google Scholar] [CrossRef]
- Lee, G.; Song, K.; Bae, J. Permanganate oxidation of arsenic(III): Reaction stoichiometry and the characterization of solid product. Geochim. Cosmochim. Acta 2011, 75, 4713–4727. [Google Scholar] [CrossRef]
- Garcia-Dominguez, E.; Mumford, A.; Rhine, E.D.; Paschal, A.; Young, L.Y. Novel autotrophic arsenite-oxidizing bacteria isolated from soil and sediments. FEMS Microbiol. Ecol. 2008, 66, 401–410. [Google Scholar] [CrossRef]
- Dong, D.; Ohtsuka, T.; Dong, D.T.; Amachi, S. Arsenite oxidation by a facultative chemolithoautotrophic Sinorhizobium sp. KGO-5 isolated from arsenic-contaminated soil. Biosci. Biotechnol. Biochem. 2014, 78, 1963–1970. [Google Scholar] [CrossRef] [PubMed]
- Bahar, M.M.; Megharaj, M.; Naidu, R. Kinetics of arsenite oxidation by Variovorax sp. MM-1 isolated from a soil and identification of arsenite oxidase gene. J. Hazard. Mater. 2013, 262, 997–1003. [Google Scholar] [CrossRef]
- Bahar, M.M.; Megharaj, M.; Naidu, R. Arsenic bioremediation potential of a new arsenite-oxidizing bacterium Stenotrophomonas sp. MM-7 isolated from soil. Biodegradation 2012, 23, 803–812. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Q.; Zhang, S.; Qin, D.; Wang, G. Phylogenetic and genome analyses of antimony-oxidizing bacteria isolated from antimony mined soil. Int. Biodeterior. Biodegrad. 2013, 76, 76–80. [Google Scholar] [CrossRef]
- Nguyen, V.K.; Lee, J.-U. Antimony-Oxidizing Bacteria Isolated from Antimony-Contaminated Sediment—A Phylogenetic Study. Geomicrobiol. J. 2015, 32, 50–58. [Google Scholar] [CrossRef]
- Xu, D.; Yang, L.; Wang, Y.; Wang, G.; Rensing, C.; Zheng, S. Proteins enriched in charged amino acids control the formation and stabilization of selenium nanoparticles in Comamonas testosteroni S44. Sci. Rep. 2018, 8, 4766. [Google Scholar] [CrossRef]




| pH | Si (mg/L) | S (mg/L) | Ca (mg/L) | Mg (mg/L) | K (mg/L) | Na (mg/L) | As (µg/L) |
|---|---|---|---|---|---|---|---|
| 6.95 | 0.921 | 9.528 | 16.518 | 3.333 | 3.297 | 18.932 | 0.954 |
| Se (µg/L) | Mo (µg/L) | Mn (µg/L) | Cu (µg/L) | Zn (µg/L) | Sb (µg/L) | Te (µg/L) | Cr (µg/L) |
| 0.055 | 2.226 | 3.230 | 1.752 | 0.815 | 0.424 | ND 1 | ND |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Won, S.; Shin, C.; Kang, H.Y. Potential Self-Attenuation of Arsenic by Indigenous Microorganisms in the Nakdong River. Microorganisms 2023, 11, 1910. https://doi.org/10.3390/microorganisms11081910
Won S, Shin C, Kang HY. Potential Self-Attenuation of Arsenic by Indigenous Microorganisms in the Nakdong River. Microorganisms. 2023; 11(8):1910. https://doi.org/10.3390/microorganisms11081910
Chicago/Turabian StyleWon, Sangmin, Chajeong Shin, and Ho Young Kang. 2023. "Potential Self-Attenuation of Arsenic by Indigenous Microorganisms in the Nakdong River" Microorganisms 11, no. 8: 1910. https://doi.org/10.3390/microorganisms11081910
APA StyleWon, S., Shin, C., & Kang, H. Y. (2023). Potential Self-Attenuation of Arsenic by Indigenous Microorganisms in the Nakdong River. Microorganisms, 11(8), 1910. https://doi.org/10.3390/microorganisms11081910






